Pokkali: A Naturally Evolved Salt-Tolerant Rice Shows a Distinguished Set of lncRNAs Possibly Contributing to the Tolerant Phenotype
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of O. sativa lncRNAs
2.2. Characterization of the Identified lncRNAs
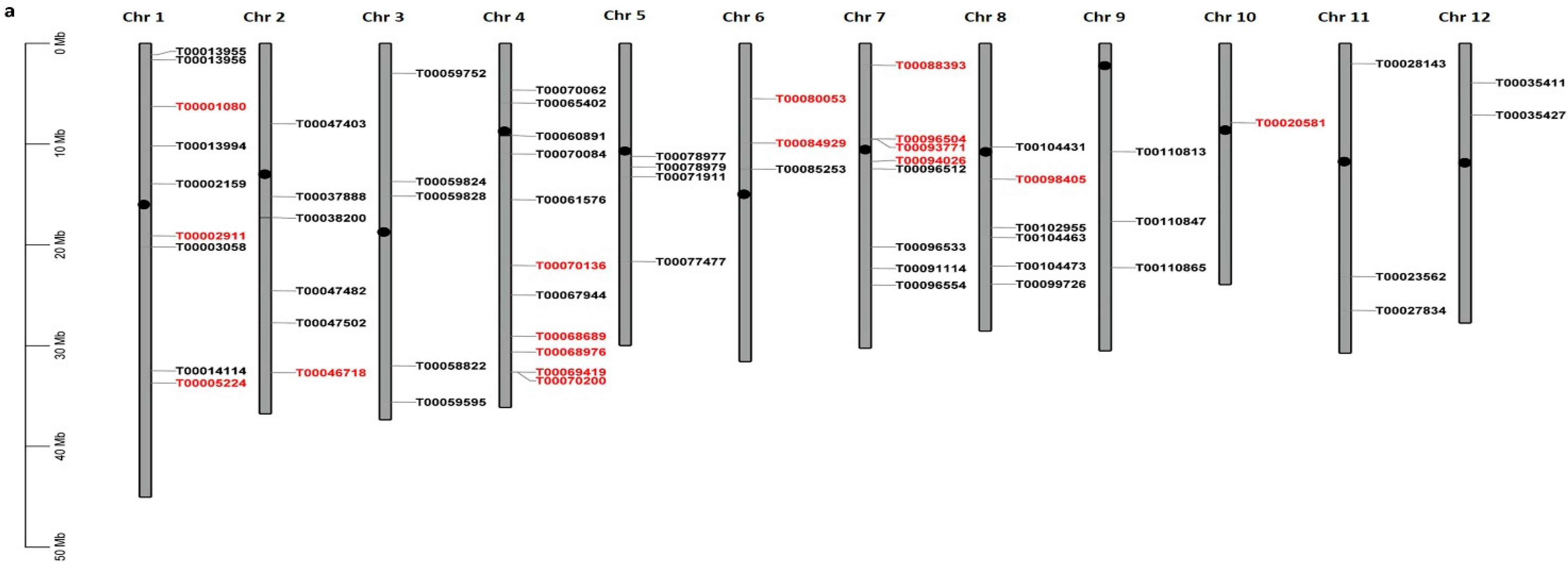
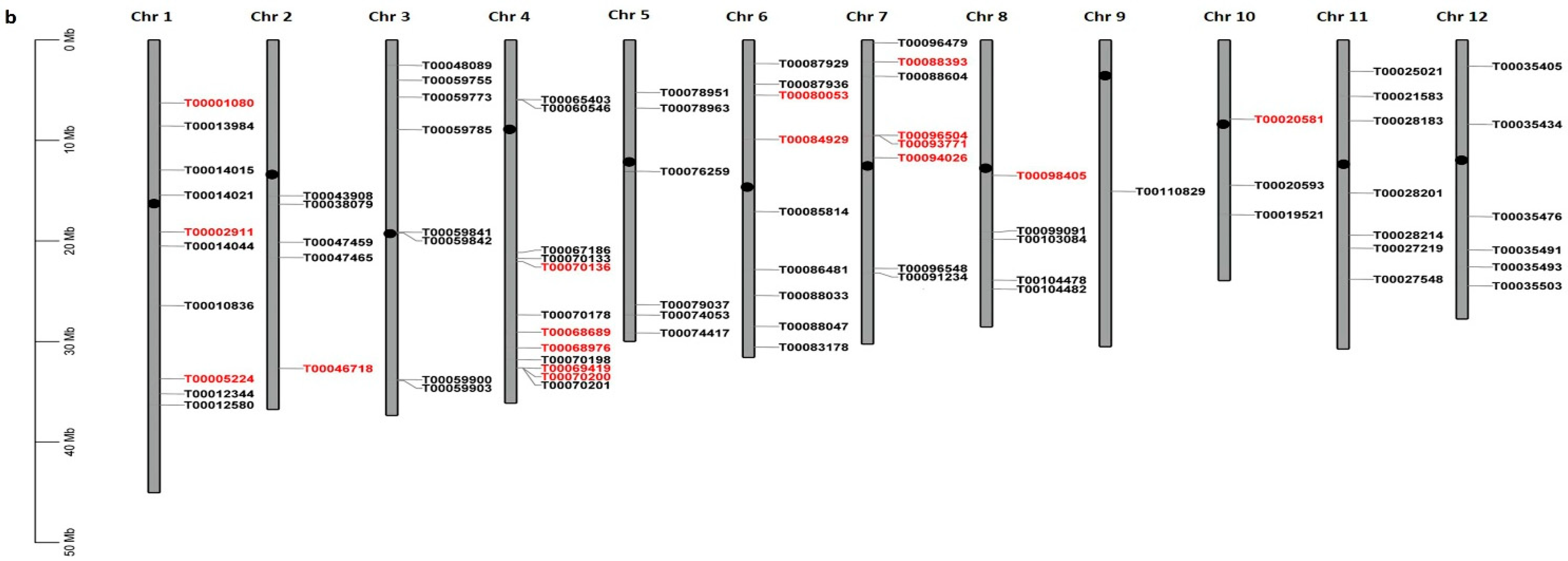
2.2.1. Chromosomal Distribution
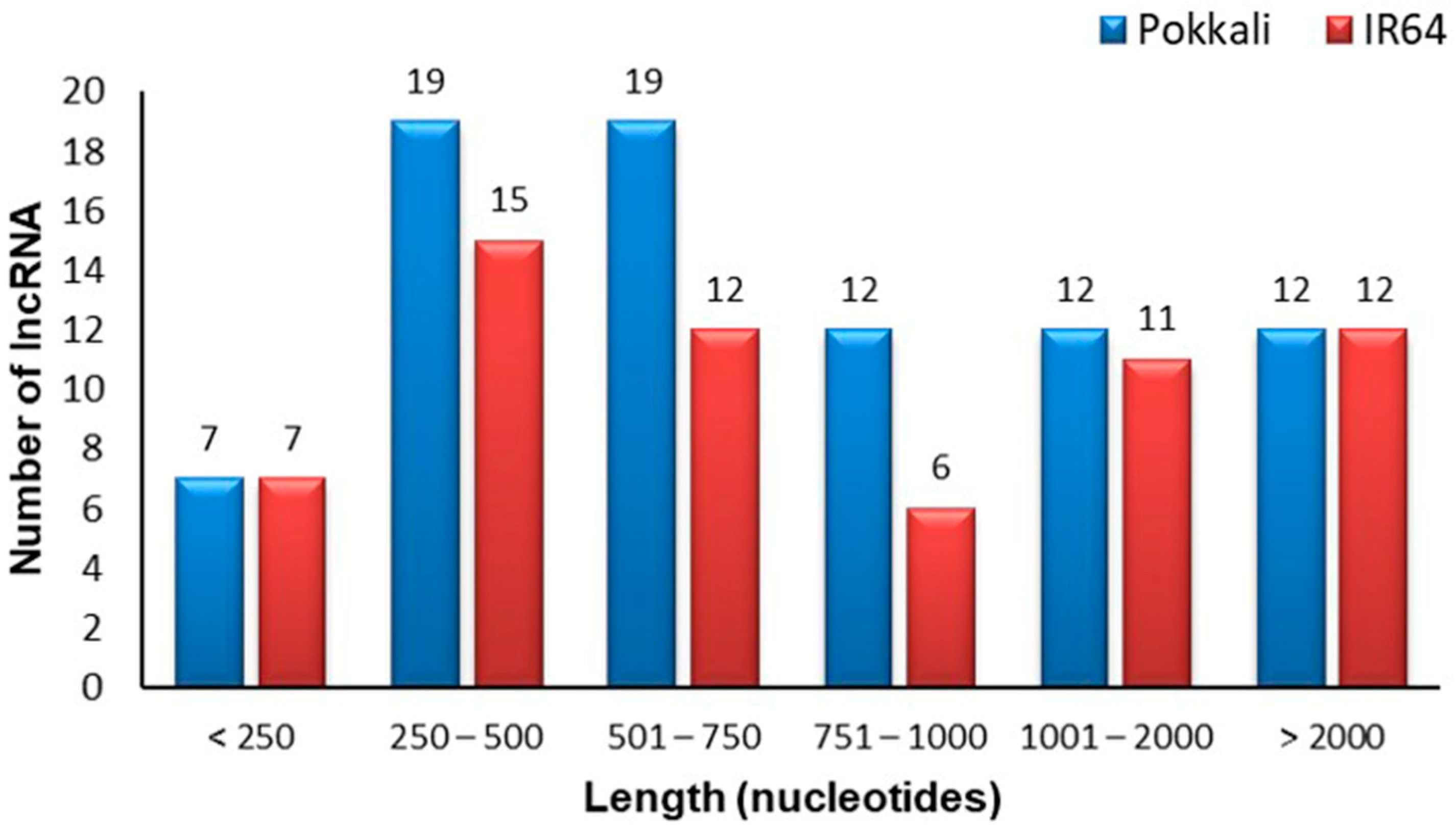
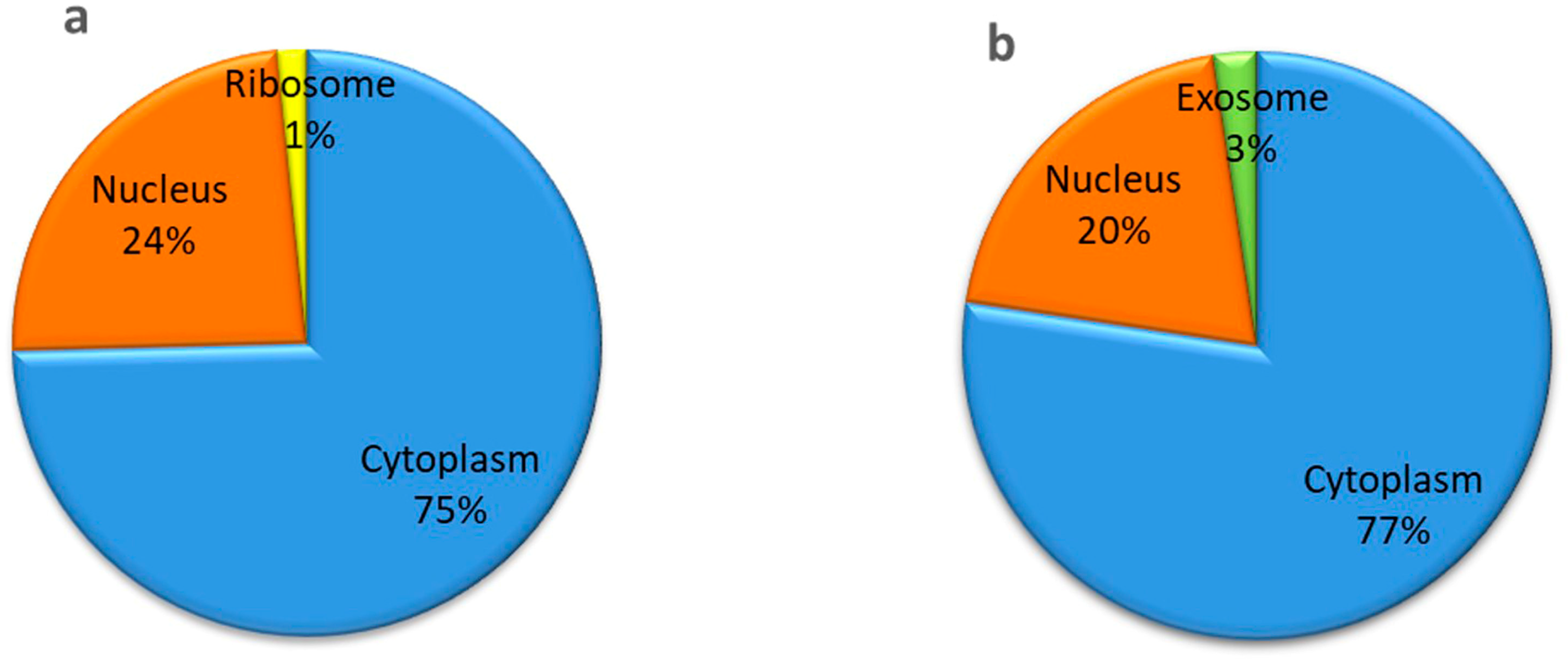
2.2.2. Length Distribution and Subcellular Localization
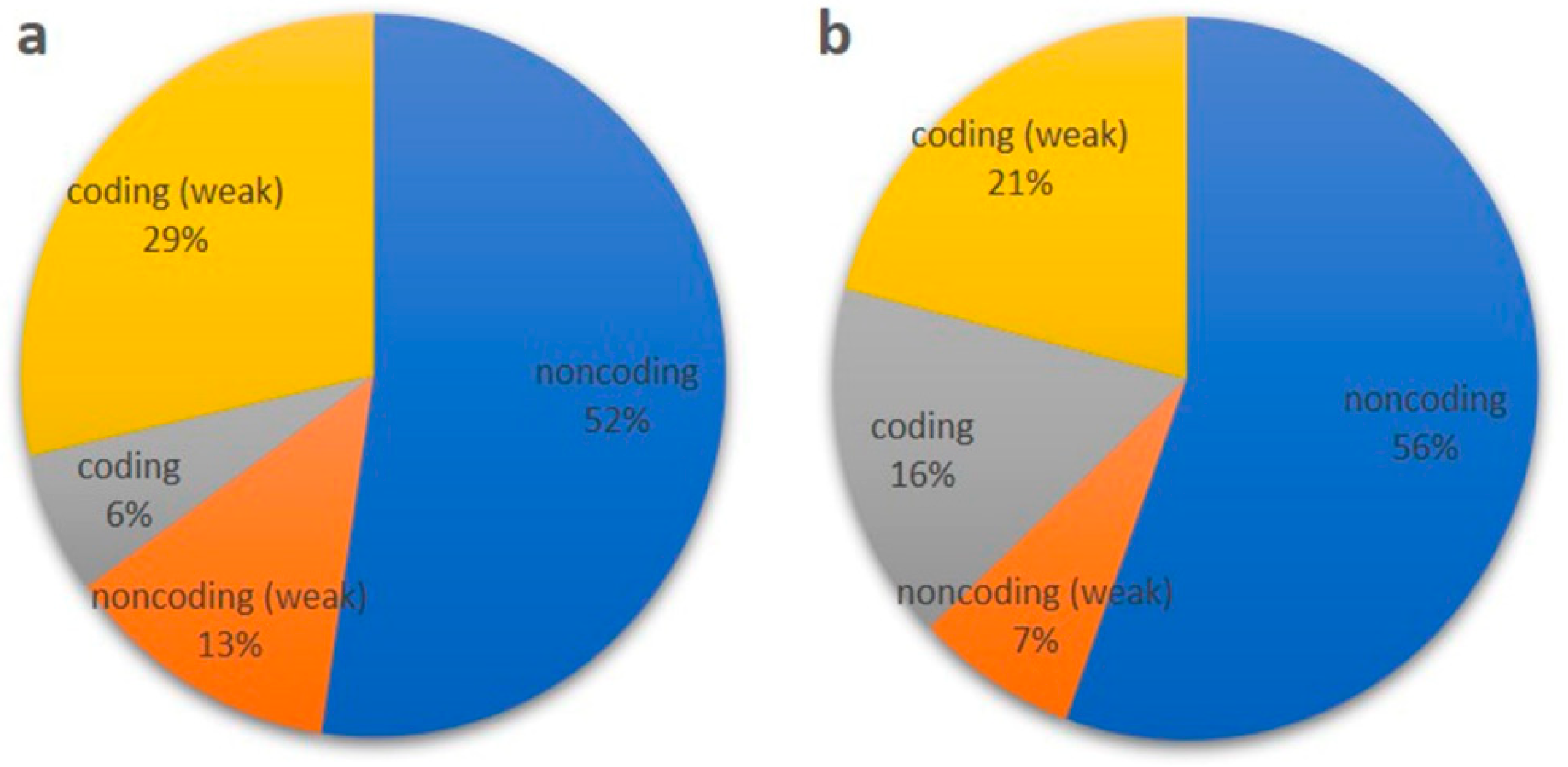
2.2.3. Classification and Other Related Traits
2.3. Expression Profile of the Identified O. sativa lncRNAs
2.4. Target Identification of the lncRNAs in O. sativa
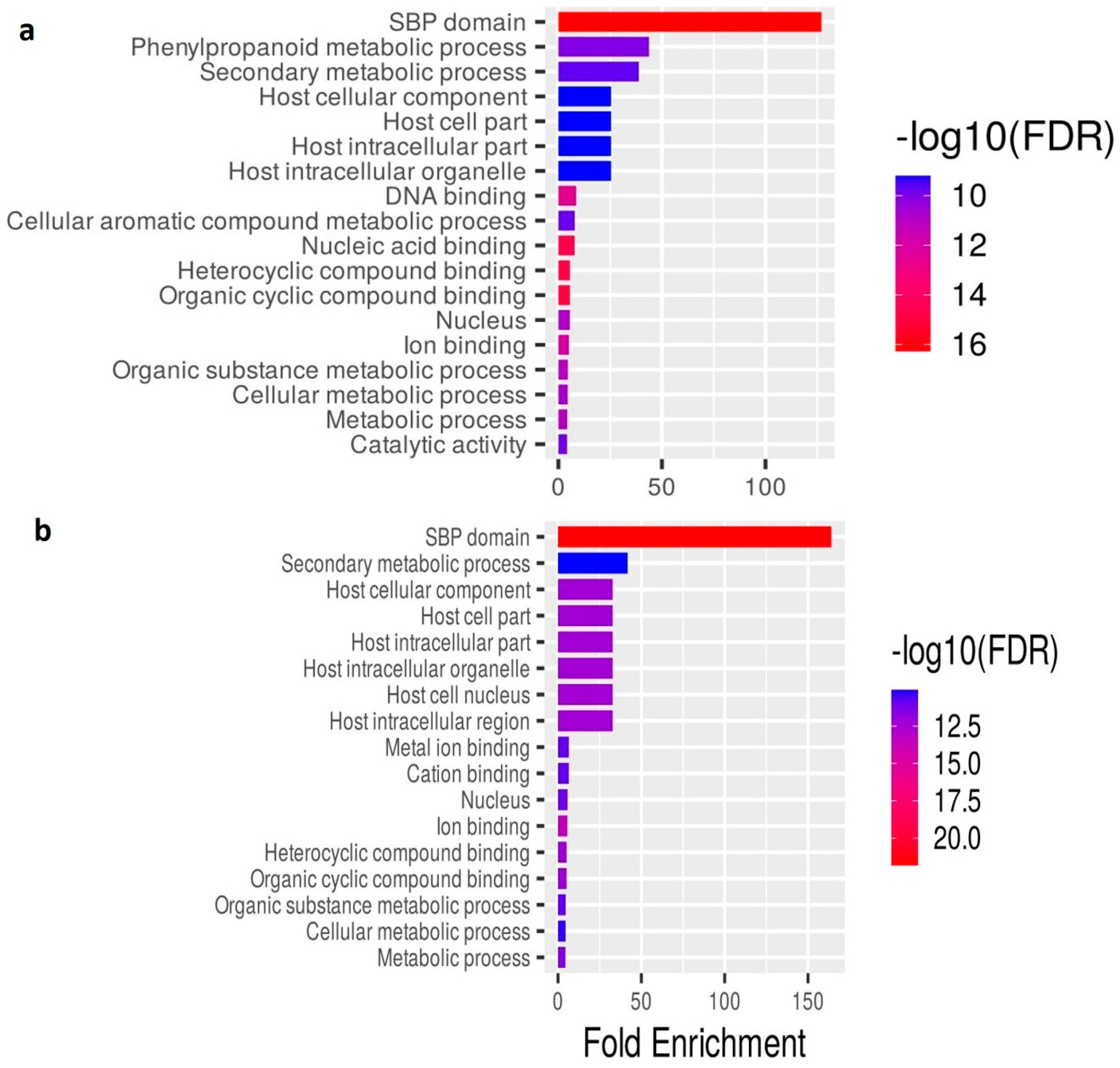
2.5. Functional Annotation of the Salt Stress-Responsive lncRNAs in O. sativa
2.6. Association of Salt-Responsive lncRNAs with Transcription Factors in O. sativa
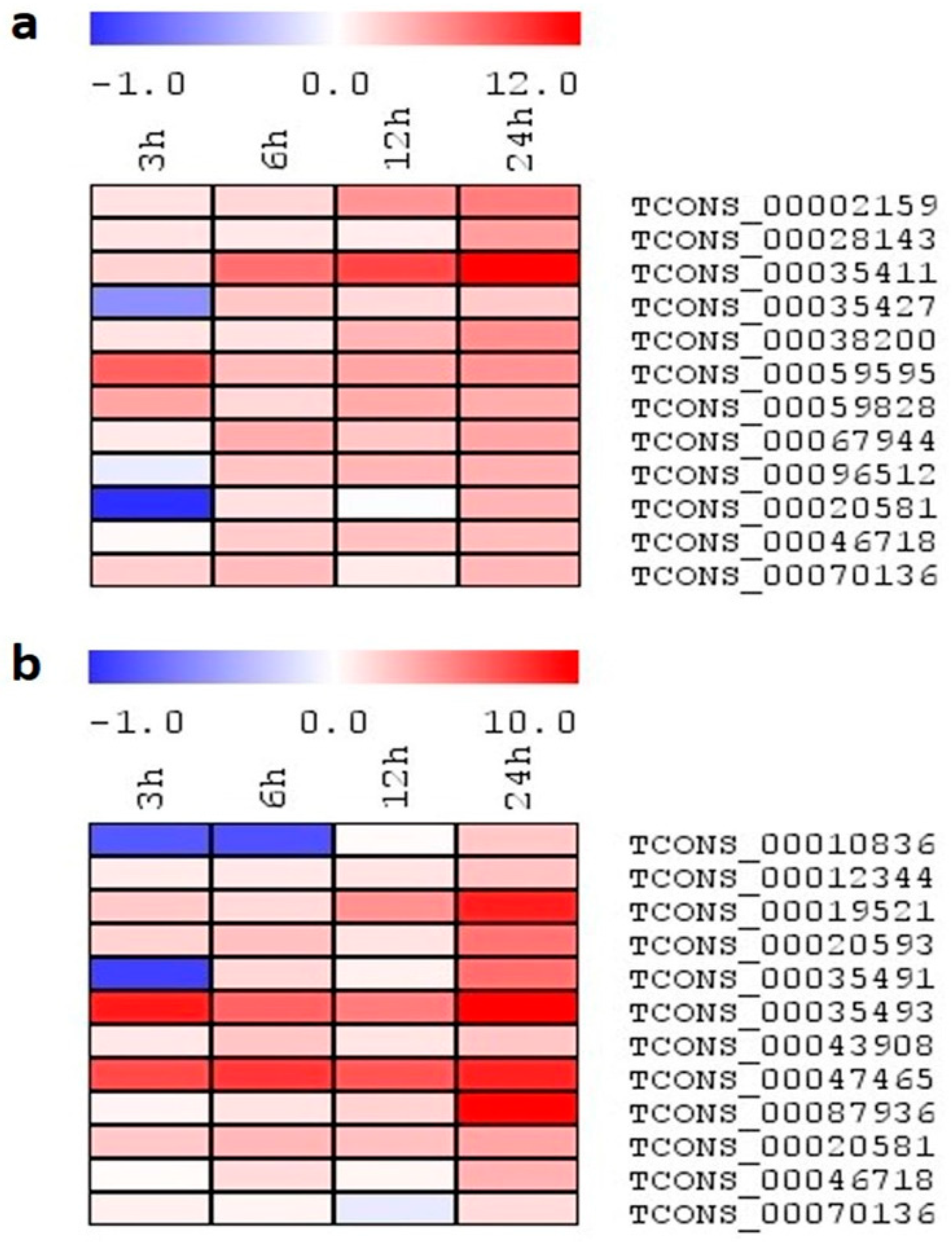
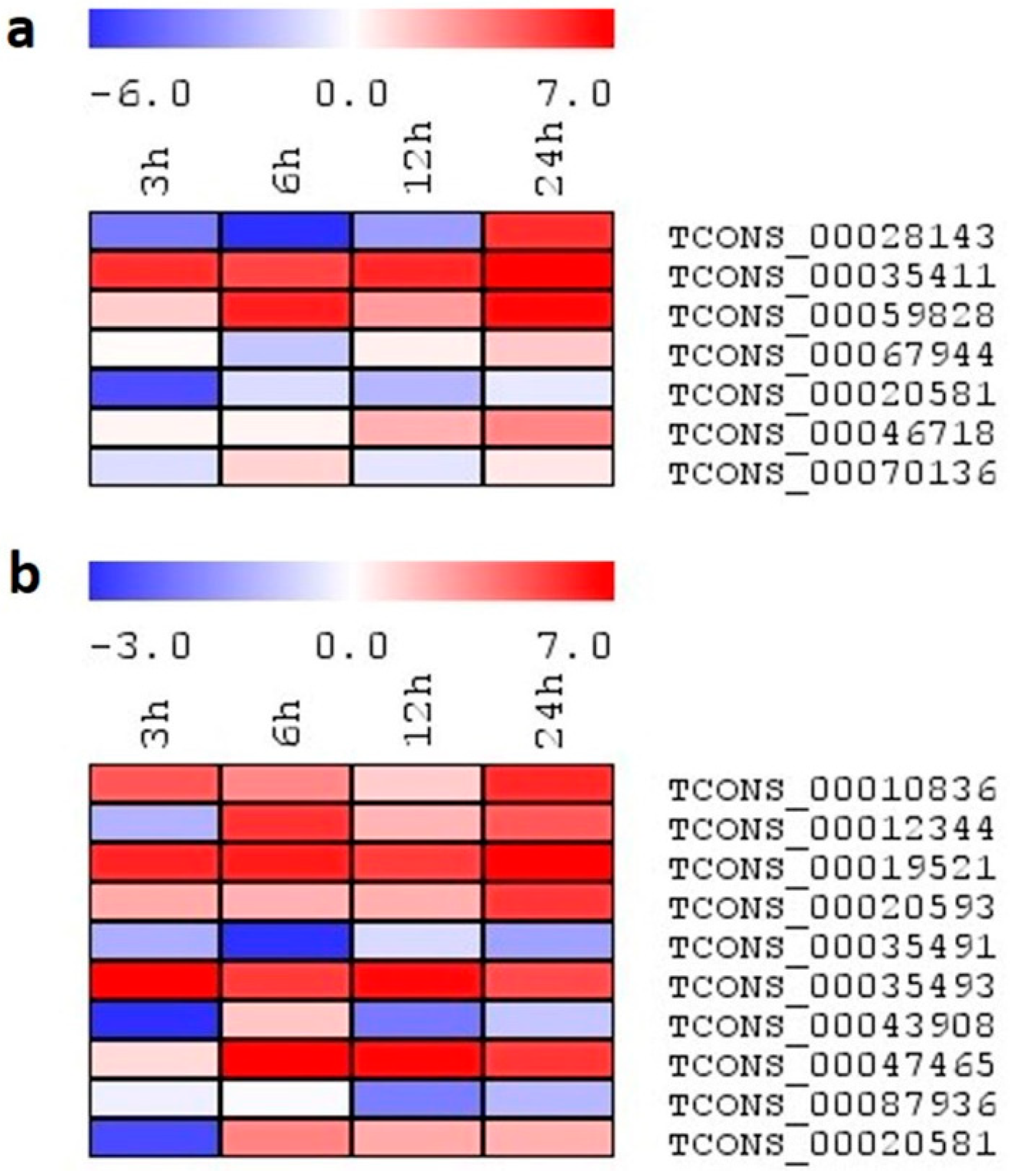
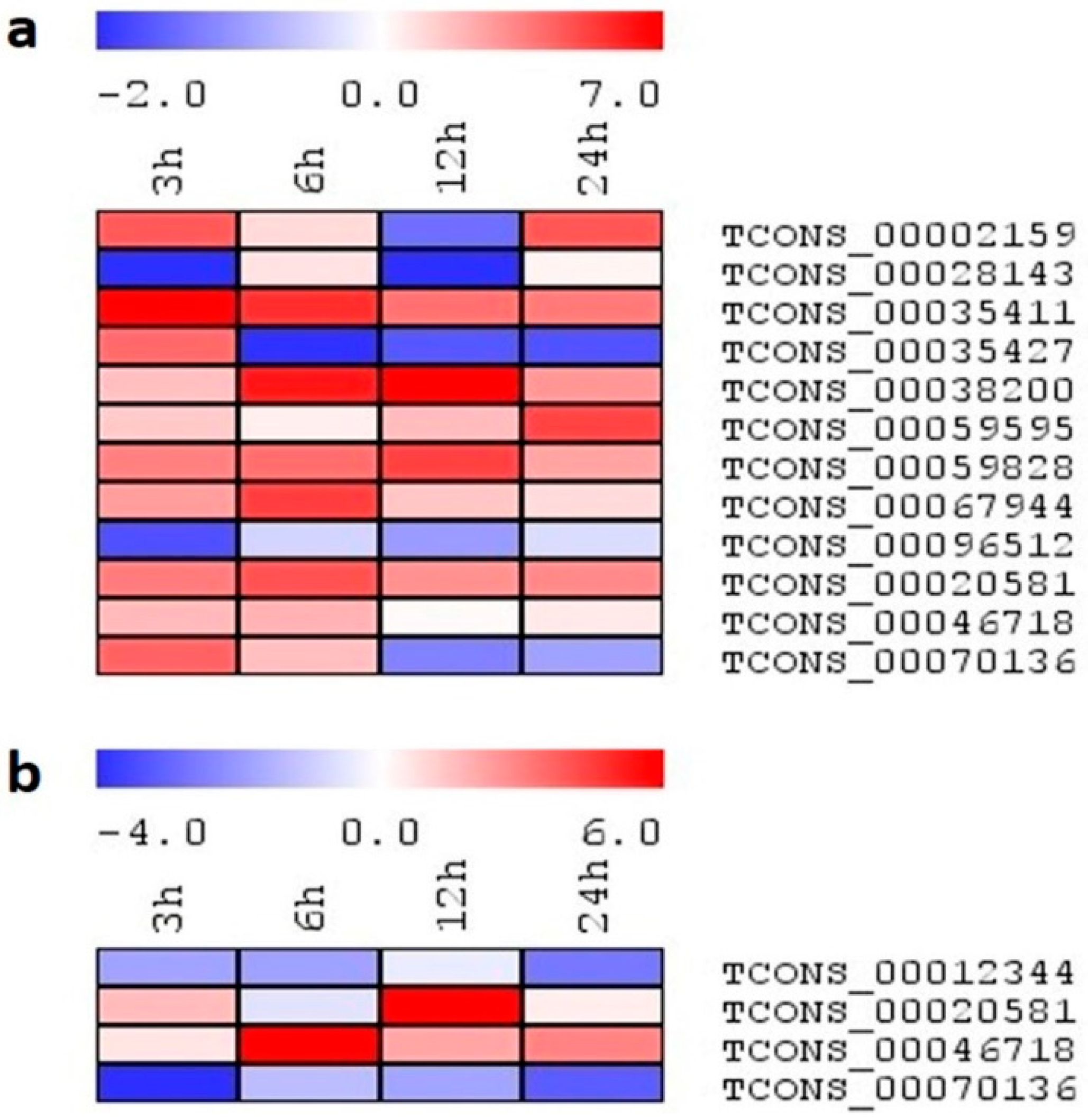
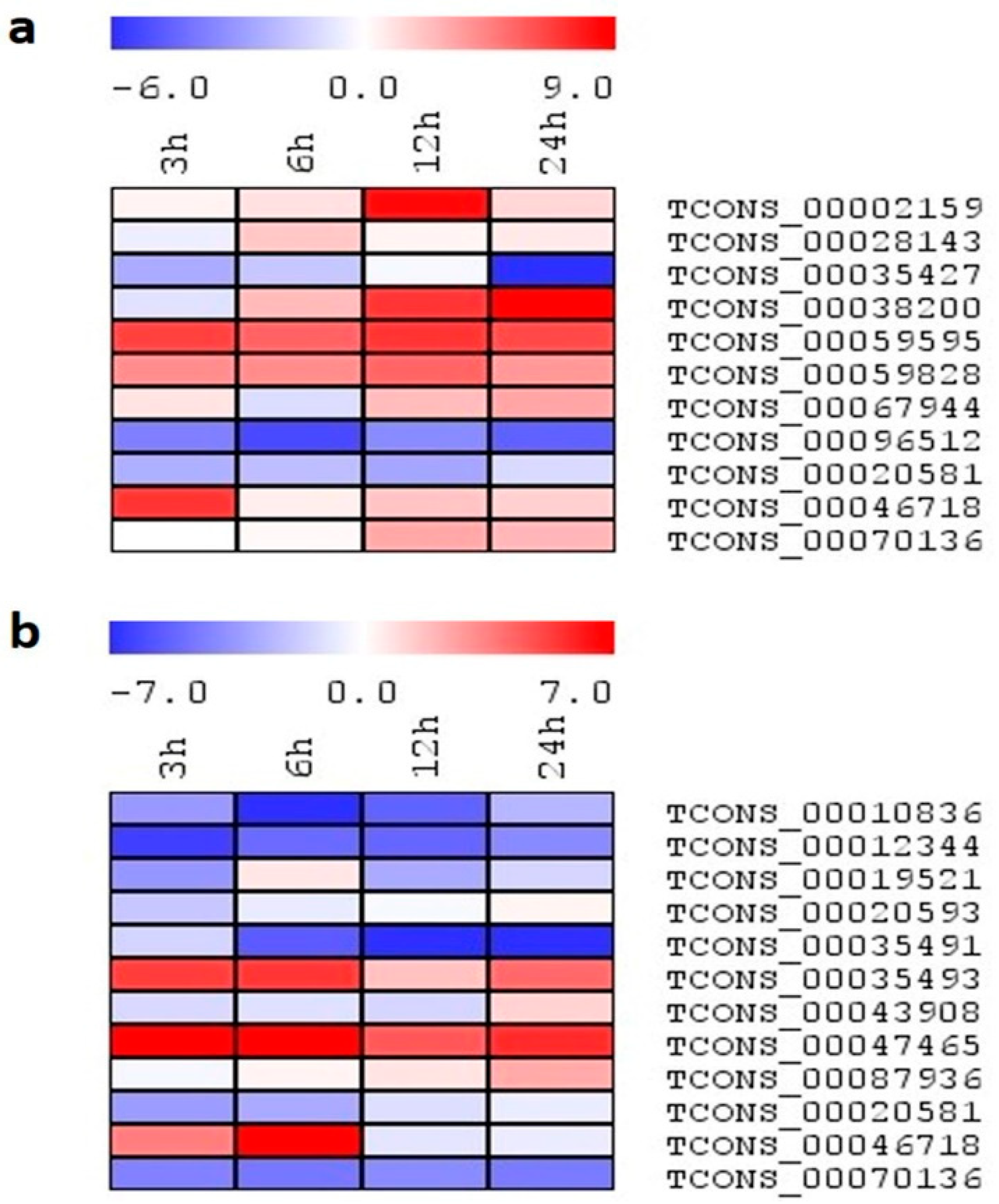
2.7. Expression Analysis of lncRNA Candidates under Salinity Stress
2.8. Expression Profiling of Select lncRNA Candidates under Different Abiotic Stresses
2.9. Expression Analysis of Selected lncRNAs on Exogenous Application of Phytohormones
3. Discussion
4. Materials and Methods
4.1. Identification of lncRNAs in O. sativa Contrasting Genotypes
4.2. Computational Approach for Characterization of Identified O. sativa lncRNAs
4.3. Association of the Identified lncRNAs with Transcription Factors (TFs)
4.4. Interaction of lncRNAs with miRNAs and mRNAs
4.5. Functional Annotation of the Identified lncRNAs
4.6. qRT-PCR-Based Expression Analysis of lncRNAs
4.6.1. Plant Materials
4.6.2. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, E.A.; Bailey-Serres, J.; Weretilnyk, E. Responses to abiotic stresses. In Biochemistry and Molecular Biology of Plants; Buchanan, B.B., Gruissem, W., Jones, R.L., Eds.; American Society of Plant Physiologists: Rockville, MD, USA, 2000; pp. 1158–1203. [Google Scholar]
- Tiwari, S.; Lata, C. Recent advancements in long noncoding RNA-mediated stress responses in rice. In Long Noncoding RNAs in Plants; Upadhyay, S.K., Ed.; Academic Press: London, UK, 2021; pp. 63–74. [Google Scholar] [CrossRef]
- Bhatia, G.; Goyal, N.; Sharma, S.; Upadhyay, S.K.; Singh, K. Present Scenario of Long Non-Coding RNAs in Plants. Non-Coding RNA 2017, 3, 16. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.-H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef]
- Kim, E.-D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Liao, J.-Y.; Li, Z.-Y.; Yu, Y.; Zhang, J.-P.; Li, Q.-F.; Qu, L.-H.; Shu, W.-S.; Chen, Y.-Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef]
- Chung, P.J.; Jung, H.; Jeong, D.-H.; Ha, S.-H.; Choi, Y.D.; Kim, J.-K. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom. 2016, 17, 563. [Google Scholar] [CrossRef]
- Singh, U.; Khemka, N.; Rajkumar, M.S.; Garg, R.; Jain, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar] [CrossRef]
- Lakra, N.; Kaur, C.; Anwar, K.; Singla-Pareek, S.L.; Pareek, A. Proteomics of contrasting rice genotypes: Identification of potential targets for raising crops for saline environment. Plant Cell Environ. 2018, 41, 947–969. [Google Scholar] [CrossRef]
- Kumari, S.; Sabharwal, V.P.N.; Kushwaha, H.R.; Sopory, S.K.; Singla-Pareek, S.L.; Pareek, A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct. Integr. Genom. 2009, 9, 109–123. [Google Scholar] [CrossRef]
- Lakra, N.; Kaur, C.; Singla-Pareek, S.L.; Pareek, A. Mapping the ‘early salinity response’ triggered proteome adaptation in contrasting rice genotypes using iTRAQ approach. Rice 2019, 12, 3. [Google Scholar] [CrossRef]
- Mishra, M.; Wungrampha, S.; Kumar, G.; Singla-Pareek, S.L.; Pareek, A. How do rice seedlings of landrace Pokkali survive in saline fields after transplantation? Physiology, biochemistry, and photosynthesis. Photosynth. Res. 2021, 150, 117–135. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.-K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genom. 2018, 19, 532. [Google Scholar] [CrossRef]
- Borah, P.; Das, A.; Milner, M.J.; Ali, A.; Bentley, A.R.; Pandey, R. Long Non-Coding RNAs as Endogenous Target Mimics and Exploration of Their Role in Low Nutrient Stress Tolerance in Plants. Genes 2018, 9, 459. [Google Scholar] [CrossRef]
- Niazi, F.; Valadkhan, S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 2012, 18, 825–843. [Google Scholar] [CrossRef]
- Yu, B.; Shan, G. Functions of long noncoding RNAs in the nucleus. Nucleus 2016, 7, 155–166. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; McClusky, W.G.; Abdelmohsen, K.; Gorospe, M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 2018, 9, e1471. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Schrumpfová, P.P.; Fojtová, M.; Fajkus, J. Telomeres in Plants and Humans: Not So Different, Not So Similar. Cells 2019, 8, 58. [Google Scholar] [CrossRef]
- Vaquero-Sedas, M.I.; Gámez-Arjona, F.M.; Vega-Palas, M.A. Arabidopsis thaliana telomeres exhibit euchromatic features. Nucleic Acids Res. 2011, 39, 2007–2017. [Google Scholar] [CrossRef]
- Vrbsky, J.; Akimcheva, S.; Watson, J.M.; Turner, T.L.; Daxinger, L.; Vyskot, B.; Aufsatz, W.; Riha, K. siRNA–mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010, 6, e1000986. [Google Scholar] [CrossRef]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-encoded peptide: Functions and predicting methods. Front. Oncol. 2021, 10, 622294. [Google Scholar] [CrossRef]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long Noncoding RNAs: Past, Present, and Future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Zhu, W.; Dong, J.; Cheng, Y.; Yin, Z.; Shen, F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 5573. [Google Scholar] [CrossRef]
- Misiak, B.; Ricceri, L.; Sąsiadek, M.M. Transposable elements and their epigenetic regulation in mental disorders: Current evidence in the field. Front. Genet. 2019, 10, 580. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, F.; Xie, J. Overexpression of miR5505 enhanced drought and salt resistance in rice (Oryza sativa). bioRxiv 2022. [Google Scholar] [CrossRef]
- Parmar, S.; Gharat, S.A.; Tagirasa, R.; Chandra, T.; Behera, L.; Dash, S.K.; Shaw, B.P. Identification and expression analysis of miRNAs and elucidation of their role in salt tolerance in rice varieties susceptible and tolerant to salinity. PLoS ONE 2020, 15, e0230958. [Google Scholar] [CrossRef]
- Arshad, M.; Gruber, M.Y.; Wall, K.; Hannoufa, A. An Insight into microRNA156 Role in Salinity Stress Responses of Alfalfa. Front. Plant Sci. 2017, 8, 356. [Google Scholar] [CrossRef]
- Goswami, K.; Mittal, D.; Gautam, B.; Sopory, S.K.; Sanan-Mishra, N. Mapping the Salt Stress-Induced Changes in the Root miRNome in Pokkali Rice. Biomolecules 2020, 10, 498. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, a SBP-box gene, plays a dual role in salt tolerance and trichome formation in rice (Oryza sativa L.). G3 Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Hou, H.; Jia, H.; Yan, Q.; Wang, X. Overexpression of a SBP-Box Gene (VpSBP16) from Chinese Wild Vitis Species in Arabidopsis Improves Salinity and Drought Stress Tolerance. Int. J. Mol. Sci. 2018, 19, 940. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef]
- Tiwari, S.; Lata, C.; Chauhan, P.S.; Prasad, V.; Prasad, M. A Functional Genomic Perspective on Drought Signalling and its Crosstalk with Phytohormone-mediated Signalling Pathways in Plants. Curr. Genom. 2017, 18, 469–482. [Google Scholar] [CrossRef]
- Tiwari, S.; Nutan, K.K.; Deshmukh, R.; Sarsu, F.; Gupta, K.J.; Singh, A.K.; Singla-Pareek, S.L.; Pareek, A. Seedling-stage salinity tolerance in rice: Decoding the role of transcription factors. Physiol. Plant 2022, 174, e13685. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, Y.; Yang, F.; Xiao, B.; Li, G. RiceLncPedia: A comprehensive database of rice long non-coding RNAs. Plant Biotechnol. J. 2021, 19, 1492–1494. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.-T.; Figueroa, C.R. Jasmonates and Plant Salt Stress: Molecular Players, Physiological Effects, and Improving Tolerance by Using Genome-Associated Tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Verma, R.; Singh, K.; Nisha, N.; Keisham, M.; Bhati, K.K.; Kim, S.T.; Gupta, R. Ethylene: A Master Regulator of Salinity Stress Tolerance in Plants. Biomolecules 2020, 10, 959. [Google Scholar] [CrossRef]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Lim, C.; Kang, K.; Shim, Y.; Sakuraba, Y.; An, G.; Paek, N.-C. Rice Ethylene Response Factor 101 Promotes Leaf Senescence through Jasmonic Acid-Mediated Regulation of OsNAP and OsMYC2. Front. Plant Sci. 2020, 11, 1096. [Google Scholar] [CrossRef]
- Tiwari, S.; Muthamilarasan, M.; Lata, C. Genome-wide identification and expression analysis of Arabidopsis GRAM-domain containing gene family in response to abiotic stresses and PGPR treatment. J. Biotechnol. 2021, 325, 7–14. [Google Scholar] [CrossRef]
- Tiwari, S.; Shweta, S.; Prasad, M.; Lata, C. Genome-wide investigation of GRAM-domain containing genes in rice reveals their role in plant-rhizobacteria interactions and abiotic stress responses. Int. J. Biol. Macromol. 2020, 156, 1243–1257. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Tyagi, A.K.; Khurana, J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006, 345, 646–651. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwari, S.; Jain, M.; Singla-Pareek, S.L.; Bhalla, P.L.; Singh, M.B.; Pareek, A. Pokkali: A Naturally Evolved Salt-Tolerant Rice Shows a Distinguished Set of lncRNAs Possibly Contributing to the Tolerant Phenotype. Int. J. Mol. Sci. 2023, 24, 11677. https://doi.org/10.3390/ijms241411677
Tiwari S, Jain M, Singla-Pareek SL, Bhalla PL, Singh MB, Pareek A. Pokkali: A Naturally Evolved Salt-Tolerant Rice Shows a Distinguished Set of lncRNAs Possibly Contributing to the Tolerant Phenotype. International Journal of Molecular Sciences. 2023; 24(14):11677. https://doi.org/10.3390/ijms241411677
Chicago/Turabian StyleTiwari, Shalini, Mukesh Jain, Sneh Lata Singla-Pareek, Prem L. Bhalla, Mohan B. Singh, and Ashwani Pareek. 2023. "Pokkali: A Naturally Evolved Salt-Tolerant Rice Shows a Distinguished Set of lncRNAs Possibly Contributing to the Tolerant Phenotype" International Journal of Molecular Sciences 24, no. 14: 11677. https://doi.org/10.3390/ijms241411677
APA StyleTiwari, S., Jain, M., Singla-Pareek, S. L., Bhalla, P. L., Singh, M. B., & Pareek, A. (2023). Pokkali: A Naturally Evolved Salt-Tolerant Rice Shows a Distinguished Set of lncRNAs Possibly Contributing to the Tolerant Phenotype. International Journal of Molecular Sciences, 24(14), 11677. https://doi.org/10.3390/ijms241411677






