Enhanced Cell Wall and Cell Membrane Activity Promotes Heat Adaptation of Enterococcus faecium
Abstract
:1. Introduction
2. Results
2.1. Analysis of Physiological and Biochemical Characteristics of RS047-wl
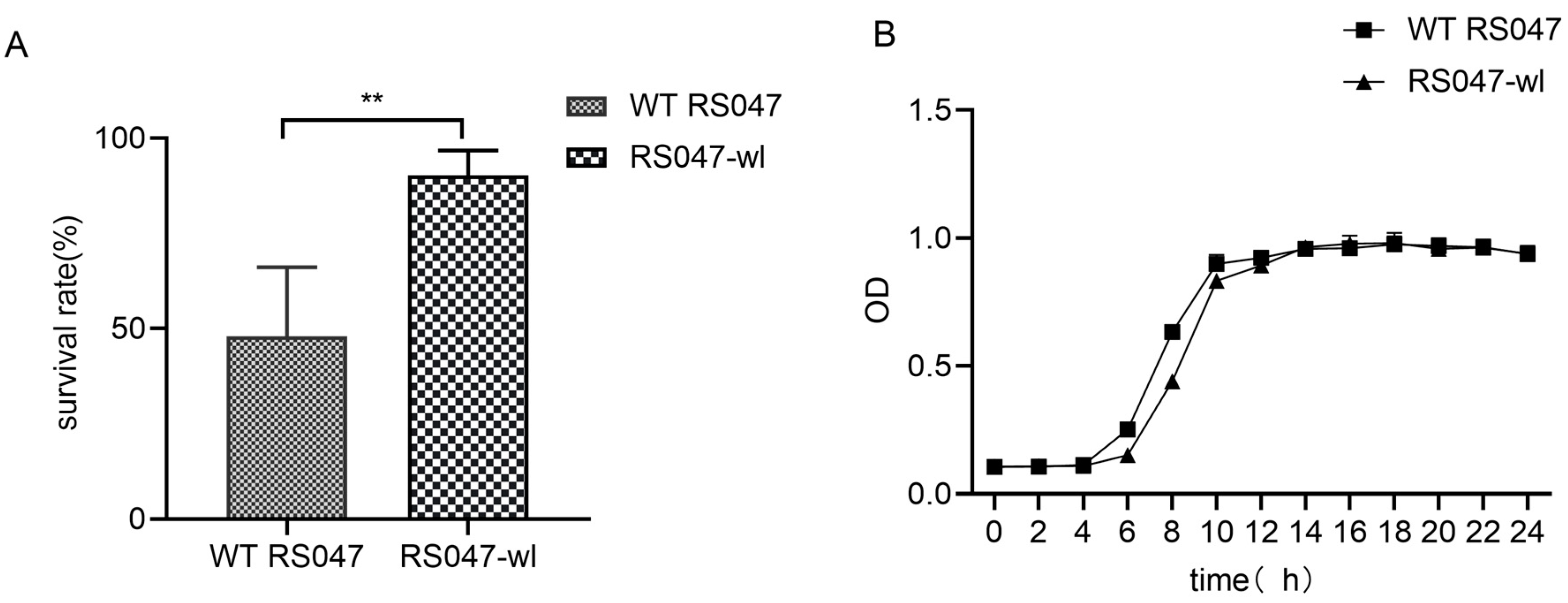
2.1.1. Evaluation of Heat Resistance of Strains for Industrial Production
2.1.2. Growth Performance of RS047-wl
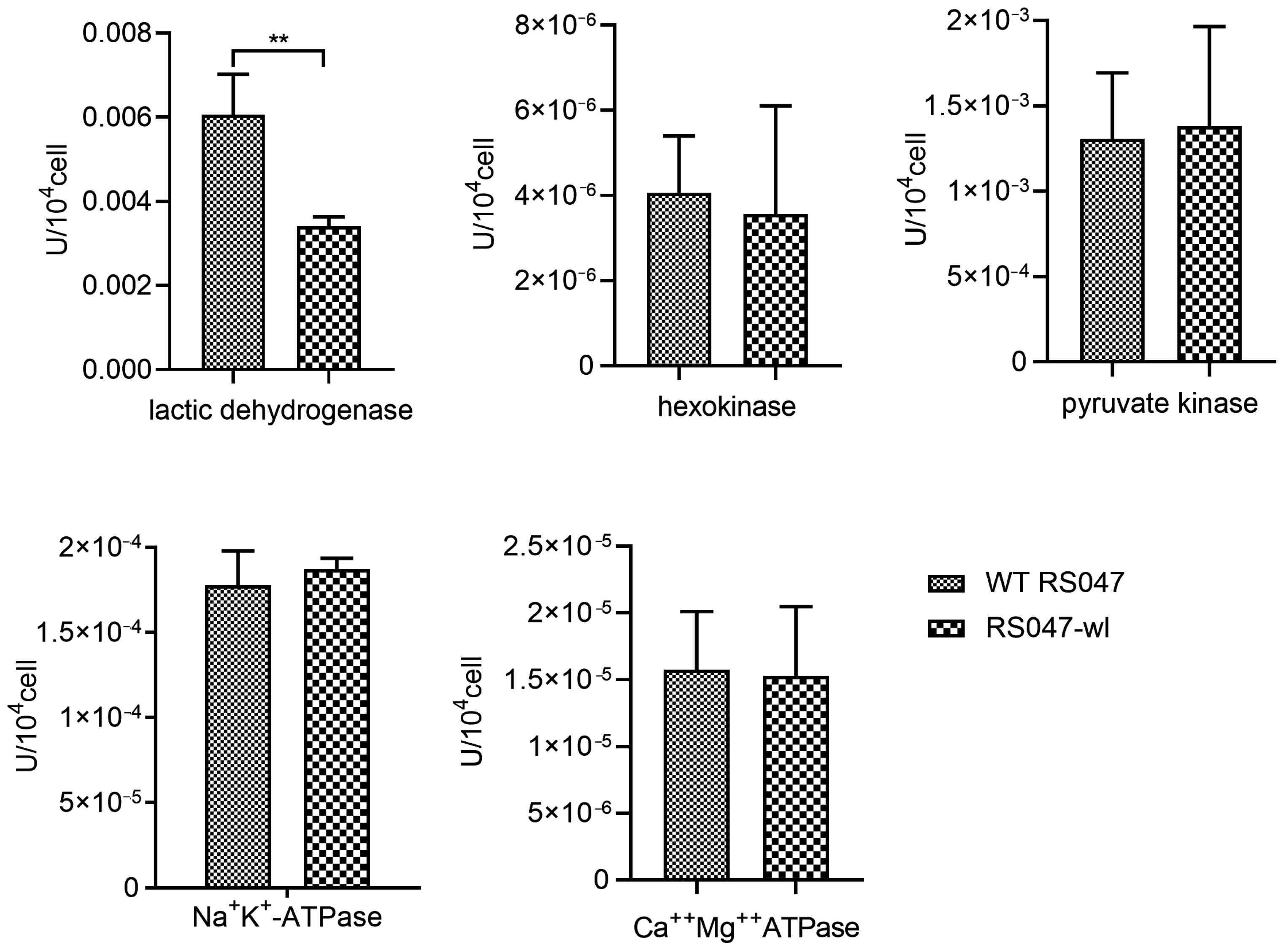
2.1.3. Determination of Key Metabolic Enzyme Activities
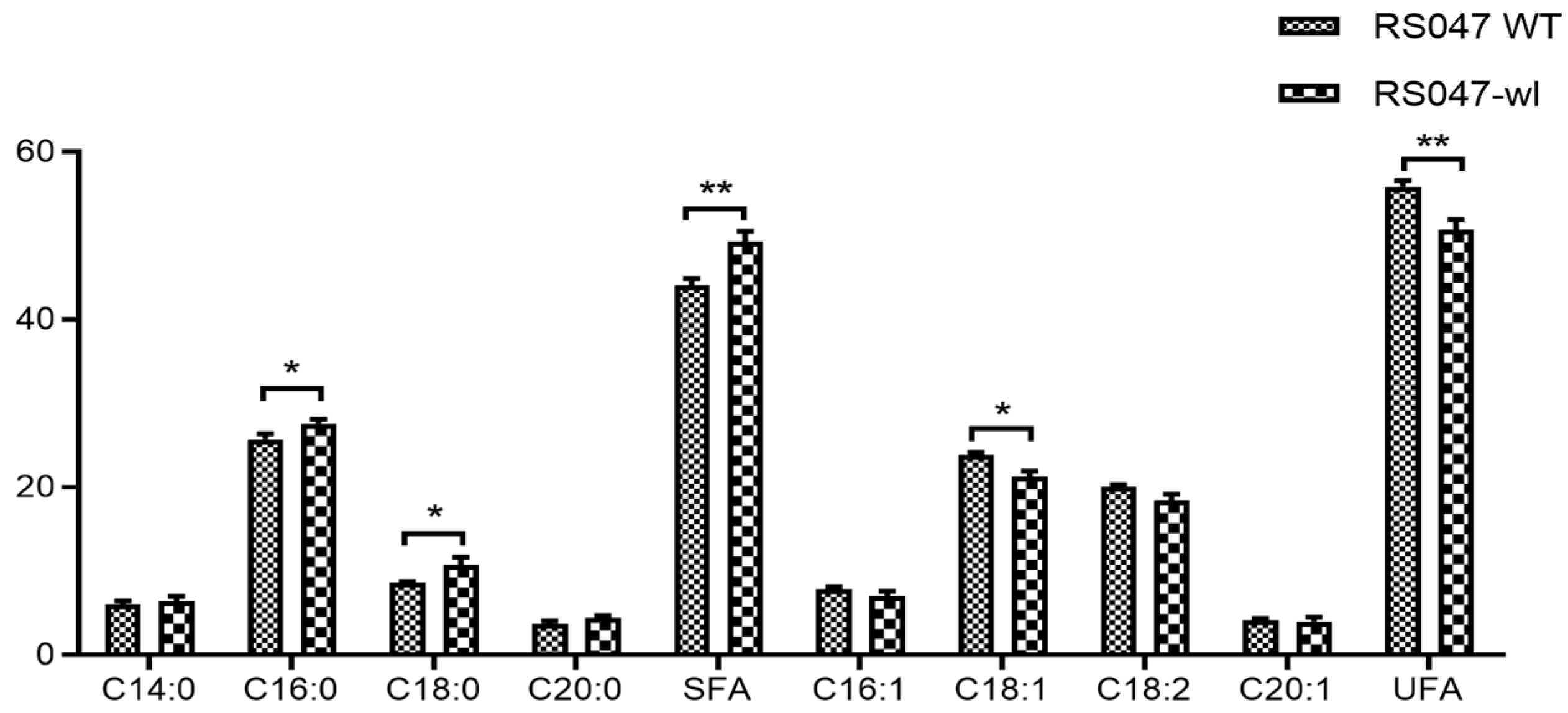
2.1.4. Fatty Acid Composition of RS047-wl
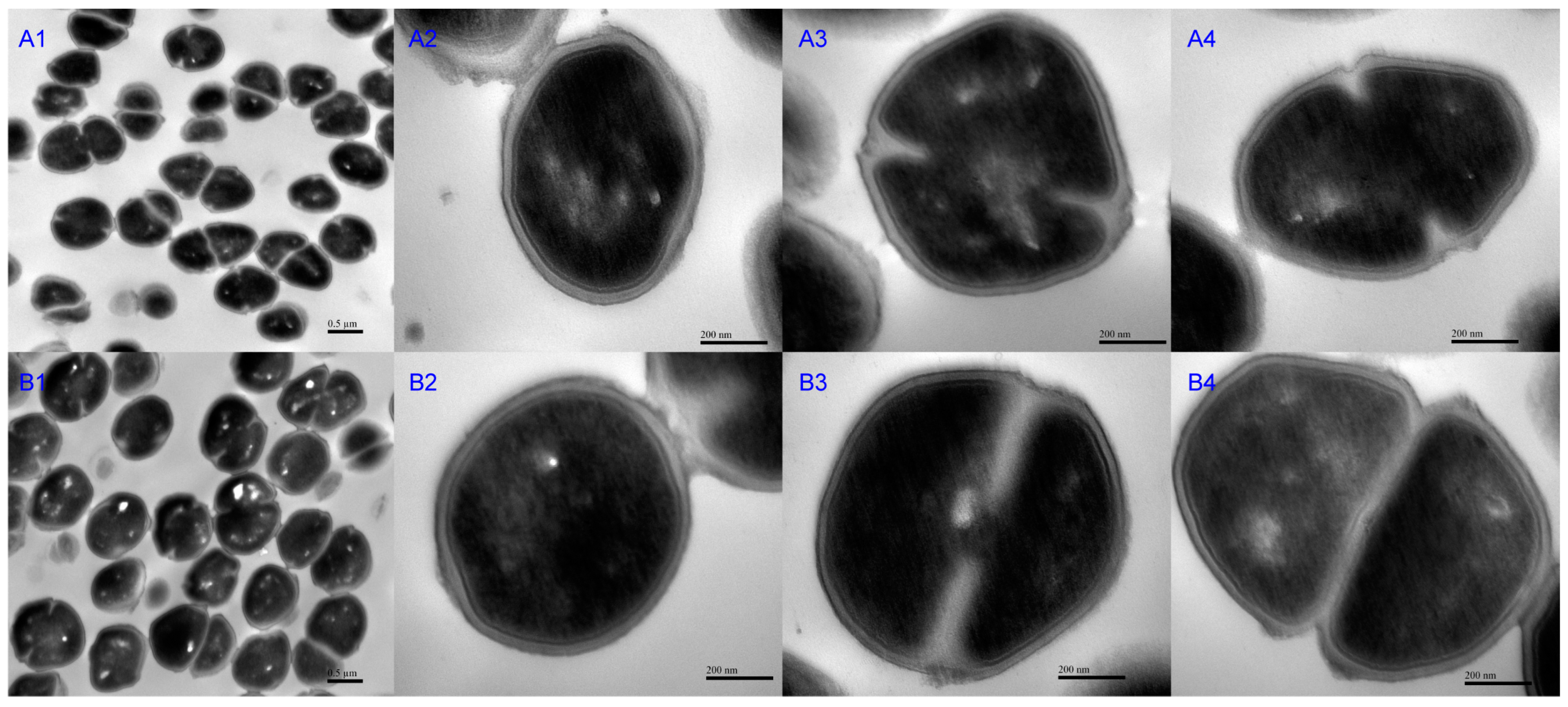
2.1.5. Morphological Observation of RS047-wl
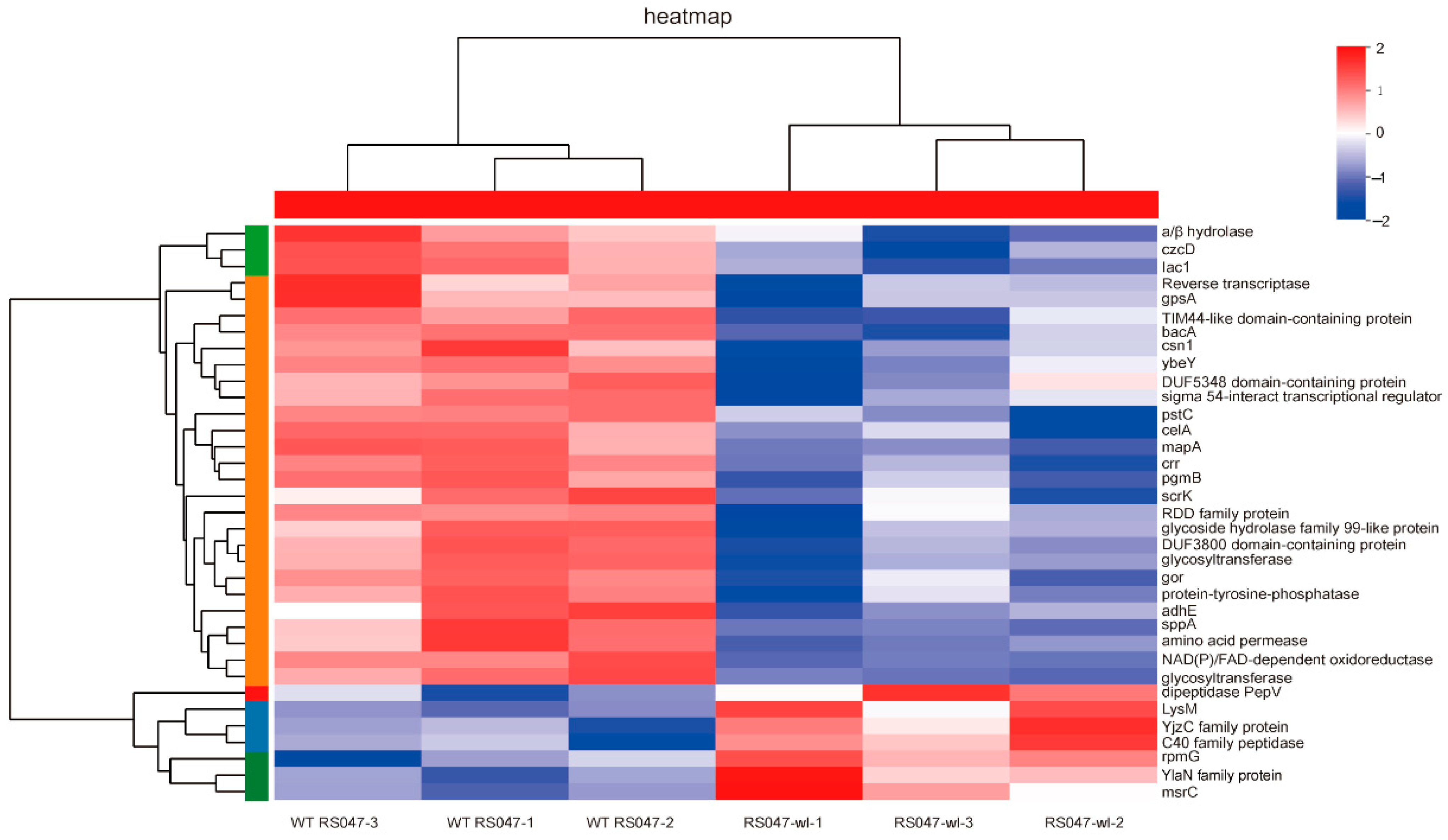
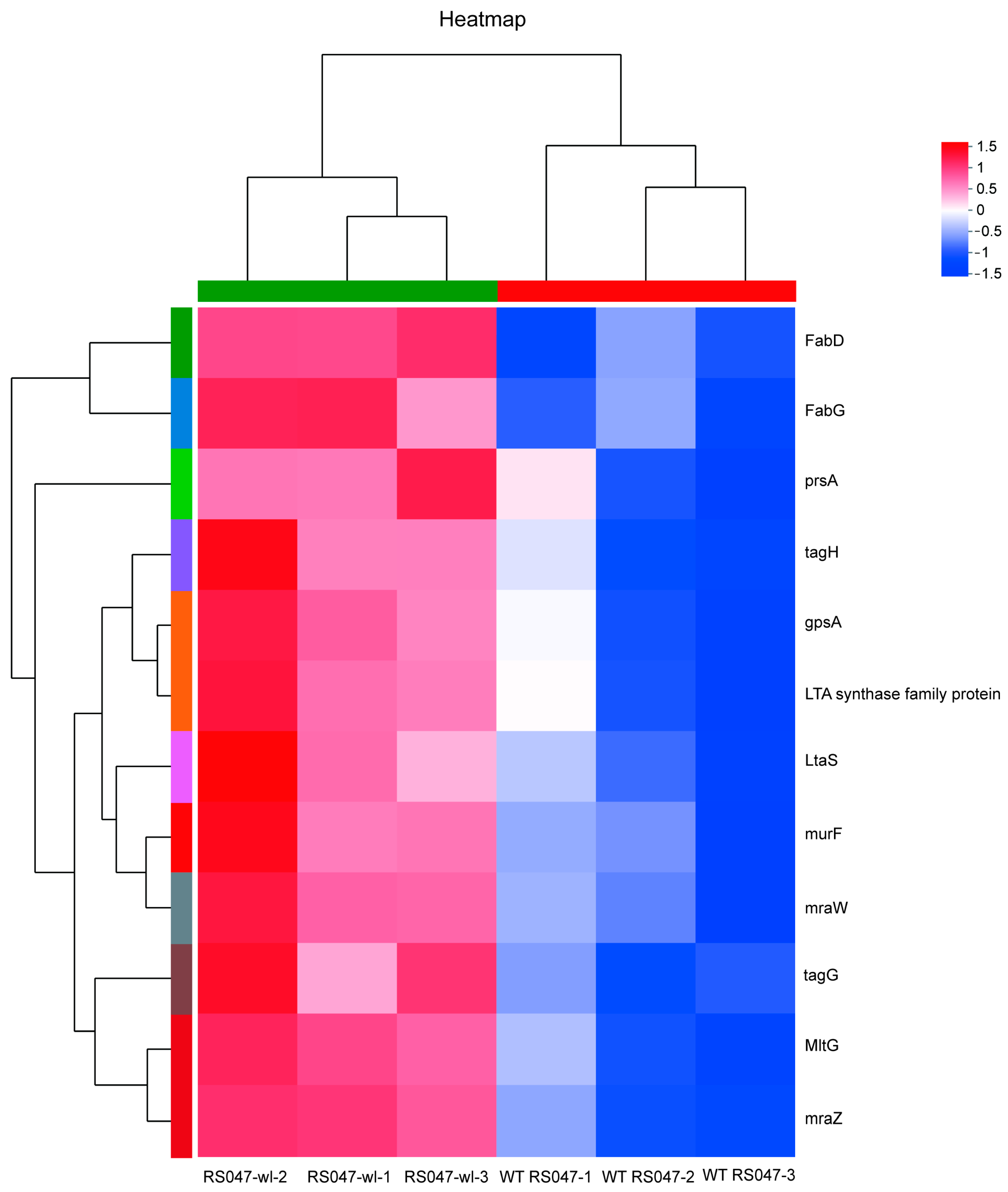
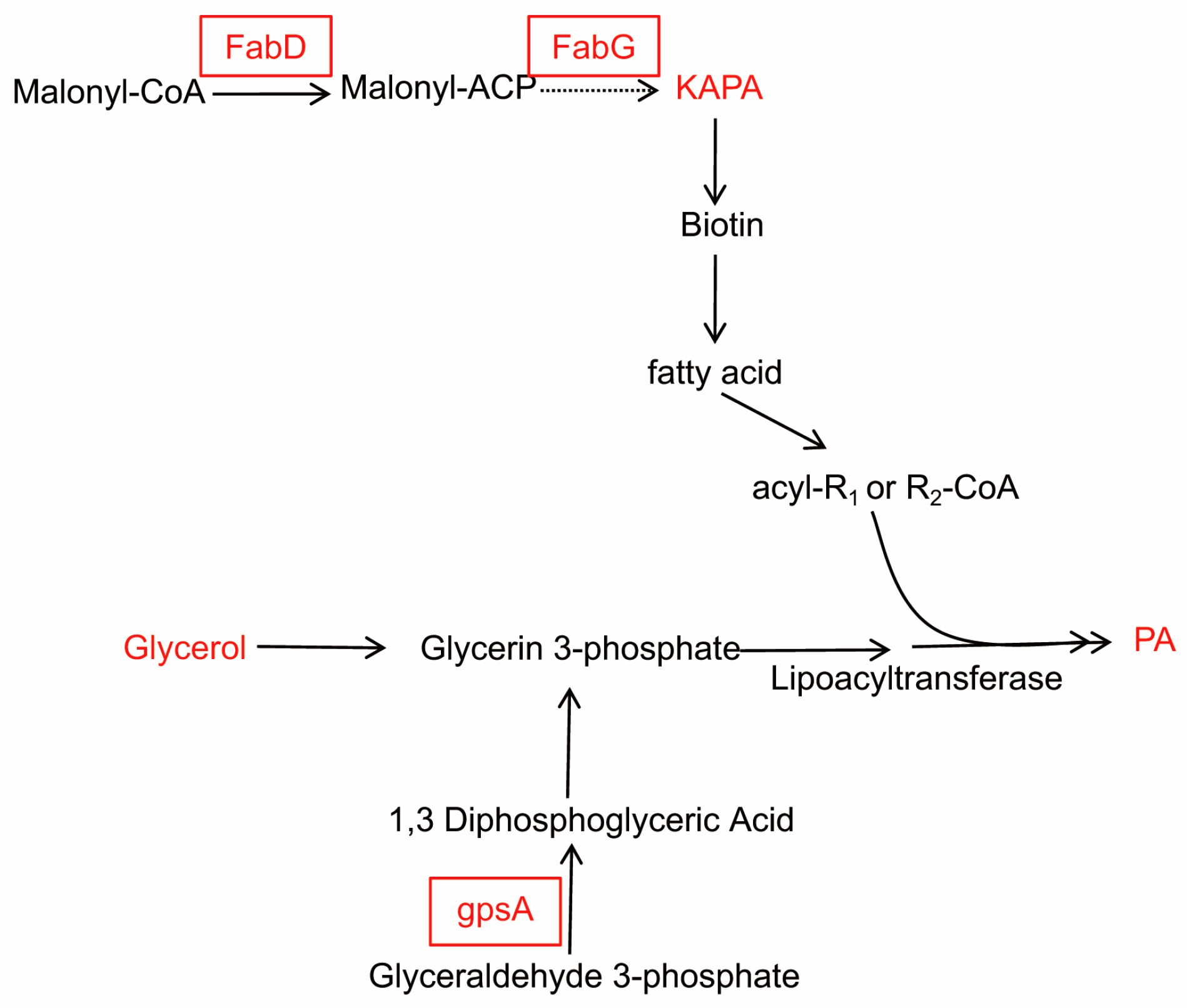
2.2. The Differential Protein Abundances of WT RS047 and RS047-wl
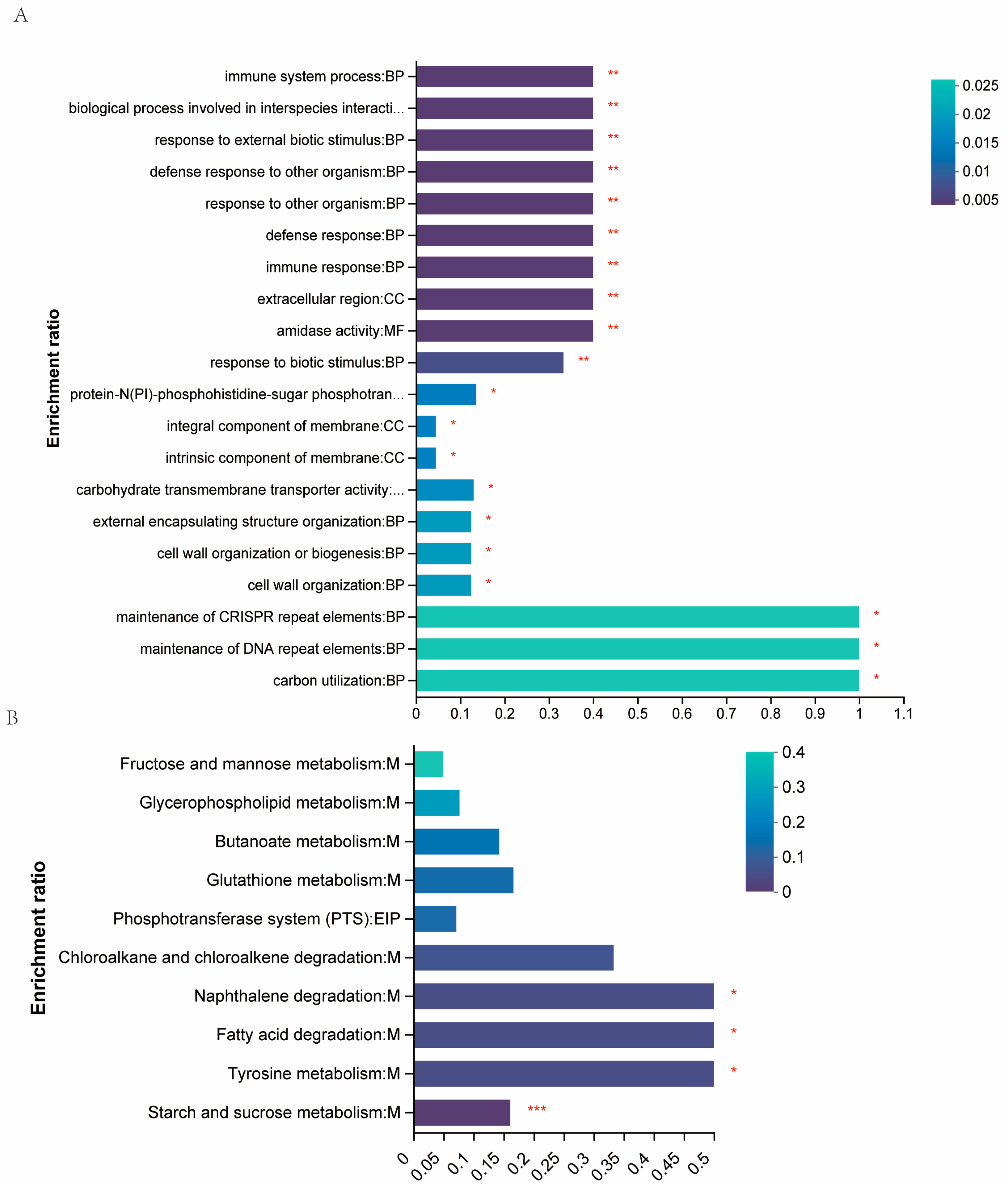
2.3. The Differential Genes Abundances between WT RS047 and RS047-wl
2.4. The Differential Metabolite Abundances of WT RS047 and RS047-wl
3. Discussion
4. Materials and Methods
4.1. Changes of Physiological and Biochemical of RS047-wl
4.1.1. Evaluation of Heat Resistance of Strains
4.1.2. Growth Performance of RS047-wl
4.1.3. Determination of Key Enzyme Activities in Metabolism
4.1.4. Membrane Fatty Acid Composition Analysis
4.1.5. Morphological Analysis of RS047-wl
4.2. Proteomic Analysis
4.2.1. Total Protein Extraction
4.2.2. Digested and Tagged of Proteins with TMT Reagent
4.2.3. Mass Spectrometry
4.2.4. Database Searching and Data Analysis
4.3. Detection of Transcriptomic and Metabolomic
4.4. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franz, C.M.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasolli, E.; De Filippis, F.; Mauriello, I.E.; Cumbo, F.; Walsh, A.M.; Leech, J.; Cotter, P.D.; Segata, N.; Ercolini, D. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020, 11, 2610. [Google Scholar] [CrossRef] [PubMed]
- Martínez, S.; López, M.; Bernardo, A. Thermal inactivation of Enterococcus faecium: Effect of growth temperature and physiological state of microbial cells. Lett. Appl. Microbiol. 2003, 37, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.; Lee, J.; Lee, J.H.; Jeong, D.W. Genomic Insight into the Salt Tolerance of Enterococcus faecium, Enterococcus faecalis and Tetragenococcus halophilus. J. Microbiol. Biotechnol. 2019, 29, 1591–1602. [Google Scholar] [CrossRef]
- Yu, Z.; Shi, D.; Liu, W.; Meng, Y.; Meng, F. Metabolome responses of Enterococcus faecium to acid shock and nitrite stress. Biotechnol. Bioeng. 2020, 117, 3559–3571. [Google Scholar] [CrossRef]
- Zhang, Y.; Shan, B.; Gong, J.; Hu, Y. Mechanism of biogenic amine synthesis of Enterococcus faecium isolated from Sanchun ham. Food Sci. Nutr. 2022, 10, 2036–2049. [Google Scholar] [CrossRef]
- Teixeira, P.; Castro, H.; Mohacsi-Farkas, C.; Kirby, R. Identification of sites of injury in Lactobacillus bulgaricus during heat stress. J. Appl. Microbiol. 1997, 83, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Abee, T.; Wouters, J.A. Microbial stress response in minimal processing. Int. J. Food. Microbiol. 1999, 50, 65–91. [Google Scholar] [CrossRef]
- Prasad, J.; McJarrow, P.; Gopal, P. Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl. Environ. Microbiol. 2003, 69, 917–925. [Google Scholar] [CrossRef] [Green Version]
- Kishimoto, T.; Iijima, L.; Tatsumi, M.; Ono, N.; Oyake, A.; Hashimoto, T.; Matsuo, M.; Okubo, M.; Suzuki, S.; Mori, K.; et al. Transition from positive to neutral in mutation fixation along with continuing rising fitness in thermal adaptive evolution. PLoS Genet. 2010, 6, e1001164. [Google Scholar] [CrossRef] [Green Version]
- Shin, Y.; Kang, C.H.; Kim, W.; So, J.S. Heat Adaptation Improved Cell Viability of Probiotic Enterococcus faecium HL7 upon Various Environmental Stresses. Probiotics Antimicrob. Proteins 2019, 11, 618–626. [Google Scholar] [CrossRef]
- Min, B.; Yoo, D.; Lee, Y.; Seo, M.; Kim, H. Complete Genomic Analysis of Enterococcus faecium Heat-Resistant Strain Developed by Two-Step Adaptation Laboratory Evolution Method. Front. Bioeng. Biotechnol. 2020, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Kim, K.; Li, V.; Cho, S.; Kim, H. Changes in Cell Membrane Fatty Acid Composition of Streptococcus thermophilus in Response to Gradually Increasing Heat Temperature. J. Microbiol. Biotechnol. 2020, 30, 739–748. [Google Scholar] [CrossRef]
- Prazdnova, E.V.; Gorovtsov, A.V.; Vasilchenko, N.G.; Kulikov, M.P.; Statsenko, V.N.; Bogdanova, A.A.; Refeld, A.G.; Brislavskiy, Y.A.; Chistyakov, V.A.; Chikindas, M.L. Quorum-Sensing Inhibition by Gram-Positive Bacteria. Microorganisms 2022, 10, 350. [Google Scholar] [CrossRef]
- Wang, L.; Qiao, L.; Li, A.; Chen, L.; He, B.; Liu, G.; Wang, W.; Fang, J. Integrative Multiomics Analysis of the Heat Stress Response of Enterococcus faecium. Biomolecules 2023, 13, 437. [Google Scholar] [CrossRef] [PubMed]
- Šajbidor, J. Effect of some environmental factors on the content and composition of microbial membrane lipids. Crit. Rev. Biotechnol. 1997, 17, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Denich, T.J.; Beaudette, L.A.; Lee, H.; Trevors, J.T. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 2003, 52, 149–182. [Google Scholar] [CrossRef]
- Lee, W.C.; Park, J.; Balasubramanian, P.K.; Kim, Y. Elucidation of the crystal structure of FabD from the multidrug-resistant bacterium Acinetobacter baumannii. Biochem. Biophys. Res. Commun. 2018, 505, 208–214. [Google Scholar] [CrossRef]
- Vickery, C.R.; Wood, B.M.; Morris, H.G.; Losick, R.; Walker, S. Reconstitution of Staphylococcus aureus Lipoteichoic Acid Synthase Activity Identifies Congo Red as a Selective Inhibitor. J. Am. Chem. Soc. 2018, 140, 876–879. [Google Scholar] [CrossRef]
- Xu, L.; Sedelnikova, S.E.; Baker, P.J.; Rice, D.W. Cloning, purification and preliminary crystallographic analysis of a conserved hypothetical protein, SA0961 (YlaN), from Staphylococcus aureus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 778–780. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Sedelnikova, S.E.; Baker, P.J.; Hunt, A.; Errington, J.; Rice, D.W. Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape. Proteins 2007, 68, 438–445. [Google Scholar] [CrossRef]
- Hunt, A.; Rawlins, J.P.; Thomaides, H.B.; Errington, J. Functional analysis of 11 putative essential genes in Bacillus subtilis. Microbiology 2006, 152, 2895–2907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.; Hernandez-Valdes, J.A.; Kuipers, O.P.; Kok, J. Lysis of a Lactococcus lactis Dipeptidase Mutant and Rescue by Mutation in the Pleiotropic Regulator CodY. Appl. Environ. Microbiol. 2020, 86, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Percy, M.G.; Gründling, A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 2014, 68, 81–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rederstorff, E.; Fatimi, A.; Sinquin, C.; Ratiskol, J.; Merceron, C.; Vinatier, C.; Weiss, P.; Colliec-Jouault, S. Sterilization of exopolysaccharides produced by deep-sea bacteria: Impact on their stability and degradation. Mar. Drugs 2011, 9, 224–241. [Google Scholar] [CrossRef] [Green Version]
- Lazarevic, V.; Karamata, D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol. Microbiol. 1995, 16, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Formstone, A.; Carballido-López, R.; Noirot, P.; Errington, J.; Scheffers, D.-J. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J. Bacteriol. 2008, 190, 1812–1821. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Liu, Y.; Gao, Y.; Dong, J.; Mu, C.; Li, D.; Yang, G. RNAIII suppresses the expression of LtaS via acting as an antisense RNA in Staphylococcus aureus. J. Basic Microbiol. 2015, 55, 255–261. [Google Scholar] [CrossRef]
- de Carvalho, C.C.; Taglialegna, A.; Rosato, A.E. Impact of PrsA on membrane lipid composition during daptomycin-resistance-mediated β-lactam sensitization in clinical MRSA strains. J. Antimicrob. Chemother. 2022, 77, 135–147. [Google Scholar] [CrossRef]
- Jousselin, A.; Renzoni, A.; Andrey, D.O.; Monod, A.; Lew, D.P.; Kelley, W.L. The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 3629–3640. [Google Scholar] [CrossRef] [Green Version]
- Jakob, R.P.; Koch, J.R.; Burmann, B.M.; Schmidpeter, P.A.; Hunkeler, M.; Hiller, S.; Schmid, F.X.; Maier, T. Dimeric structure of the bacterial extracellular foldase PrsA. J. Biol. Chem. 2015, 290, 3278–3292. [Google Scholar] [CrossRef] [Green Version]
- Yunck, R.; Cho, H.; Bernhardt, T.G. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol. 2016, 99, 700–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [Green Version]
- Bohrhunter, J.L.; Rohs, P.D.A.; Torres, G.; Yunck, R.; Bernhardt, T.G. MltG activity antagonizes cell wall synthesis by both types of peptidoglycan polymerases in Escherichia coli. Mol. Microbiol. 2021, 115, 1170–1180. [Google Scholar] [CrossRef]
- Selby, K.; Mascher, G.; Somervuo, P.; Lindstrom, M.; Korkeala, H. Heat shock and prolonged heat stress attenuate neurotoxin and sporulation gene expression in group I Clostridium botulinum strain ATCC 3502. PLoS ONE 2017, 12, e0176944. [Google Scholar] [CrossRef] [PubMed]
- Kanshin, E.; Kubiniok, P.; Thattikota, Y.; D’Amours, D.; Thibault, P. Phosphoproteome dynamics of Saccharomyces cerevisiae under heat shock and cold stress. Mol. Syst. Biol. 2015, 11, 813. [Google Scholar] [CrossRef] [PubMed]
- Garcés, R.; Mancha, M. One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal. Biochem. 1993, 211, 139–143. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, A.; Fang, J.; Wang, Y.; Chen, L.; Qiao, L.; Wang, W. Enhanced Cell Wall and Cell Membrane Activity Promotes Heat Adaptation of Enterococcus faecium. Int. J. Mol. Sci. 2023, 24, 11822. https://doi.org/10.3390/ijms241411822
Wang L, Li A, Fang J, Wang Y, Chen L, Qiao L, Wang W. Enhanced Cell Wall and Cell Membrane Activity Promotes Heat Adaptation of Enterococcus faecium. International Journal of Molecular Sciences. 2023; 24(14):11822. https://doi.org/10.3390/ijms241411822
Chicago/Turabian StyleWang, Li, Aike Li, Jun Fang, Yongwei Wang, Lixian Chen, Lin Qiao, and Weiwei Wang. 2023. "Enhanced Cell Wall and Cell Membrane Activity Promotes Heat Adaptation of Enterococcus faecium" International Journal of Molecular Sciences 24, no. 14: 11822. https://doi.org/10.3390/ijms241411822




