Pluripotent Stem Cell-Derived Hepatocyte-like Cells: Induction Methods and Applications
Abstract
:1. Introduction
2. Advantages of iPSCs in the Treatment of Liver Disease
3. Development of the Liver in the Embryo
4. The HLCs Induced from iPSCs
4.1. Role of Cytokines/Small Molecules in Inducing iPSC Differentiation into HLCs
4.2. Role of Other Additives in Inducing iPSC Differentiation into HLCs
4.3. The Three-Dimensional (3D) Culture System Recreates the Liver Microenvironment
4.3.1. Induction of Differentiation Using Specific Extracellular Matrices or Culture Platforms
4.3.2. Coculture with Other Cells
4.3.3. 3D Bioprinting
5. Identification Methods of HLCs Differentiated from iPSCs
6. Applications of HLCs Differentiated from iPSCs
6.1. Modeling Liver Disease by Human iPSCs
6.2. Hepatoprotective Drug Screening via Human iPSCs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiao, J.; Wang, F.; Wong, N.K.; He, J.; Zhang, R.; Sun, R.; Xu, Y.; Liu, Y.; Li, W.; Koike, K.; et al. Global liver disease burdens and research trends: Analysis from a Chinese perspective. J. Hepatol. 2019, 71, 212–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, F.S.; Fan, J.G.; Zhang, Z.; Gao, B.; Wang, H.Y. The global burden of liver disease: The major impact of China. Hepatology 2014, 60, 2099–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef]
- Kuse, Y.; Taniguchi, H. Present and Future Perspectives of Using Human-Induced Pluripotent Stem Cells and Organoid against Liver Failure. Cell Transplant. 2019, 28 (Suppl. S1), 160S–165S. [Google Scholar] [CrossRef] [Green Version]
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Soltys, K.A.; Setoyama, K.; Tafaleng, E.N.; Soto Gutierrez, A.; Fong, J.; Fukumitsu, K.; Nishikawa, T.; Nagaya, M.; Sada, R.; Haberman, K.; et al. Host conditioning and rejection monitoring in hepatocyte transplantation in humans. J. Hepatol. 2017, 66, 987–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De, D.; Karmakar, P.; Bhattacharya, D. Stem Cell Aging and Regenerative Medicine. Adv. Exp. Med. Biol. 2021, 1326, 11–37. [Google Scholar]
- Hosseini, S.; Taghiyar, L.; Safari, F.; Baghaban Eslaminejad, M. Regenerative Medicine Applications of Mesenchymal Stem Cells. Adv. Exp. Med. Biol. 2018, 1089, 115–141. [Google Scholar]
- Yamanaka, S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef]
- Muraro, P.A.; Martin, R.; Mancardi, G.L.; Nicholas, R.; Sormani, M.P.; Saccardi, R. Autologous haematopoietic stem cell transplantation for treatment of multiple sclerosis. Nat. Rev. Neurol. 2017, 13, 391–405. [Google Scholar] [CrossRef]
- Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Induced Pluripotent Stem Cells (iPSCs)-Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells 2021, 10, 2319. [Google Scholar] [CrossRef]
- Tapia, N.; Scholer, H.R. Molecular Obstacles to Clinical Translation of iPSCs. Cell Stem Cell 2016, 19, 298–309. [Google Scholar] [CrossRef] [Green Version]
- Takeishi, K.; Collin de l’Hortet, A.; Wang, Y.; Handa, K.; Guzman-Lepe, J.; Matsubara, K.; Morita, K.; Jang, S.; Haep, N.; Florentino, R.M.; et al. Assembly and Function of a Bioengineered Human Liver for Transplantation Generated Solely from Induced Pluripotent Stem Cells. Cell Rep. 2020, 31, 107711. [Google Scholar] [CrossRef]
- Tilson, S.G.; Morell, C.M.; Lenaerts, A.S.; Park, S.B.; Hu, Z.; Jenkins, B.; Koulman, A.; Liang, T.J.; Vallier, L. Modeling PNPLA3-Associated NAFLD Using Human-Induced Pluripotent Stem Cells. Hepatology 2021, 74, 2998–3017. [Google Scholar] [CrossRef]
- Nikokiraki, C.; Psaraki, A.; Roubelakis, M.G. The Potential Clinical Use of Stem/Progenitor Cells and Organoids in Liver Diseases. Cells 2022, 11, 1410. [Google Scholar] [CrossRef]
- Adam, R.; Karam, V.; Cailliez, V.; Grady, J.G.O.; Mirza, D.; Cherqui, D.; Klempnauer, J.; Salizzoni, M.; Pratschke, J.; Jamieson, N.; et al. 2018 Annual Report of the European Liver Transplant Registry (ELTR)—50-year evolution of liver transplantation. Transpl. Int. 2018, 31, 1293–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luce, E.; Messina, A.; Duclos-Vallee, J.C.; Dubart-Kupperschmitt, A. Advanced Techniques and Awaited Clinical Applications for Human Pluripotent Stem Cell Differentiation into Hepatocytes. Hepatology 2021, 74, 1101–1116. [Google Scholar] [CrossRef] [PubMed]
- Godoy, P.; Schmidt-Heck, W.; Hellwig, B.; Nell, P.; Feuerborn, D.; Rahnenfuhrer, J.; Kattler, K.; Walter, J.; Bluthgen, N.; Hengstler, J.G. Assessment of stem cell differentiation based on genome-wide expression profiles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rombaut, M.; Boeckmans, J.; Rodrigues, R.M.; van Grunsven, L.A.; Vanhaecke, T.; De Kock, J. Direct reprogramming of somatic cells into induced hepatocytes: Cracking the Enigma code. J. Hepatol. 2021, 75, 690–705. [Google Scholar] [CrossRef] [PubMed]
- Grandy, R.; Tomaz, R.A.; Vallier, L. Modeling Disease with Human Inducible Pluripotent Stem Cells. Annu. Rev. Pathol. 2019, 14, 449–468. [Google Scholar] [CrossRef] [PubMed]
- Raasch, M.; Fritsche, E.; Kurtz, A.; Bauer, M.; Mosig, A.S. Microphysiological systems meet hiPSC technology—New tools for disease modeling of liver infections in basic research and drug development. Adv. Drug Deliv. Rev. 2019, 140, 51–67. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Blaszkiewicz, J.; Duncan, S.A. Advancements in Disease Modeling and Drug Discovery Using iPSC-Derived Hepatocyte-like Cells. Genes 2022, 13, 573. [Google Scholar] [CrossRef]
- Hirschi, K.K.; Li, S.; Roy, K. Induced pluripotent stem cells for regenerative medicine. Annu. Rev. Biomed. Eng. 2014, 16, 277–294. [Google Scholar] [CrossRef] [Green Version]
- Rossant, J.; Tam, P.P.L. Early human embryonic development: Blastocyst formation to gastrulation. Dev. Cell 2022, 57, 152–165. [Google Scholar] [CrossRef]
- Baker, C.L.; Pera, M.F. Capturing Totipotent Stem Cells. Cell Stem Cell 2018, 22, 25–34. [Google Scholar] [CrossRef] [Green Version]
- McLin, V.A.; Rankin, S.A.; Zorn, A.M. Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development 2007, 134, 2207–2217. [Google Scholar] [CrossRef] [Green Version]
- Prior, N.; Inacio, P.; Huch, M. Liver organoids: From basic research to therapeutic applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef] [Green Version]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Ober, E.A.; Lemaigre, F.P. Development of the liver: Insights into organ and tissue morphogenesis. J. Hepatol. 2018, 68, 1049–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachinidis, A.; Albrecht, W.; Nell, P.; Cherianidou, A.; Hewitt, N.J.; Edlund, K.; Hengstler, J.G. Road Map for Development of Stem Cell-Based Alternative Test Methods. Trends Mol. Med. 2019, 25, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Zorn, A.M. Liver development. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2008. [Google Scholar]
- Gordillo, M.; Evans, T.; Gouon-Evans, V. Orchestrating liver development. Development 2015, 142, 2094–2108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimbrel, E.A.; Lanza, R. Current status of pluripotent stem cells: Moving the first therapies to the clinic. Nat. Rev. Drug Discov. 2015, 14, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Maepa, S.W.; Ndlovu, H. Advances in generating liver cells from pluripotent stem cells as a tool for modeling liver diseases. Stem Cells 2020, 38, 606–612. [Google Scholar] [CrossRef] [Green Version]
- Mun, S.J.; Ryu, J.S.; Lee, M.O.; Son, Y.S.; Oh, S.J.; Cho, H.S.; Son, M.Y.; Kim, D.S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef]
- Shinozawa, T.; Kimura, M.; Cai, Y.; Saiki, N.; Yoneyama, Y.; Ouchi, R.; Koike, H.; Maezawa, M.; Zhang, R.R.; Dunn, A.; et al. High-Fidelity Drug-Induced Liver Injury Screen Using Human Pluripotent Stem Cell-Derived Organoids. Gastroenterology 2021, 160, 831–846.e10. [Google Scholar] [CrossRef]
- Ramli, M.N.B.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e12. [Google Scholar] [CrossRef]
- Wu, F.; Wu, D.; Ren, Y.; Huang, Y.; Feng, B.; Zhao, N.; Zhang, T.; Chen, X.; Chen, S.; Xu, A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019, 70, 1145–1158. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Jiang, C.; Kim, M.; Yang, W.; Zhu, K.; Guan, D.; Lv, W.; Xiao, Y.; Wilson, J.R.; Rader, D.J.; et al. Individual-specific functional epigenomics reveals genetic determinants of adverse metabolic effects of glucocorticoids. Cell Metab. 2021, 33, 1592–1609.e7. [Google Scholar] [CrossRef] [PubMed]
- Overeem, A.W.; Klappe, K.; Parisi, S.; Kloters-Planchy, P.; Matakovic, L.; du Teil Espina, M.; Drouin, C.A.; Weiss, K.H.; van, I.S.C.D. Pluripotent stem cell-derived bile canaliculi-forming hepatocytes to study genetic liver diseases involving hepatocyte polarity. J. Hepatol. 2019, 71, 344–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertero, A.; Madrigal, P.; Galli, A.; Hubner, N.C.; Moreno, I.; Burks, D.; Brown, S.; Pedersen, R.A.; Gaffney, D.; Mendjan, S.; et al. Activin/nodal signaling and NANOG orchestrate human embryonic stem cell fate decisions by controlling the H3K4me3 chromatin mark. Genes. Dev. 2015, 29, 702–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, Z.; Niu, B.; Duan, K.; Si, C.; Wang, S.; Xiang, L.; Zhu, X.; Zhu, Q.; Feng, C.; Yin, Y.; et al. Modulation of Wnt and Activin/Nodal supports efficient derivation, cloning and suspension expansion of human pluripotent stem cells. Biomaterials 2020, 249, 120015. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Steichen, C.; Dianat, N.; Weber, A.; Dubart-Kupperschmitt, A. The potential of induced pluripotent stem cell derived hepatocytes. J. Hepatol. 2016, 65, 182–199. [Google Scholar] [CrossRef] [Green Version]
- Bloise, E.; Ciarmela, P.; Dela Cruz, C.; Luisi, S.; Petraglia, F.; Reis, F.M. Activin A in Mammalian Physiology. Physiol. Rev. 2019, 99, 739–780. [Google Scholar] [CrossRef]
- Wang, Z.; Li, W.; Chen, T.; Yang, J.; Wen, Z.; Yan, X.; Shen, T.; Liang, R. Activin A can induce definitive endoderm differentiation from human parthenogenetic embryonic stem cells. Biotechnol. Lett. 2015, 37, 1711–1717. [Google Scholar] [CrossRef]
- Tafaleng, E.N.; Chakraborty, S.; Han, B.; Hale, P.; Wu, W.; Soto-Gutierrez, A.; Feghali-Bostwick, C.A.; Wilson, A.A.; Kotton, D.N.; Nagaya, M.; et al. Induced pluripotent stem cells model personalized variations in liver disease resulting from alpha1-antitrypsin deficiency. Hepatology 2015, 62, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Tolosa, L.; Caron, J.; Hannoun, Z.; Antoni, M.; Lopez, S.; Burks, D.; Castell, J.V.; Weber, A.; Gomez-Lechon, M.J.; Dubart-Kupperschmitt, A. Transplantation of hESC-derived hepatocytes protects mice from liver injury. Stem Cell Res. Ther. 2015, 6, 246. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; von Meyenn, F.; Santos, F.; Chen, Y.; Reik, W.; Bertone, P.; Smith, A.; Nichols, J. Naive Pluripotent Stem Cells Derived Directly from Isolated Cells of the Human Inner Cell Mass. Stem Cell Rep. 2016, 6, 437–446. [Google Scholar] [CrossRef] [Green Version]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell 2015, 162, 452–453. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic Identification of Culture Conditions for Induction and Maintenance of Naive Human Pluripotency. Cell Stem Cell 2014, 15, 524–526. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xie, G.; Singh, M.; Ghanbarian, A.T.; Rasko, T.; Szvetnik, A.; Cai, H.; Besser, D.; Prigione, A.; Fuchs, N.V.; et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature 2014, 516, 405–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Liu, B.; Xu, J.; Wang, J.; Wu, J.; Shi, C.; Xu, Y.; Dong, J.; Wang, C.; Lai, W.; et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell 2017, 169, 243–257.e25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dao Thi, V.L.; Wu, X.; Belote, R.L.; Andreo, U.; Takacs, C.N.; Fernandez, J.P.; Vale-Silva, L.A.; Prallet, S.; Decker, C.C.; Fu, R.M.; et al. Stem cell-derived polarized hepatocytes. Nat. Commun. 2020, 11, 1677. [Google Scholar] [CrossRef] [Green Version]
- Czysz, K.; Minger, S.; Thomas, N. DMSO efficiently down regulates pluripotency genes in human embryonic stem cells during definitive endoderm derivation and increases the proficiency of hepatic differentiation. PLoS ONE 2015, 10, e0117689. [Google Scholar] [CrossRef]
- Touboul, T.; Chen, S.; To, C.C.; Mora-Castilla, S.; Sabatini, K.; Tukey, R.H.; Laurent, L.C. Stage-specific regulation of the WNT/beta-catenin pathway enhances differentiation of hESCs into hepatocytes. J. Hepatol. 2016, 64, 1315–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, T.; Uchiyama, S.; Tanaka, H.; Kataoka, H. Hepatocyte Growth Factor Activator: A Proteinase Linking Tissue Injury with Repair. Int. J. Mol. Sci. 2018, 19, 3435. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Jiang, M.; Wang, G.; Li, B.; Jia, X.; Ai, Y.; Chen, S.; Tang, P.; Liu, A.; Yuan, Q.; et al. IL6 supports long-term expansion of hepatocytes in vitro. Nat. Commun. 2022, 13, 7345. [Google Scholar] [CrossRef]
- Lv, L.; Han, Q.; Chu, Y.; Zhang, M.; Sun, L.; Wei, W.; Jin, C.; Li, W. Self-renewal of hepatoblasts under chemically defined conditions by iterative growth factor and chemical screening. Hepatology 2015, 61, 337–347. [Google Scholar] [CrossRef]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayama, K.; Hagihara, Y.; Toba, Y.; Sekiguchi, K.; Sakurai, F.; Mizuguchi, H. Enrichment of high-functioning human iPS cell-derived hepatocyte-like cells for pharmaceutical research. Biomaterials 2018, 161, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Peaslee, C.; Esteva-Font, C.; Su, T.; Munoz-Howell, A.; Duwaerts, C.C.; Liu, Z.; Rao, S.; Liu, K.; Medina, M.; Sneddon, J.B.; et al. Doxycycline Significantly Enhances Induction of Induced Pluripotent Stem Cells to Endoderm by Enhancing Survival through Protein Kinase B Phosphorylation. Hepatology 2021, 74, 2102–2117. [Google Scholar] [CrossRef]
- Zhou, M.; Li, P.; Tan, L.; Qu, S.; Ying, Q.L.; Song, H. Differentiation of mouse embryonic stem cells into hepatocytes induced by a combination of cytokines and sodium butyrate. J. Cell Biochem. 2010, 109, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhao, Y.; Liu, Y.; Ye, F.; Song, Z.; Qin, H.; Meng, S.; Chen, Y.; Zhou, R.; Song, X.; et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 2007, 45, 1229–1239. [Google Scholar] [CrossRef]
- Chen, Y.F.; Tseng, C.Y.; Wang, H.W.; Kuo, H.C.; Yang, V.W.; Lee, O.K. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology 2012, 55, 1193–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Chen, S.; Duo, S.; Xiang, C.; Jia, J.; Guo, M.; Lai, W.; Lu, S.; Deng, H. Promotion of the efficient metabolic maturation of human pluripotent stem cell-derived hepatocytes by correcting specification defects. Cell Res. 2013, 23, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Lou, Y.R.; Leung, A.W. Next generation organoids for biomedical research and applications. Biotechnol. Adv. 2018, 36, 132–149. [Google Scholar] [CrossRef]
- Yiangou, L.; Ross, A.D.B.; Goh, K.J.; Vallier, L. Human Pluripotent Stem Cell-Derived Endoderm for Modeling Development and Clinical Applications. Cell Stem Cell 2018, 22, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Cameron, K.; Tan, R.; Schmidt-Heck, W.; Campos, G.; Lyall, M.J.; Wang, Y.; Lucendo-Villarin, B.; Szkolnicka, D.; Bates, N.; Kimber, S.J.; et al. Recombinant Laminins Drive the Differentiation and Self-Organization of hESC-Derived Hepatocytes. Stem Cell Rep. 2015, 5, 1250–1262. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, K.; Owens, D.J.; Raju, R.; Firpo, M.; O’Brien, T.D.; Verfaillie, C.M.; Hu, W.S. Spheroid culture for enhanced differentiation of human embryonic stem cells to hepatocyte-like cells. Stem Cells Dev. 2014, 23, 124–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, D.R.; Ware, B.R.; Davidson, M.D.; Allsup, S.R.; Khetani, S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology 2015, 61, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Gieseck, R.L., 3rd; Hannan, N.R.; Bort, R.; Hanley, N.A.; Drake, R.A.; Cameron, G.W.; Wynn, T.A.; Vallier, L. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS ONE 2014, 9, e86372. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.; Kim, H.; Park, J.Y.; Kim, G.; Han, J.; Chung, S.; Yang, J.H.; Jeon, J.S.; Woo, D.H.; Han, C.; et al. Generation of uniform liver spheroids from human pluripotent stem cells for imaging-based drug toxicity analysis. Biomaterials 2021, 269, 120529. [Google Scholar] [CrossRef]
- Vyas, D.; Baptista, P.M.; Brovold, M.; Moran, E.; Gaston, B.; Booth, C.; Samuel, M.; Atala, A.; Soker, S. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2018, 67, 750–761. [Google Scholar] [CrossRef] [Green Version]
- Ouchi, R.; Togo, S.; Kimura, M.; Shinozawa, T.; Koido, M.; Koike, H.; Thompson, W.; Karns, R.A.; Mayhew, C.N.; McGrath, P.S.; et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 2019, 30, 374–384.e6. [Google Scholar] [CrossRef] [PubMed]
- Vallverdu, J.; Martinez Garcia de la Torre, R.A.; Mannaerts, I.; Verhulst, S.; Smout, A.; Coll, M.; Arino, S.; Rubio-Tomas, T.; Aguilar-Bravo, B.; Martinez-Sanchez, C.; et al. Directed differentiation of human induced pluripotent stem cells to hepatic stellate cells. Nat. Protoc. 2021, 16, 2542–2563. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Toprakhisar, B.; Van Haele, M.; Antoranz, A.; Boon, R.; Chesnais, F.; De Smedt, J.; Tricot, T.; Idoype, T.I.; Canella, M.; et al. A fully defined matrix to support a pluripotent stem cell derived multi-cell-liver steatohepatitis and fibrosis model. Biomaterials 2021, 276, 121006. [Google Scholar] [CrossRef]
- Szkolnicka, D.; Hay, D.C. Concise Review: Advances in Generating Hepatocytes from Pluripotent Stem Cells for Translational Medicine. Stem Cells 2016, 34, 1421–1426. [Google Scholar] [CrossRef] [Green Version]
- Martini, T.; Naef, F.; Tchorz, J.S. Spatiotemporal Metabolic Liver Zonation and Consequences on Pathophysiology. Annu. Rev. Pathol. 2023, 18, 439–466. [Google Scholar] [CrossRef]
- Kim, D.S.; Ryu, J.W.; Son, M.Y.; Oh, J.H.; Chung, K.S.; Lee, S.; Lee, J.J.; Ahn, J.H.; Min, J.S.; Ahn, J.; et al. A liver-specific gene expression panel predicts the differentiation status of in vitro hepatocyte models. Hepatology 2017, 66, 1662–1674. [Google Scholar] [CrossRef]
- Pareja, E.; Gomez-Lechon, M.J.; Tolosa, L. Induced pluripotent stem cells for the treatment of liver diseases: Challenges and perspectives from a clinical viewpoint. Ann. Transl. Med. 2020, 8, 566. [Google Scholar] [CrossRef]
- Cheng, Z.; He, Z.; Cai, Y.; Zhang, C.; Fu, G.; Li, H.; Sun, W.; Liu, C.; Cui, X.; Ning, B.; et al. Conversion of hepatoma cells to hepatocyte-like cells by defined hepatocyte nuclear factors. Cell Res. 2019, 29, 124–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knolle, P.A.; Wohlleber, D. Immunological functions of liver sinusoidal endothelial cells. Cell Mol. Immunol. 2016, 13, 347–353. [Google Scholar] [CrossRef] [Green Version]
- Maslak, E.; Gregorius, A.; Chlopicki, S. Liver sinusoidal endothelial cells (LSECs) function and NAFLD.; NO-based therapy targeted to the liver. Pharmacol. Rep. 2015, 67, 689–694. [Google Scholar] [CrossRef]
- Poisson, J.; Lemoinne, S.; Boulanger, C.; Durand, F.; Moreau, R.; Valla, D.; Rautou, P.E. Liver sinusoidal endothelial cells: Physiology and role in liver diseases. J. Hepatol. 2017, 66, 212–227. [Google Scholar] [CrossRef] [Green Version]
- Heymann, F.; Tacke, F. Immunology in the liver--from homeostasis to disease. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 88–110. [Google Scholar] [CrossRef] [PubMed]
- Sufletel, R.T.; Melincovici, C.S.; Gheban, B.A.; Toader, Z.; Mihu, C.M. Hepatic stellate cells—From past till present: Morphology, human markers, human cell lines, behavior in normal and liver pathology. Rom. J. Morphol. Embryol. 2020, 61, 615–642. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, H.J.; Choi, D. Cell sources, liver support systems and liver tissue engineering: Alternatives to liver transplantation. Int. J. Stem Cells 2015, 8, 36–47. [Google Scholar] [CrossRef]
- Goulart, E.; de Caires-Junior, L.C.; Telles-Silva, K.A.; Araujo, B.H.S.; Rocco, S.A.; Sforca, M.; de Sousa, I.L.; Kobayashi, G.S.; Musso, C.M.; Assoni, A.F.; et al. 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication 2019, 12, 015010. [Google Scholar] [CrossRef]
- Ma, X.; Qu, X.; Zhu, W.; Li, Y.S.; Yuan, S.; Zhang, H.; Liu, J.; Wang, P.; Lai, C.S.; Zanella, F.; et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc. Natl. Acad. Sci. USA 2016, 113, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Matsuzaki, J.; Katsuda, T.; Saito, Y.; Saito, H.; Ochiya, T. Generation of functional human hepatocytes in vitro: Current status and future prospects. Inflamm. Regen. 2019, 39, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwartz, R.E.; Fleming, H.E.; Khetani, S.R.; Bhatia, S.N. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnol. Adv. 2014, 32, 504–513. [Google Scholar] [CrossRef] [Green Version]
- Laemmle, A.; Poms, M.; Hsu, B.; Borsuk, M.; Rufenacht, V.; Robinson, J.; Sadowski, M.C.; Nuoffer, J.M.; Haberle, J.; Willenbring, H. Aquaporin 9 induction in human iPSC-derived hepatocytes facilitates modeling of ornithine transcarbamylase deficiency. Hepatology 2022, 76, 646–659. [Google Scholar] [CrossRef]
- Passier, R.; Orlova, V.; Mummery, C. Complex Tissue and Disease Modeling using hiPSCs. Cell Stem Cell 2016, 18, 309–321. [Google Scholar] [CrossRef] [Green Version]
- Roy-Chowdhury, N.; Wang, X.; Guha, C.; Roy-Chowdhury, J. Hepatocyte-like cells derived from induced pluripotent stem cells. Hepatol. Int. 2017, 11, 54–69. [Google Scholar] [CrossRef]
- Palakkan, A.A.; Nanda, J.; Ross, J.A. Pluripotent stem cells to hepatocytes, the journey so far. Biomed. Rep. 2017, 6, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Duan, L.; He, X.; Li, S.; Wu, Y.; Xiang, G.; Bao, F.; Yang, L.; Shi, H.; Gao, M.; et al. A Combined Model of Human iPSC-Derived Liver Organoids and Hepatocytes Reveals Ferroptosis in DGUOK Mutant mtDNA Depletion Syndrome. Adv. Sci. 2021, 8, 2004680. [Google Scholar] [CrossRef]
- Zabaleta, N.; Hommel, M.; Salas, D.; Gonzalez-Aseguinolaza, G. Genetic-Based Approaches to Inherited Metabolic Liver Diseases. Hum. Gene Ther. 2019, 30, 1190–1203. [Google Scholar] [CrossRef] [PubMed]
- Benam, K.H.; Dauth, S.; Hassell, B.; Herland, A.; Jain, A.; Jang, K.J.; Karalis, K.; Kim, H.J.; MacQueen, L.; Mahmoodian, R.; et al. Engineered in vitro disease models. Annu. Rev. Pathol. 2015, 10, 195–262. [Google Scholar] [CrossRef] [Green Version]
- DeBoever, C.; Li, H.; Jakubosky, D.; Benaglio, P.; Reyna, J.; Olson, K.M.; Huang, H.; Biggs, W.; Sandoval, E.; D’Antonio, M.; et al. Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell 2017, 20, 533–546.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pashos, E.E.; Park, Y.; Wang, X.; Raghavan, A.; Yang, W.; Abbey, D.; Peters, D.T.; Arbelaez, J.; Hernandez, M.; Kuperwasser, N.; et al. Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell 2017, 20, 558–570.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, C.R.; O’Sullivan, J.F.; Friesen, M.; Becker, C.E.; Zhang, X.; Liu, P.; Wakabayashi, Y.; Morningstar, J.E.; Shi, X.; Choi, J.; et al. Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell 2017, 20, 547–557.e7. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [Green Version]
- Pournasr, B.; Duncan, S.A. Modeling Inborn Errors of Hepatic Metabolism Using Induced Pluripotent Stem Cells. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1994–1999. [Google Scholar] [CrossRef] [Green Version]
- Rashid, S.T.; Corbineau, S.; Hannan, N.; Marciniak, S.J.; Miranda, E.; Alexander, G.; Huang-Doran, I.; Griffin, J.; Ahrlund-Richter, L.; Skepper, J.; et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Investig. 2010, 120, 3127–3136. [Google Scholar] [CrossRef] [Green Version]
- Cayo, M.A.; Cai, J.; DeLaForest, A.; Noto, F.K.; Nagaoka, M.; Clark, B.S.; Collery, R.F.; Si-Tayeb, K.; Duncan, S.A. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology 2012, 56, 2163–2171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Oikonomopoulos, A.; Sayed, N.; Wu, J.C. Modeling human diseases with induced pluripotent stem cells: From 2D to 3D and beyond. Development 2018, 145, dev156166. [Google Scholar] [CrossRef] [Green Version]
- McCauley, H.A.; Wells, J.M. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Development 2017, 144, 958–962. [Google Scholar] [CrossRef] [Green Version]
- Dutta, D.; Heo, I.; Clevers, H. Disease Modeling in Stem Cell-Derived 3D Organoid Systems. Trends Mol. Med. 2017, 23, 393–410. [Google Scholar] [CrossRef]
- Lee, P.C.; Truong, B.; Vega-Crespo, A.; Gilmore, W.B.; Hermann, K.; Angarita, S.A.; Tang, J.K.; Chang, K.M.; Wininger, A.E.; Lam, A.K.; et al. Restoring Ureagenesis in Hepatocytes by CRISPR/Cas9-mediated Genomic Addition to Arginase-deficient Induced Pluripotent Stem Cells. Mol. Ther. Nucleic Acids 2016, 5, e394. [Google Scholar] [CrossRef] [PubMed]
- Yoshitoshi-Uebayashi, E.Y.; Toyoda, T.; Yasuda, K.; Kotaka, M.; Nomoto, K.; Okita, K.; Yasuchika, K.; Okamoto, S.; Takubo, N.; Nishikubo, T.; et al. Modelling urea-cycle disorder citrullinemia type 1 with disease-specific iPSCs. Biochem. Biophys. Res. Commun. 2017, 486, 613–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Choi, J.Y.; Lee, S.H.; Lee, B.H.; Yoo, H.W.; Han, Y.M. Malfunction in Mitochondrial beta-Oxidation Contributes to Lipid Accumulation in Hepatocyte-Like Cells Derived from Citrin Deficiency-Induced Pluripotent Stem Cells. Stem Cells Dev. 2016, 25, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Corbett, J.L.; Duncan, S.A. iPSC-Derived Hepatocytes as a Platform for Disease Modeling and Drug Discovery. Front. Med. 2019, 6, 265. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.P. Application of hepatocyte-like cells to enhance hepatic safety risk assessment in drug discovery. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373, 20170228. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, W.; Kappenberg, F.; Brecklinghaus, T.; Stoeber, R.; Marchan, R.; Zhang, M.; Ebbert, K.; Kirschner, H.; Grinberg, M.; Leist, M.; et al. Prediction of human drug-induced liver injury (DILI) in relation to oral doses and blood concentrations. Arch. Toxicol. 2019, 93, 1609–1637. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Schmidt-Heck, W.; Natarajan, K.; Lucendo-Villarin, B.; Szkolnicka, D.; Asplund, A.; Bjorquist, P.; Widera, A.; Stober, R.; Campos, G.; et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 2015, 63, 934–942. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Chen, X.; Wu, M.; Zhu, W.; Arslan, A.; Takeda, S.; Nguyen, M.H.; Majeti, R.; Thomas, D.; Zheng, M.; et al. The phosphatidylethanolamine biosynthesis pathway provides a new target for cancer chemotherapy. J. Hepatol. 2020, 72, 746–760. [Google Scholar] [CrossRef]
- Medvinsky, A.; Livesey, F.J. On human development: Lessons from stem cell systems. Development 2015, 142, 17–20. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Liu, M.; Pan, X.; Zhu, H. Tumorigenicity risk of iPSCs in vivo: Nip it in the bud. Precis. Clin. Med. 2022, 5, pbac004. [Google Scholar] [CrossRef]


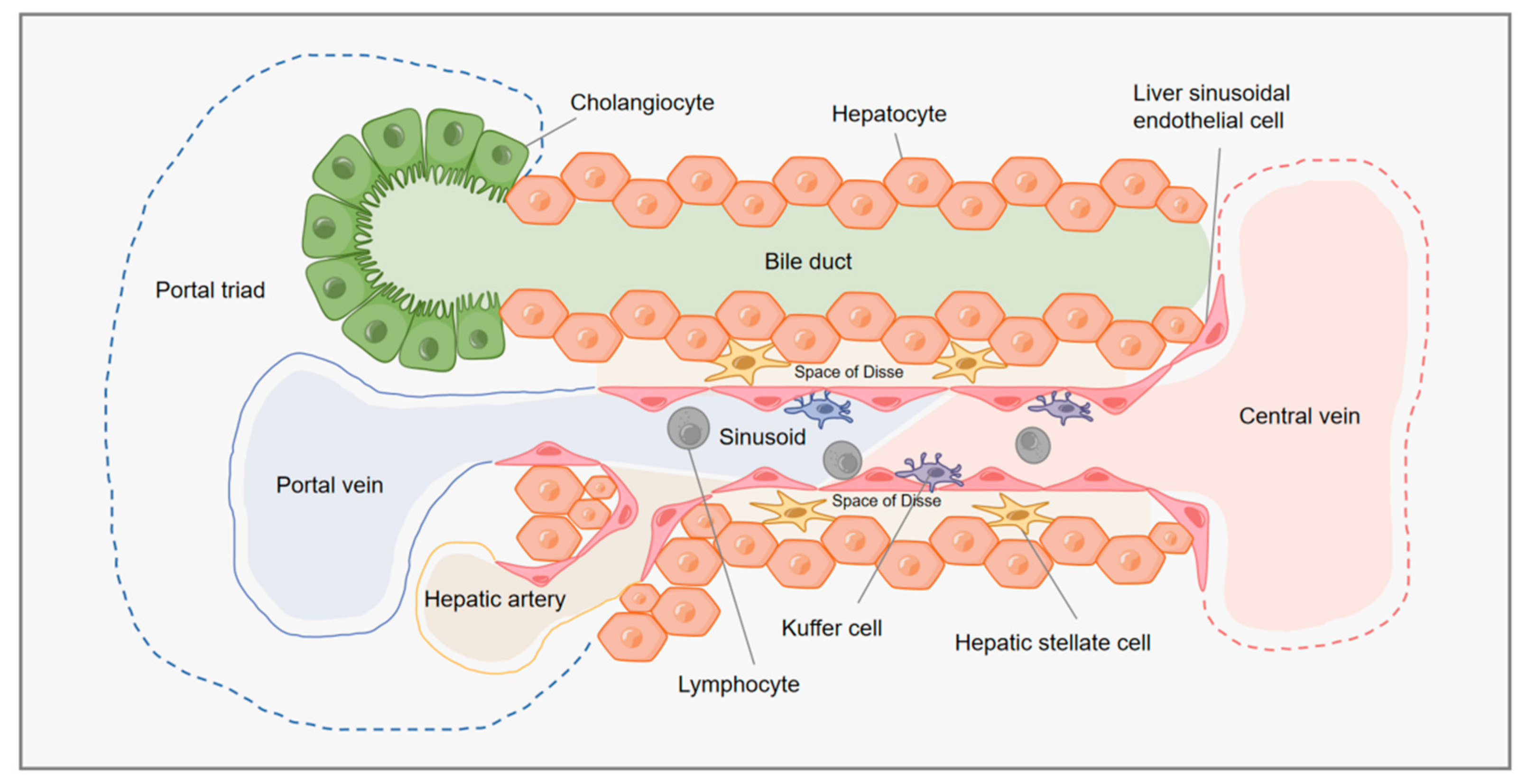
| Liver Cells | Types | Mass Percentage (%) | Functions | References |
|---|---|---|---|---|
| Parenchymal cells (PCs) | Hepatocytes and biliary tract cells | 70–80 |
| [81,82,83,84] |
| Nonparenchymal cells (NPCs) | Liver sinusoidal endothelial cells (LSECs) | 15–20 |
| [85,86,87] |
| Kupffer cells (KCs) | 15 |
| [21,85,88] | |
| Hepatic stellate cells (HSCs) | 15 |
| [89] |
| Strategy | Abstract | Disease Model | Conclusions | References |
|---|---|---|---|---|
| 2D differentiation strategy | Investigated whether iPSCs from α1-antitrypsin deficiency (ATD) individuals with or without severe liver disease could model these personalized variations in hepatic disease phenotypes. | Liver disease resulting from ATD |
| [49] |
| A library of human iPSCs lines were generated from individuals with a range of inherited metabolic disorders (IMDs), with a focus on 3 of the diseases, and hepatocytes were derived using human iPSCs from affected patients. | IMDs of the liver |
| [107] | |
| 3D differentiation strategy | Generated hepatic organoids that comprise different parenchymal liver cell types and have structural features of the liver using human pluripotent stem cells. | Nonalcoholic steatohepatitis (NASH) |
| [40] |
| Using 11 different healthy and diseased pluripotent stem cell lines, a reproducible method was developed to obtain multicellular human liver organs composed of hepatocytes, stellate cells, and Kupffer-like cells. | Steatohepatitis |
| [77] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Q.; Wang, N.; Que, H.; Mai, E.; Hu, Y.; Tan, R.; Gu, J.; Gong, P. Pluripotent Stem Cell-Derived Hepatocyte-like Cells: Induction Methods and Applications. Int. J. Mol. Sci. 2023, 24, 11592. https://doi.org/10.3390/ijms241411592
Luo Q, Wang N, Que H, Mai E, Hu Y, Tan R, Gu J, Gong P. Pluripotent Stem Cell-Derived Hepatocyte-like Cells: Induction Methods and Applications. International Journal of Molecular Sciences. 2023; 24(14):11592. https://doi.org/10.3390/ijms241411592
Chicago/Turabian StyleLuo, Qiulin, Nan Wang, Hanyun Que, Erziya Mai, Yanting Hu, Rui Tan, Jian Gu, and Puyang Gong. 2023. "Pluripotent Stem Cell-Derived Hepatocyte-like Cells: Induction Methods and Applications" International Journal of Molecular Sciences 24, no. 14: 11592. https://doi.org/10.3390/ijms241411592
APA StyleLuo, Q., Wang, N., Que, H., Mai, E., Hu, Y., Tan, R., Gu, J., & Gong, P. (2023). Pluripotent Stem Cell-Derived Hepatocyte-like Cells: Induction Methods and Applications. International Journal of Molecular Sciences, 24(14), 11592. https://doi.org/10.3390/ijms241411592




