Effective Healing of Staphylococcus aureus-Infected Wounds in Pig Cathelicidin Protegrin-1-Overexpressing Transgenic Mice
Abstract
1. Introduction
2. Results and Discussion
2.1. Enhanced Wound Healing in Staphylococcus aureus-Infected Wounds in PG1 Tg Mice
2.2. Bacterial Clearance in Infected Wounds of PG1 Tg Mice
2.3. Efficient Formation of Neo-epithelium in PG1 Tg Mice with Infected Wounds
2.4. Successful Production of Recombinant PG1 Using a Bacterial Expression System
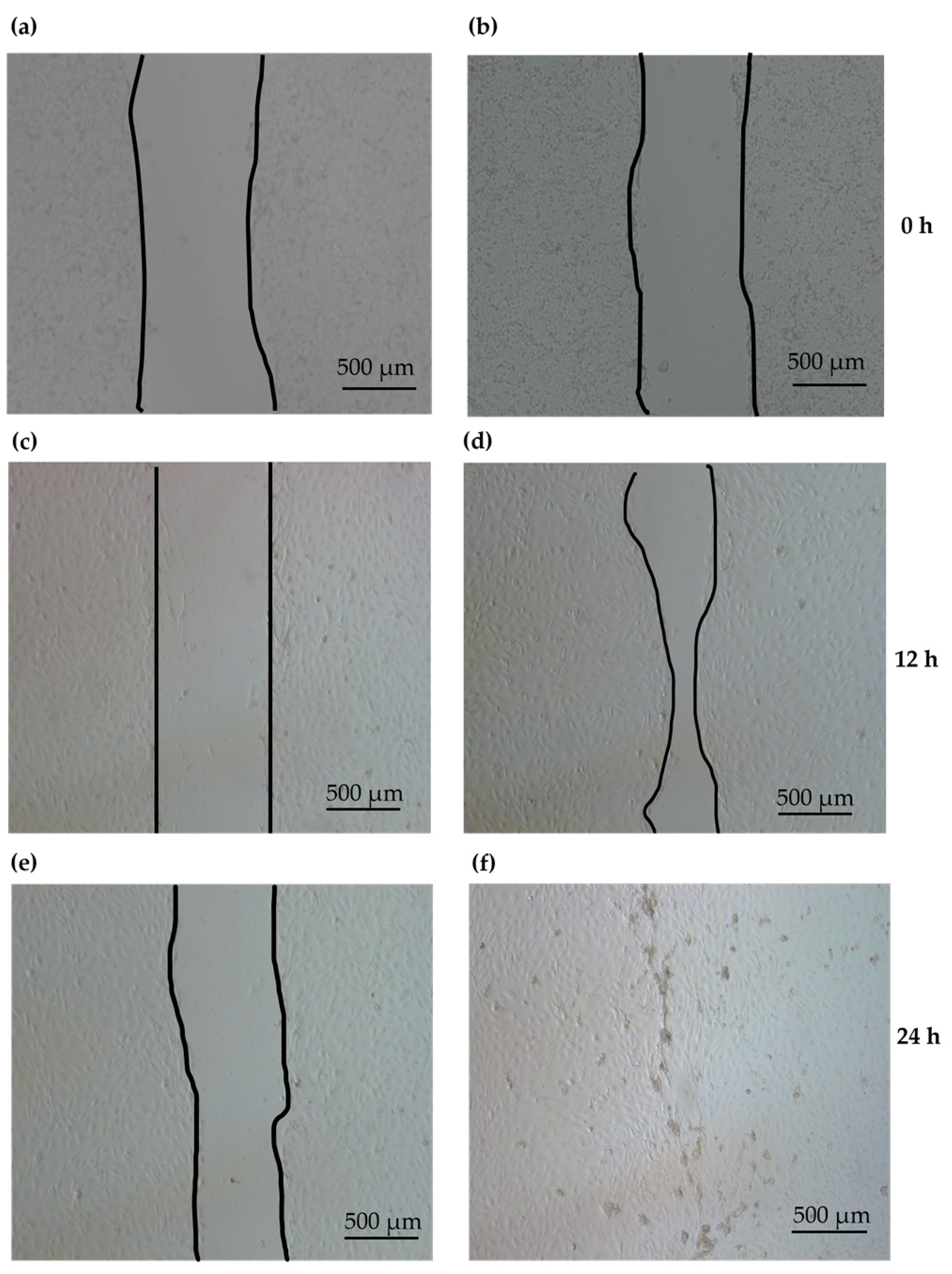
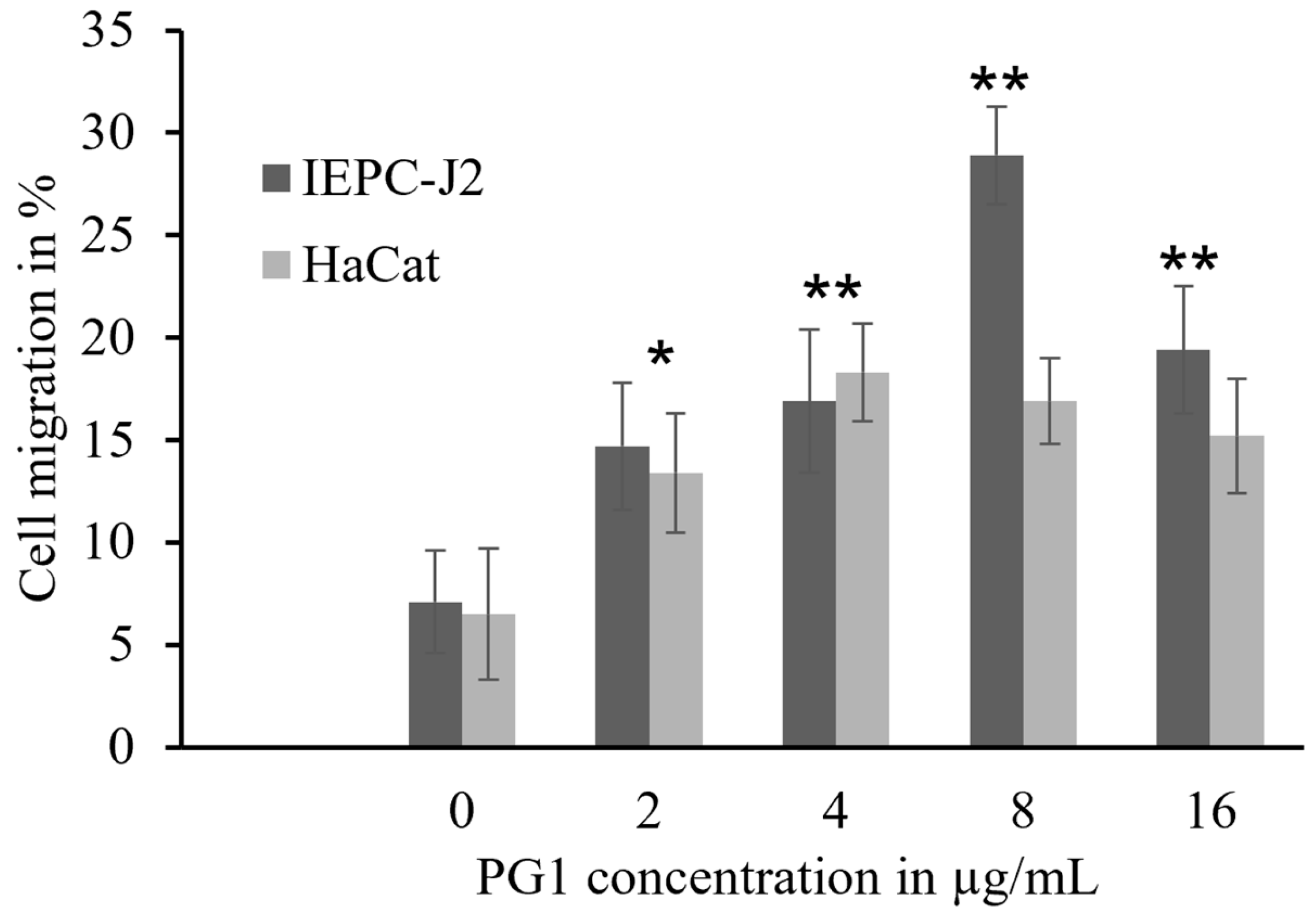
2.5. Enhancement of Epidermal Keratinocyte Migration by PG1
2.6. Immunomodulatory Effects of PG1 on Epidermal KCs
3. Materials and Methods
3.1. Animal Experiments
3.2. Determination of Bacteria Load in Wound Biopsies
3.3. Histological Analyses
3.4. KC Isolation
3.5. Production of Recombinant PG1 Peptides
3.6. Determination of PG1 Antimicrobial Activity
3.7. Cell Scratch Assay
3.8. Migration Assay
3.9. Real-Time Quantitative PCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nguyen, A.V.; Soulika, A.M. The Dynamics of the Skin’s Immune System. Int. J. Mol. Sci. 2019, 20, 1811. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.S.; Maan, Z.N.; Wu, J.C.; Rennert, R.C.; Hong, W.X.; Lai, T.S.; Cheung, A.T.; Walmsley, G.G.; Chung, M.T.; McArdle, A.; et al. Tissue engineering and regenerative repair in wound healing. Ann. Biomed. Eng. 2014, 42, 1494–1507. [Google Scholar] [CrossRef] [PubMed]
- Larouche, J.; Sheoran, S.; Maruyama, K.; Martino, M.M. Immune Regulation of Skin Wound Healing: Mechanisms and Novel Therapeutic Targets. Adv. Wound Care 2018, 7, 209–231. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Taylor, P.K.; Yeung, A.T.; Hancock, R.E. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J. Biotechnol. 2014, 191, 121–130. [Google Scholar] [CrossRef]
- Mancl, K.A.; Kirsner, R.S.; Ajdic, D. Wound biofilms: Lessons learned from oral biofilms. Wound Repair Regen. Off. Publ. Wound Health Soc. Eur. Tissue Repair Soc. 2013, 21, 352–362. [Google Scholar] [CrossRef]
- Del Pozo, J.L.; Patel, R. The challenge of treating biofilm-associated bacterial infections. Clin. Pharmacol. Ther. 2007, 82, 204–209. [Google Scholar] [CrossRef]
- Wilson, M.A. Skin and soft-tissue infections: Impact of resistant gram-positive bacteria. Am. J. Surg. 2003, 186, 35S–41S, discussion 42S–43S, 61S–64S. [Google Scholar] [CrossRef]
- Minasyan, H. Sepsis: Mechanisms of bacterial injury to the patient. Scand. J. Trauma Resusc. Emerg. Med. 2019, 27, 19. [Google Scholar] [CrossRef]
- Zadeh, M.; Khan, M.W.; Goh, Y.J.; Selle, K.; Owen, J.L.; Klaenhammer, T.; Mohamadzadeh, M. Induction of intestinal pro-inflammatory immune responses by lipoteichoic acid. J. Inflamm. 2012, 9, 7. [Google Scholar] [CrossRef]
- Zanetti, M. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 2004, 75, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Tollin, M.; Bergman, P.; Svenberg, T.; Jörnvall, H.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides in the first line defence of human colon mucosa. Peptides 2003, 24, 523–530. [Google Scholar] [CrossRef]
- Nagasundarapandian, S.; Cho, H.S.; Prathap, S.; Kang, M.; Choi, M.; Lee, Y.; Jeon, H.; Song, H.; Kim, J.H.; Park, C. Cathelicidin ΔPb-CATH4 derived from Python bivittatus accelerates the healing of Staphylococcus aureus-infected wounds in mice. Amino Acids 2021, 53, 313–317. [Google Scholar] [CrossRef]
- Turner, J.; Cho, Y.; Dinh, N.N.; Waring, A.J.; Lehrer, R.I. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 1998, 42, 2206–2214. [Google Scholar] [CrossRef]
- Penney, J.; Li, J. Protegrin 1 Enhances Innate Cellular Defense via the Insulin-Like Growth Factor 1 Receptor Pathway. Front. Cell. Infect. Microbiol. 2018, 8, 331. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.S.; Yum, J.; Larivière, A.; Lévêque, N.; Le, Q.V.C.; Ahn, B.; Jeon, H.; Hong, K.; Soundrarajan, N.; Kim, J.H.; et al. Opossum Cathelicidins Exhibit Antimicrobial Activity Against a Broad Spectrum of Pathogens Including West Nile Virus. Front. Immunol. 2020, 11, 347. [Google Scholar] [CrossRef] [PubMed]
- Mi, B.; Liu, J.; Liu, Y.; Hu, L.; Liu, Y.; Panayi, A.C.; Zhou, W.; Liu, G. The Designer Antimicrobial Peptide A-hBD-2 Facilitates Skin Wound Healing by Stimulating Keratinocyte Migration and Proliferation. Cell. Physiol. Biochem. 2018, 51, 647–663. [Google Scholar] [CrossRef]
- Koczulla, R.; von Degenfeld, G.; Kupatt, C.; Krötz, F.; Zahler, S.; Gloe, T.; Issbrücker, K.; Unterberger, P.; Zaiou, M.; Lebherz, C.; et al. An angiogenic role for the human peptide antibiotic LL-37/hCAP-18. J. Clin. Investig. 2003, 111, 1665–1672. [Google Scholar] [CrossRef]
- Heilborn, J.D.; Nilsson, M.F.; Kratz, G.; Weber, G.; Sørensen, O.; Borregaard, N.; Ståhle-Bäckdahl, M. The cathelicidin anti-microbial peptide LL-37 is involved in re-epithelialization of human skin wounds and is lacking in chronic ulcer epithelium. J. Investig. Dermatol. 2003, 120, 379–389. [Google Scholar] [CrossRef]
- Tomasinsig, L.; Zanetti, M. The cathelicidins--structure, function and evolution. Curr. Protein Pept. Sci. 2005, 6, 23–34. [Google Scholar] [CrossRef]
- Zanetti, M.; Gennaro, R.; Scocchi, M.; Skerlavaj, B. Structure and biology of cathelicidins. Adv. Exp. Med. Biol. 2000, 479, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T. Cathelicidins: A family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 2002, 9, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Duplantier, A.J.; van Hoek, M.L. The Human Cathelicidin Antimicrobial Peptide LL-37 as a Potential Treatment for Polymicrobial Infected Wounds. Front. Immunol. 2013, 4, 143. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.-K.; Le, M.T.; Cho, H.; Soundrarajan, N.; Jeon, H.; Park, C.K.; Cha, S.Y.; Kim, J.-H.; Seo, K.; Park, C. Defining the genetic relationship of protegrin-related sequences and the in vivo expression of protegrins. FEBS J. 2014, 281, 5420–5431. [Google Scholar] [CrossRef]
- Kościuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzałkowska, N.; Jóźwik, A.; Horbańczuk, J.; Krzyżewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: Family of antimicrobial peptides. A review. Mol. Biol. Rep. 2012, 39, 10957–10970. [Google Scholar] [CrossRef]
- Dorschner, R.A.; Pestonjamasp, V.K.; Tamakuwala, S.; Ohtake, T.; Rudisill, J.; Nizet, V.; Agerberth, B.; Gudmundsson, G.H.; Gallo, R.L. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J. Investig. Dermatol. 2001, 117, 91–97. [Google Scholar] [CrossRef]
- Ramos, R.; Silva, J.P.; Rodrigues, A.C.; Costa, R.; Guardao, L.; Schmitt, F.; Soares, R.; Vilanova, M.; Domingues, L.; Gama, M. Wound healing activity of the human antimicrobial peptide LL37. Peptides 2011, 32, 1469–1476. [Google Scholar] [CrossRef]
- Cao, X.; Wang, Y.; Wu, C.; Li, X.; Fu, Z.; Yang, M.; Bian, W.; Wang, S.; Song, Y.; Tang, J.; et al. Cathelicidin-OA1, a novel antioxidant peptide identified from an amphibian, accelerates skin wound healing. Sci. Rep. 2018, 8, 943. [Google Scholar] [CrossRef]
- Chromek, M.; Slamová, Z.; Bergman, P.; Kovács, L.; Podracká, L.; Ehrén, I.; Hökfelt, T.; Gudmundsson, G.H.; Gallo, R.L.; Agerberth, B.; et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006, 12, 636–641. [Google Scholar] [CrossRef]
- Lowry, M.B.; Guo, C.; Zhang, Y.; Fantacone, M.L.; Logan, I.E.; Campbell, Y.; Zhang, W.; Le, M.; Indra, A.K.; Ganguli-Indra, G.; et al. A mouse model for vitamin D-induced human cathelicidin antimicrobial peptide gene expression. J. Steroid Biochem. Mol. Biol. 2020, 198, 105552. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.H.; Ohtake, T.; Zaiou, M.; Murakami, M.; Rudisill, J.A.; Lin, K.H.; Gallo, R.L. Expression of an additional cathelicidin antimicrobial peptide protects against bacterial skin infection. Proc. Natl. Acad. Sci. USA 2005, 102, 3750–3755. [Google Scholar] [CrossRef]
- Wu, G.; Deng, X.; Wu, P.; Shen, Z.; Xu, H. Subacute toxicity of antimicrobial peptide S-thanatin in ICR mice. Peptides 2012, 36, 109–113. [Google Scholar] [CrossRef]
- Wei, D.; Zhang, X. Biosynthesis, bioactivity, biotoxicity and applications of antimicrobial peptides for human health. Biosaf. Health 2022, 4, 118–134. [Google Scholar] [CrossRef]
- Soundrarajan, N.; Park, S.; Le Van Chanh, Q.; Cho, H.S.; Raghunathan, G.; Ahn, B.; Song, H.; Kim, J.H.; Park, C. Protegrin-1 cytotoxicity towards mammalian cells positively correlates with the magnitude of conformational changes of the unfolded form upon cell interaction. Sci. Rep. 2019, 9, 11569. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, L.; Lehrer, R.I. Identification of a new member of the protegrin family by cDNA cloning. FEBS Lett. 1994, 346, 285–288. [Google Scholar] [CrossRef]
- Maystrenko, A.; Feng, Y.; Akhtar, N.; Li, J. The Addition of a Synthetic LPS-Targeting Domain Improves Serum Stability While Maintaining Antimicrobial, Antibiofilm, and Cell Stimulating Properties of an Antimicrobial Peptide. Biomolecules 2020, 10, 1014. [Google Scholar] [CrossRef]
- Choi, M.K.; Le, M.T.; Cho, H.S.; Lee, J.; Jeon, H.; Cha, S.Y.; Na, M.; Chun, T.; Kim, J.H.; Song, H.; et al. Transgenic Mice Overexpressing PG1 Display Corneal Opacity and Severe Inflammation in the Eye. Int. J. Mol. Sci. 2021, 22, 1586. [Google Scholar] [CrossRef]
- Cheung, Q.C.K.; Turner, P.V.; Song, C.; Wu, D.; Cai, H.Y.; MacInnes, J.I.; Li, J. Enhanced Resistance to Bacterial Infection in Protegrin-1 Transgenic Mice. Antimicrob. Agents Chemother. 2008, 52, 1812–1819. [Google Scholar] [CrossRef][Green Version]
- Soundrarajan, N.; Cho, H.-s.; Ahn, B.; Choi, M.; Thong, L.M.; Choi, H.; Cha, S.-Y.; Kim, J.-H.; Park, C.-K.; Seo, K.; et al. Green fluorescent protein as a scaffold for high efficiency production of functional bacteriotoxic proteins in Escherichia coli. Sci. Rep. 2016, 6, 20661. [Google Scholar] [CrossRef] [PubMed]
- Seeger, M.A.; Paller, A.S. The Roles of Growth Factors in Keratinocyte Migration. Adv. Wound Care 2015, 4, 213–224. [Google Scholar] [CrossRef]
- Niyonsaba, F.; Ushio, H.; Nakano, N.; Ng, W.; Sayama, K.; Hashimoto, K.; Nagaoka, I.; Okumura, K.; Ogawa, H. Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Investig. Dermatol. 2007, 127, 594–604. [Google Scholar] [CrossRef]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. Growth factors and cytokines in wound healing. Wound Repair Regen. Off. Publ. Wound Health Soc. Eur. Tissue Repair Soc. 2008, 16, 585–601. [Google Scholar] [CrossRef]
- Werner, S.; Grose, R. Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 2003, 83, 835–870. [Google Scholar] [CrossRef] [PubMed]
- Klemke, R.L.; Cai, S.; Giannini, A.L.; Gallagher, P.J.; de Lanerolle, P.; Cheresh, D.A. Regulation of cell motility by mitogen-activated protein kinase. J. Cell Biol. 1997, 137, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, M.S.; Jung, S.J.; Kim, D.; Park, H.J.; Cho, D. ERK activating peptide, AES16-2M promotes wound healing through accelerating migration of keratinocytes. Sci. Rep. 2018, 8, 14398. [Google Scholar] [CrossRef] [PubMed]
- Schanzer, J.M.; Wartha, K.; Moessner, E.; Hosse, R.J.; Moser, S.; Croasdale, R.; Trochanowska, H.; Shao, C.; Wang, P.; Shi, L.; et al. XGFR*, a novel affinity-matured bispecific antibody targeting IGF-1R and EGFR with combined signaling inhibition and enhanced immune activation for the treatment of pancreatic cancer. MAbs 2016, 8, 811–827. [Google Scholar] [CrossRef]
- Smith, T.J. Insulin-like growth factor-I regulation of immune function: A potential therapeutic target in autoimmune diseases? Pharmacol. Rev. 2010, 62, 199–236. [Google Scholar] [CrossRef]
- Kim, D.J.; Lee, Y.W.; Park, M.K.; Shin, J.R.; Lim, K.J.; Cho, J.H.; Kim, S.C. Efficacy of the designer antimicrobial peptide SHAP1 in wound healing and wound infection. Amino Acids 2014, 46, 2333–2343. [Google Scholar] [CrossRef]
- Mangoni, M.L.; McDermott, A.M.; Zasloff, M. Antimicrobial peptides and wound healing: Biological and therapeutic considerations. Exp. Derm. 2016, 25, 167–173. [Google Scholar] [CrossRef]
- Kim, D.; Soundrarajan, N.; Lee, J.; Cho, H.S.; Choi, M.; Cha, S.Y.; Ahn, B.; Jeon, H.; Le, M.T.; Song, H.; et al. Genomewide Analysis of the Antimicrobial Peptides in Python bivittatus and Characterization of Cathelicidins with Potent Antimicrobial Activity and Low Cytotoxicity. Antimicrob. Agents Chemother. 2017, 61, 9. [Google Scholar] [CrossRef]
- Fujimura, S.; Fuse, K.; Takane, H.; Nakano, Y.; Watanabe, A.; Fuse, K.; Gomi, K.; Kikuchi, T. Antibacterial effects of brand-name teicoplanin and generic products against clinical isolates of methicillin-resistant Staphylococcus aureus. J. Infect. Chemother. 2011, 17, 30–33. [Google Scholar] [CrossRef]
- Steinberg, D.A.; Hurst, M.A.; Fujii, C.A.; Kung, A.H.; Ho, J.F.; Cheng, F.C.; Loury, D.J.; Fiddes, J.C. Protegrin-1: A broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 1997, 41, 1738–1742. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Adase, C.A.; Zhang, L.J. Isolation and Culture of Primary Mouse Keratinocytes from Neonatal and Adult Mouse Skin. J. Vis. Exp. JoVE 2017, 125, 56027. [Google Scholar] [CrossRef]
- CLSI Supplement M100-S28; Performance Standards for Antimicrobial Susceptibility Testing, 28th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Choi, M.; Cho, H.S.; Ahn, B.; Prathap, S.; Nagasundarapandian, S.; Park, C. Genomewide Analysis and Biological Characterization of Cathelicidins with Potent Antimicrobial Activity and Low Cytotoxicity from Three Bat Species. Antibiotics 2022, 11, 989. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
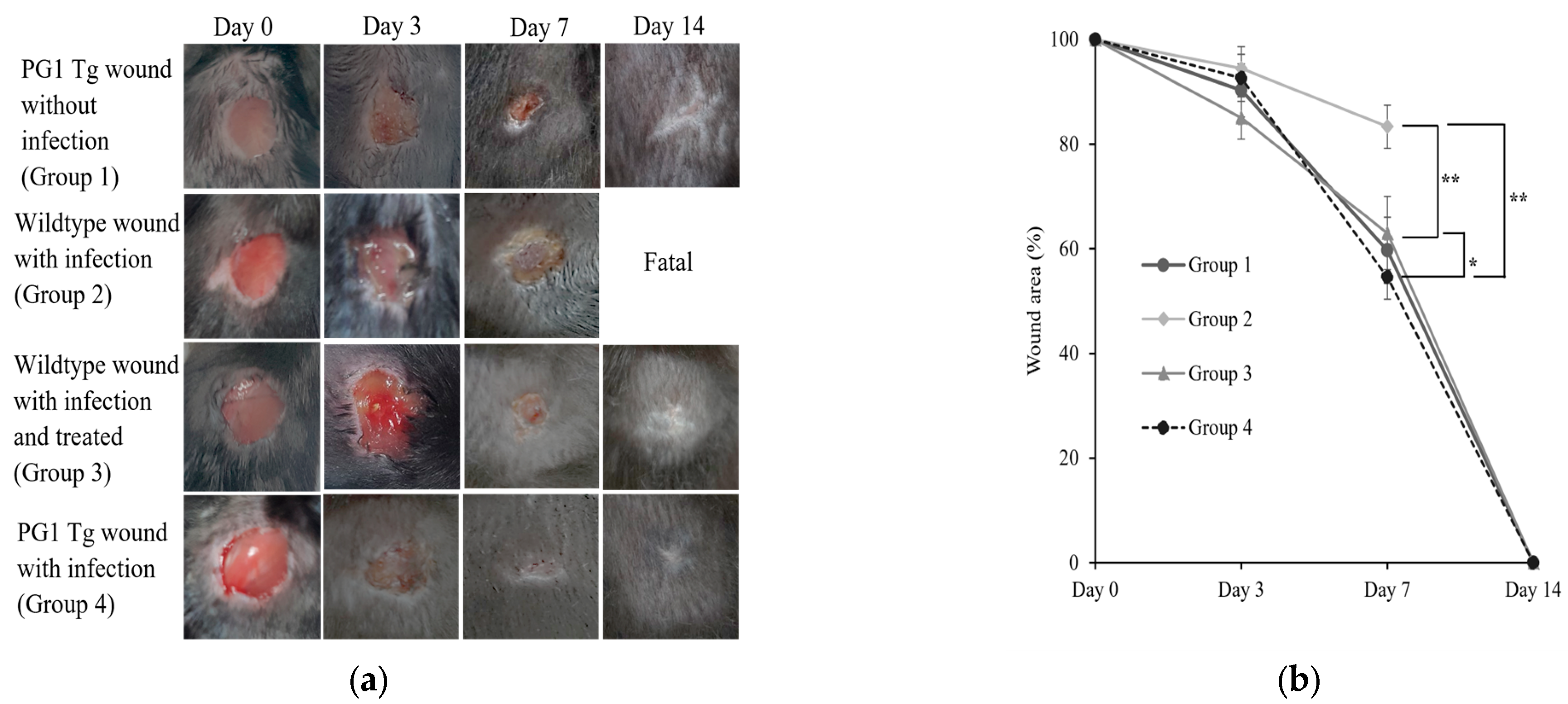
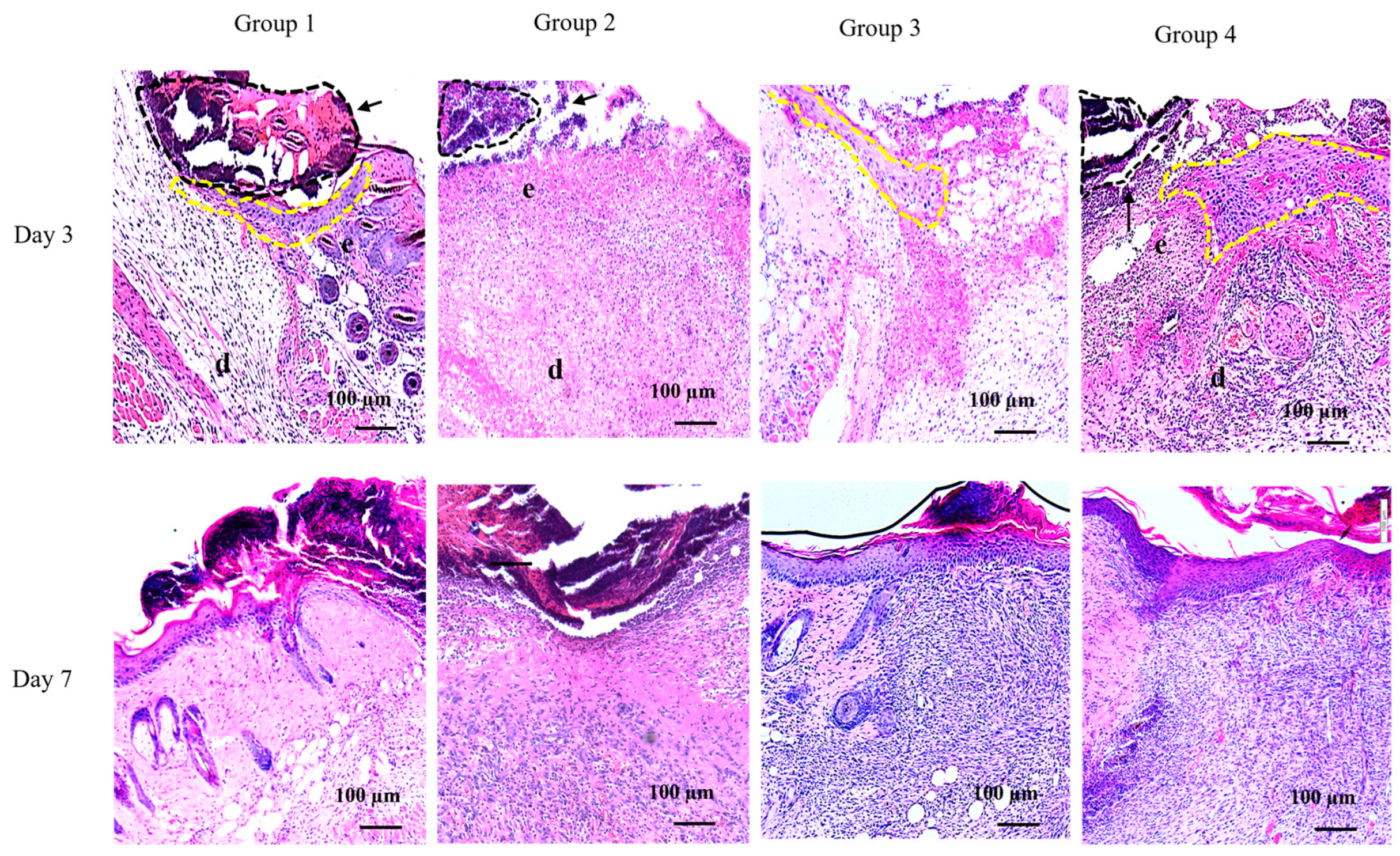
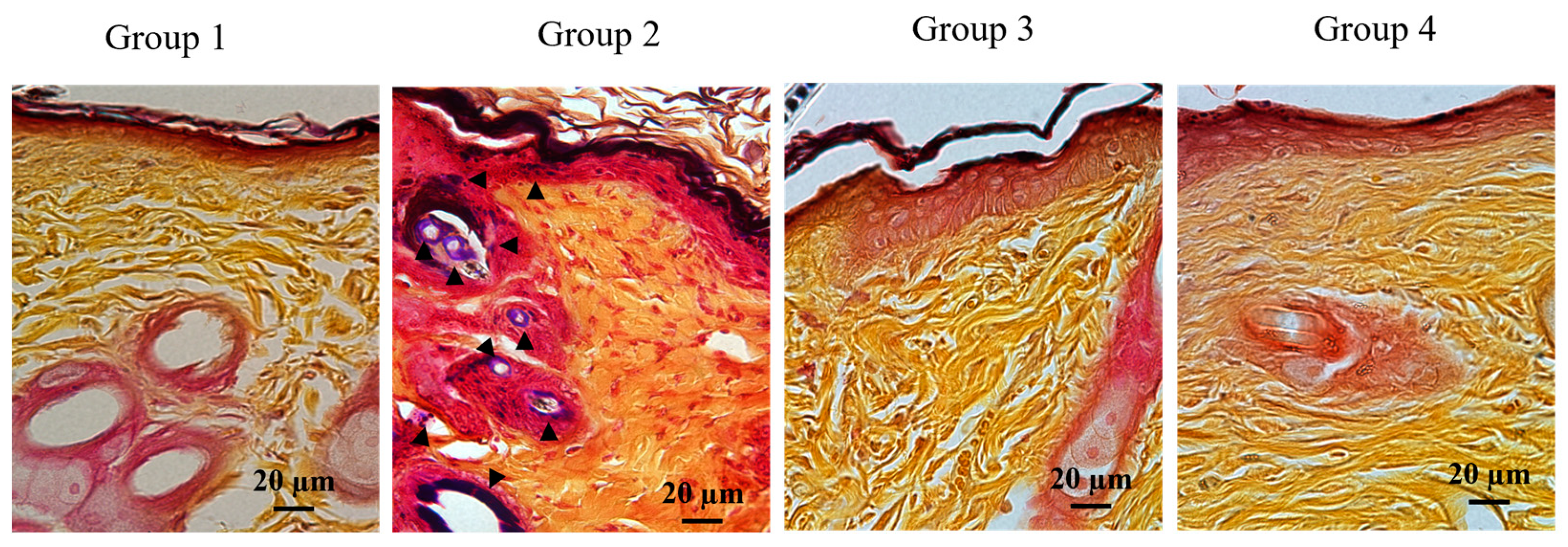

| Experimental Conditions | Day 0 (CFUs/g) | Day 3 (CFUs/g) | Day 7 (CFUs/g) | Day 14 (CFUs/g) |
|---|---|---|---|---|
| PG1 mice with uninfected wounds (group 1) 1 | 0 | 0 | 3 ± 4.3 | 5 ± 5.1 |
| Wild-type mice with S. aureus-infected wounds (group 2) 1 | 1.85 ± 0.8 × 106 * | 8.21 ± 3.4 × 108 | 8.75 ± 3.2 × 108 | Fatal |
| Wild-type mice with S. aureus-infected wounds treated with gentamycin for 3 days (group 3) 1 | 1.85 ± 0.8 × 106 * | 4.89 ± 2.3 × 105 | 1.41 ± 0.9 × 102 | 41 ± 3.8 |
| PG1 transgenic mouse with S. aureus-infected wounds that were not subjected to antibiotic treatment (group 4) 1 | 1.85 ± 0.8 × 106 * | 6.5 ± 2.7 × 104 | 1.39 ± 2.4 × 102 | 31 ± 4.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soundrarajan, N.; Somasundaram, P.; Kim, D.; Cho, H.-S.; Jeon, H.; Ahn, B.; Kang, M.; Song, H.; Park, C. Effective Healing of Staphylococcus aureus-Infected Wounds in Pig Cathelicidin Protegrin-1-Overexpressing Transgenic Mice. Int. J. Mol. Sci. 2023, 24, 11658. https://doi.org/10.3390/ijms241411658
Soundrarajan N, Somasundaram P, Kim D, Cho H-S, Jeon H, Ahn B, Kang M, Song H, Park C. Effective Healing of Staphylococcus aureus-Infected Wounds in Pig Cathelicidin Protegrin-1-Overexpressing Transgenic Mice. International Journal of Molecular Sciences. 2023; 24(14):11658. https://doi.org/10.3390/ijms241411658
Chicago/Turabian StyleSoundrarajan, Nagasundarapandian, Prathap Somasundaram, Dohun Kim, Hye-Sun Cho, Hyoim Jeon, Byeonyong Ahn, Mingue Kang, Hyuk Song, and Chankyu Park. 2023. "Effective Healing of Staphylococcus aureus-Infected Wounds in Pig Cathelicidin Protegrin-1-Overexpressing Transgenic Mice" International Journal of Molecular Sciences 24, no. 14: 11658. https://doi.org/10.3390/ijms241411658
APA StyleSoundrarajan, N., Somasundaram, P., Kim, D., Cho, H.-S., Jeon, H., Ahn, B., Kang, M., Song, H., & Park, C. (2023). Effective Healing of Staphylococcus aureus-Infected Wounds in Pig Cathelicidin Protegrin-1-Overexpressing Transgenic Mice. International Journal of Molecular Sciences, 24(14), 11658. https://doi.org/10.3390/ijms241411658







