Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells
Abstract
1. Introduction
2. Results
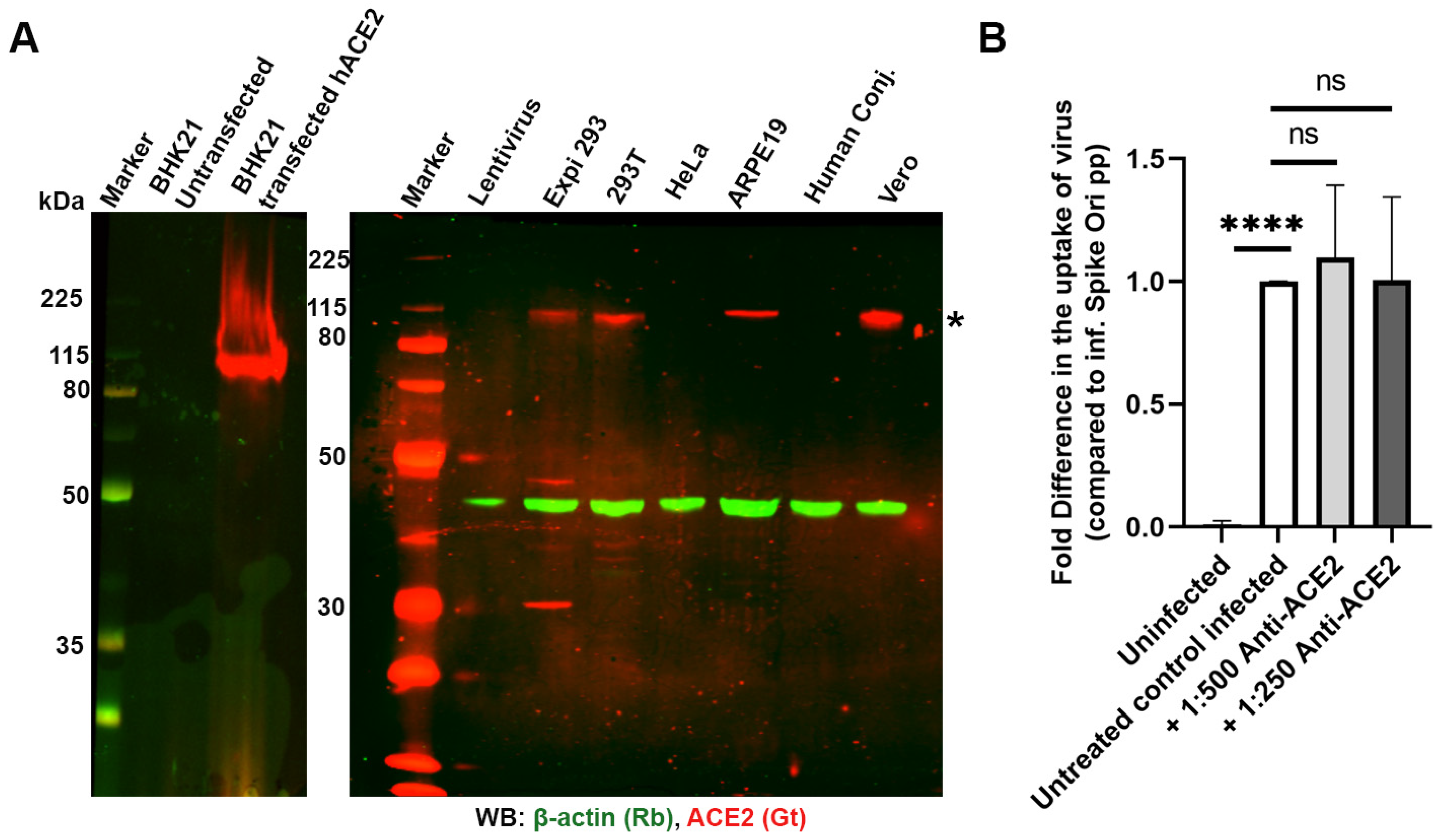
2.1. Spike Pseudovirions Can Infect ARPE-19 Independently of ACE2
2.2. Perturbation of Cholesterol Homeostasis Inhibits Spike-Pseudovirion Infection of ARPE-19
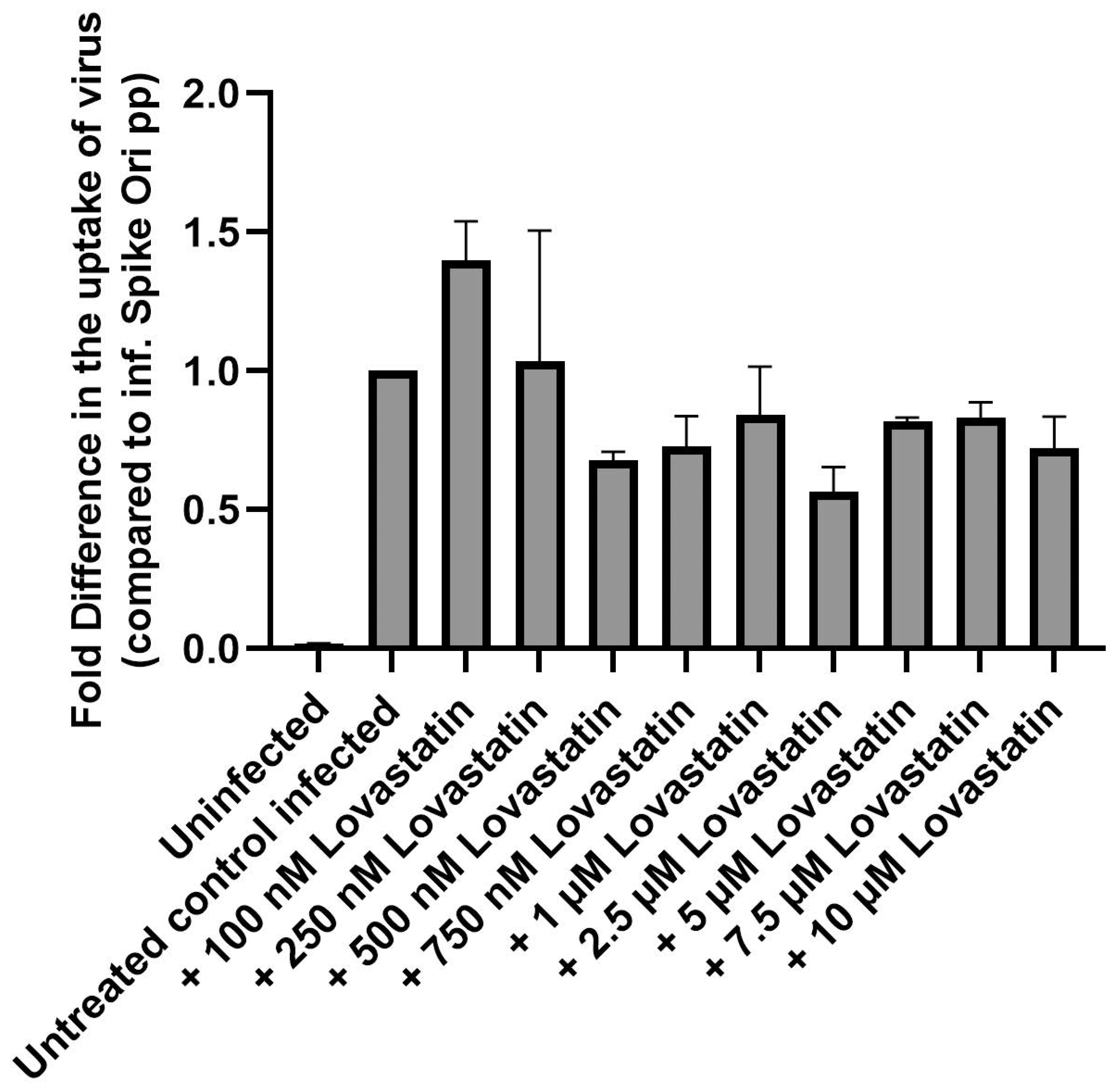
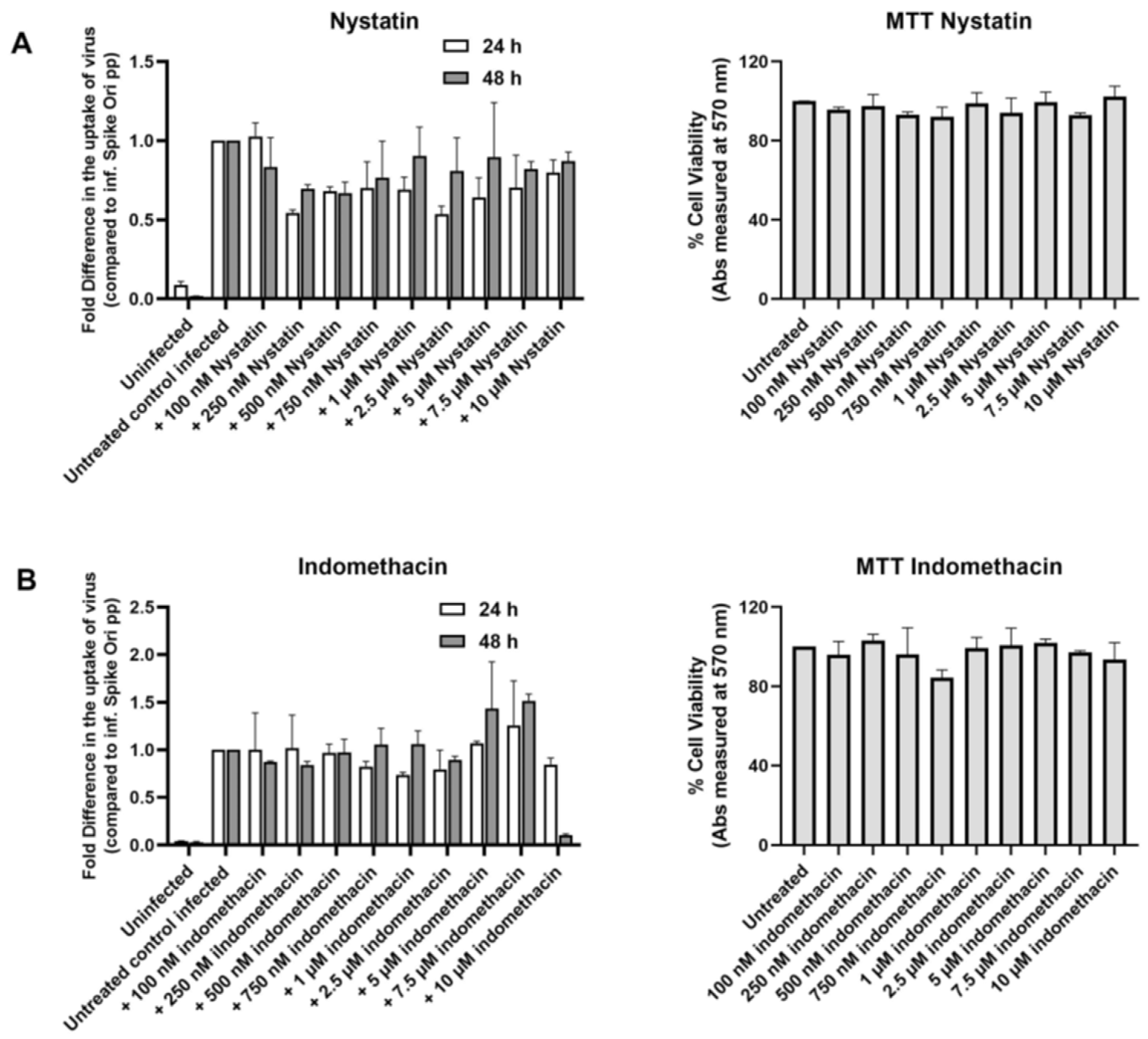
2.2.1. Cyclodextrins
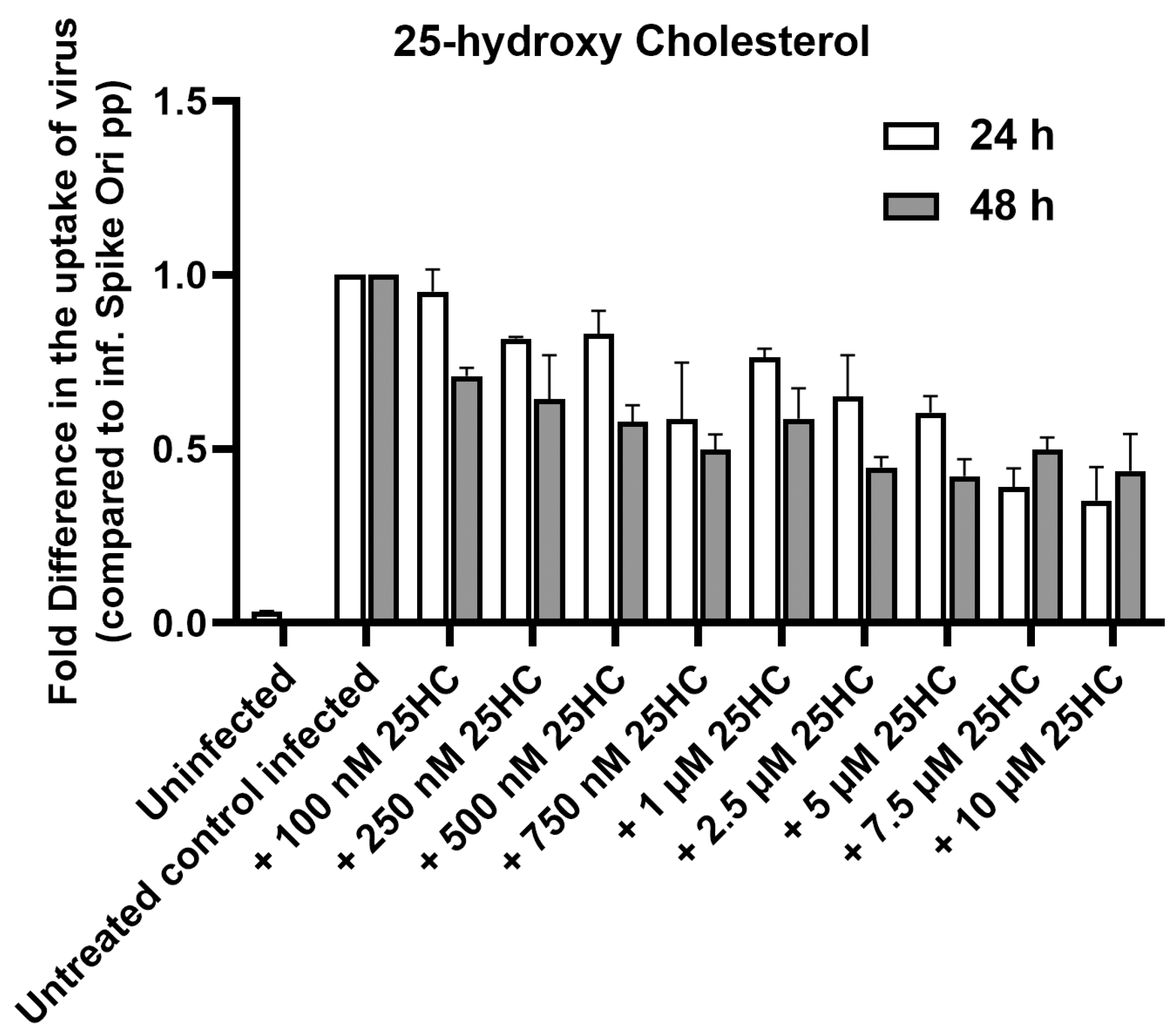
2.2.2. Oxysterol 25-HC
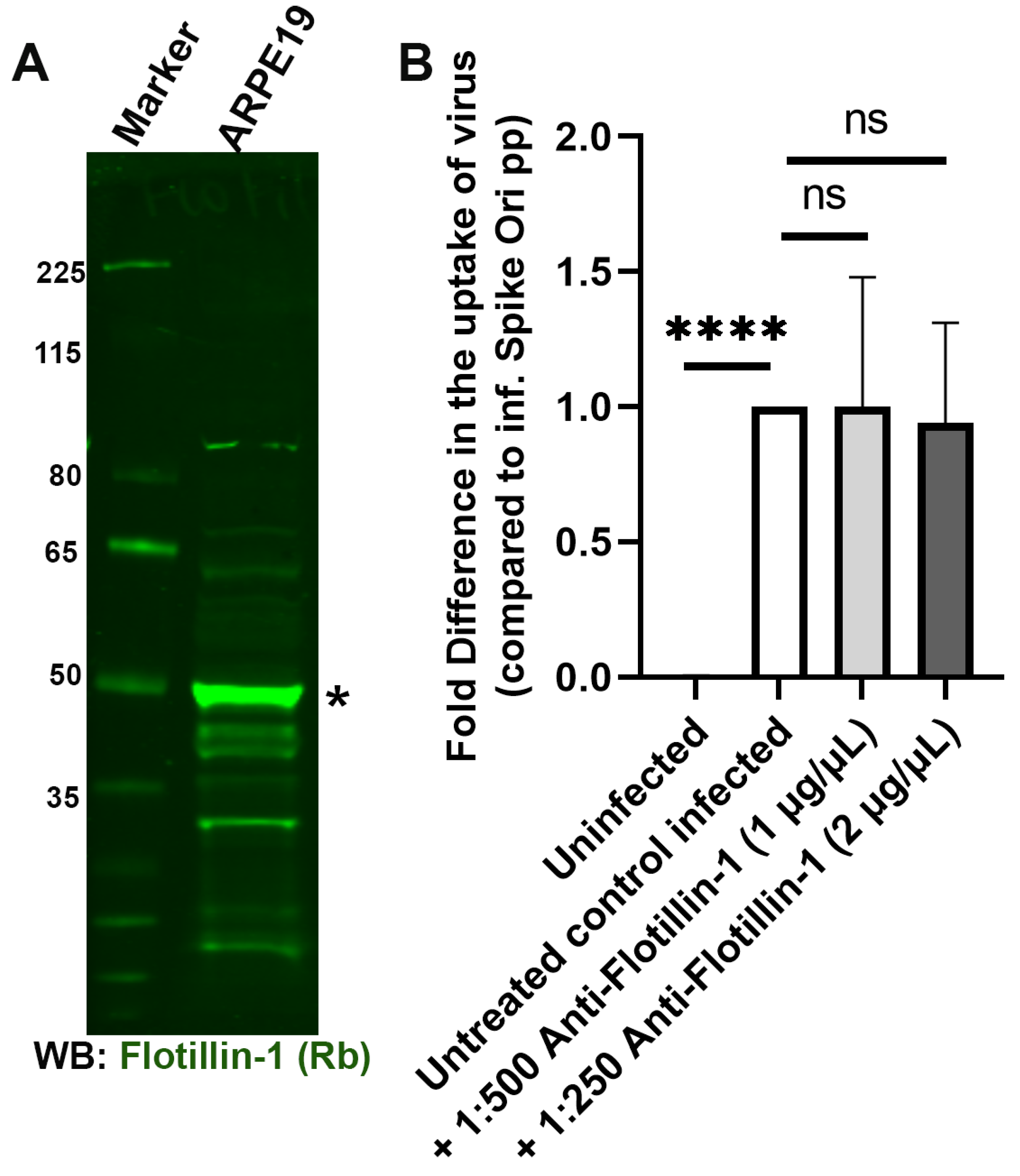
2.3. Clathrin-Dependent and Flotillin-Dependent Endocytic Pathways Are Not Major Pathways in Internalization of SARS-CoV-2 Spike Pseudotyped Lentiviral Particles
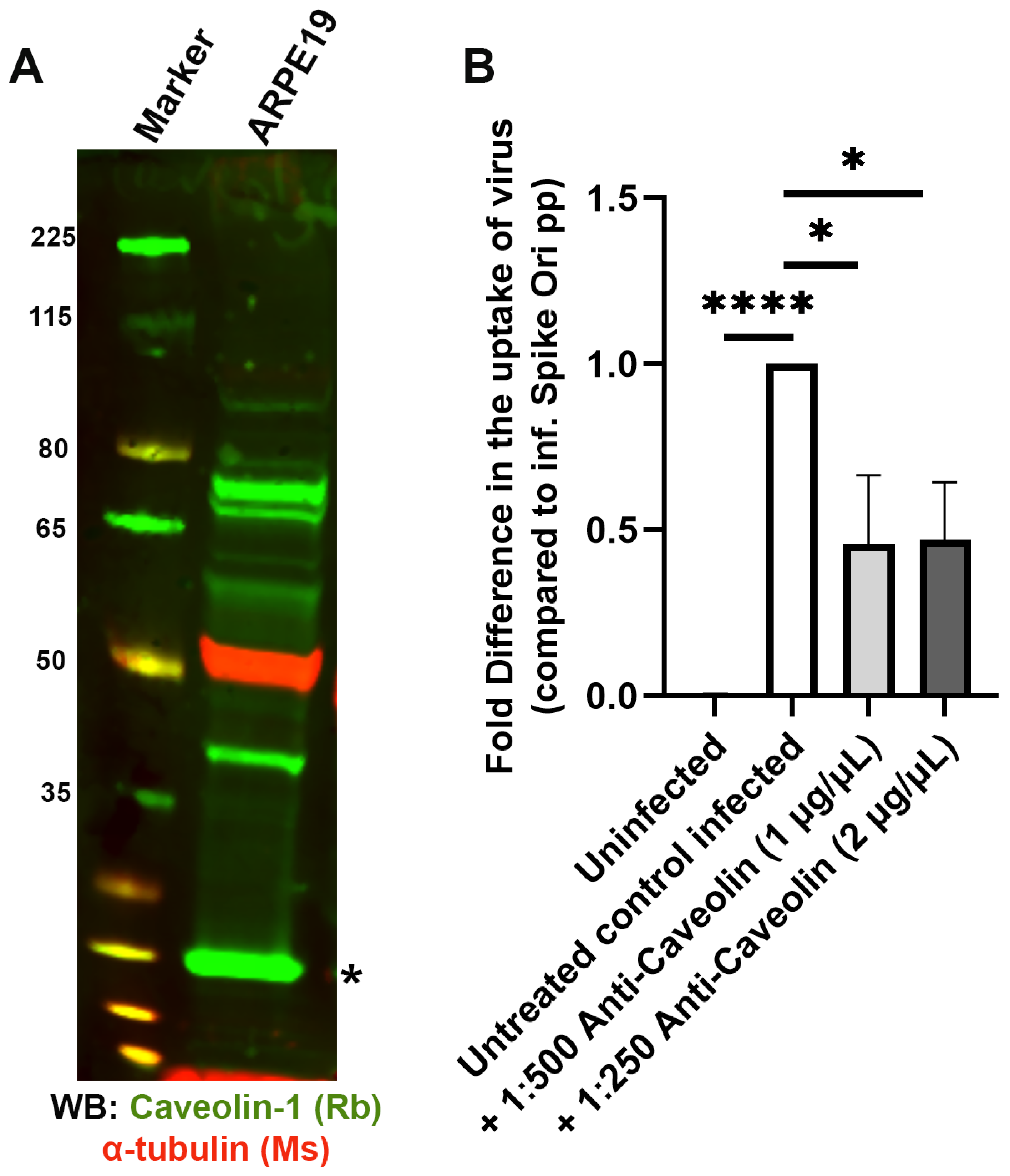
2.4. Caveolae-Mediated LDL Transcytosis Is Highjacked by Spike Pseudotyped Lentivral Particles to Infect ARPE19 Cells in Our SARS-CoV-2 Infection Model
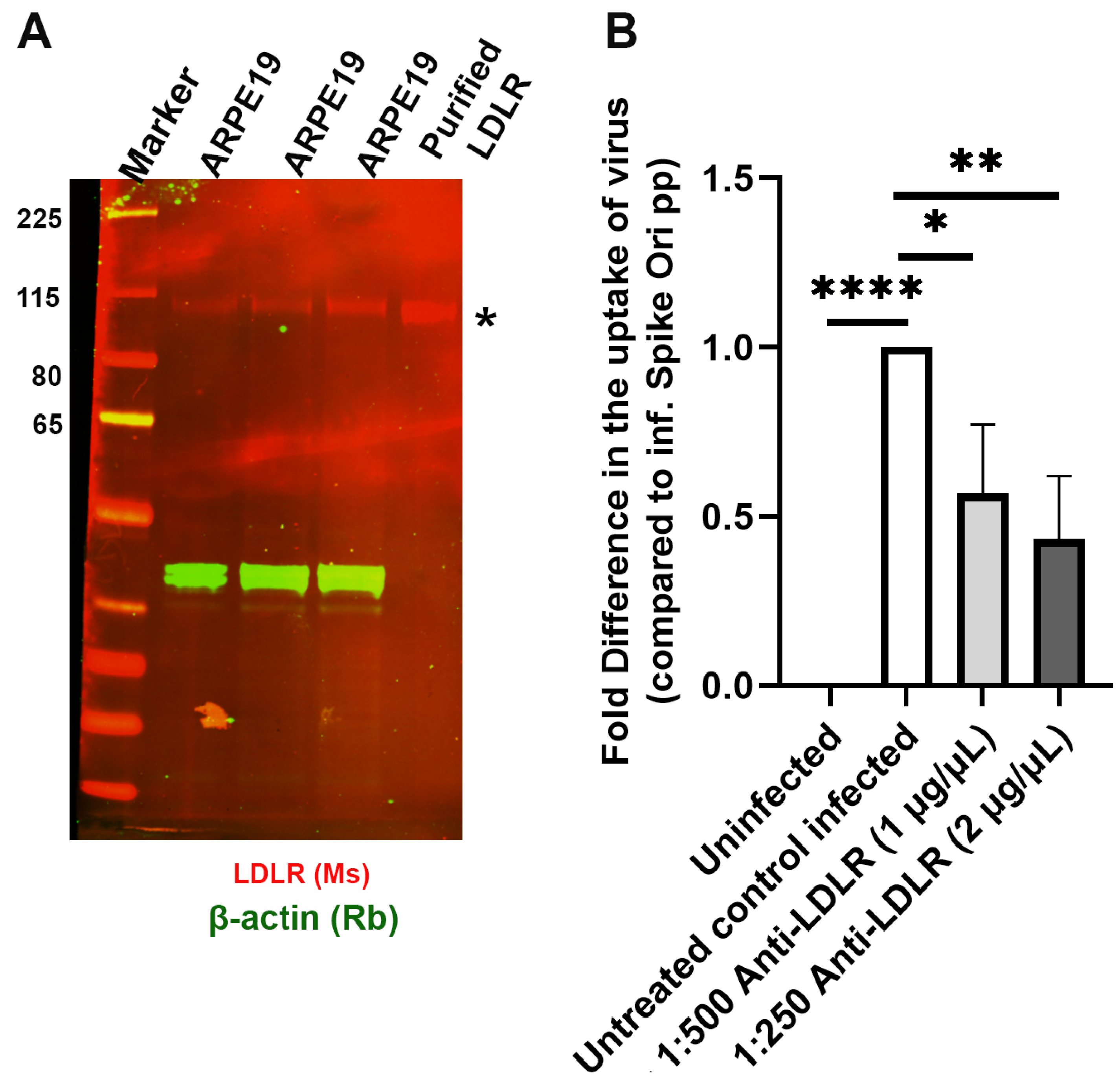
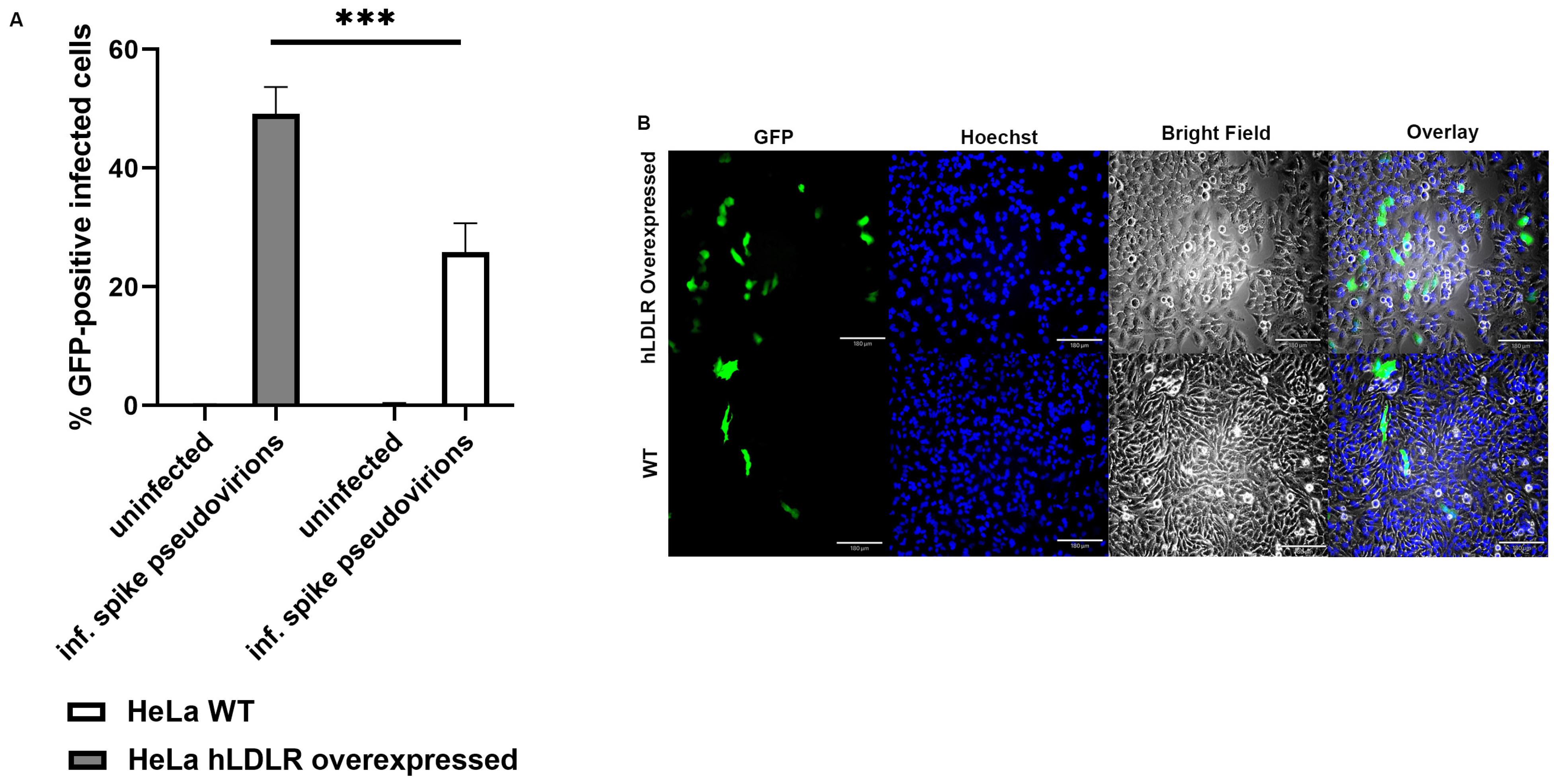
2.5. LDL Receptor Overexpression in HeLa (hLDLR-HeLa) Greatly Increased Spike Pseudotyped Lentivral Particles Uptake in Our Model of SARS-CoV-2 Infection
2.6. Modes of Evolution of ACE2 and LDLR
3. Discussion
4. Materials and Methods
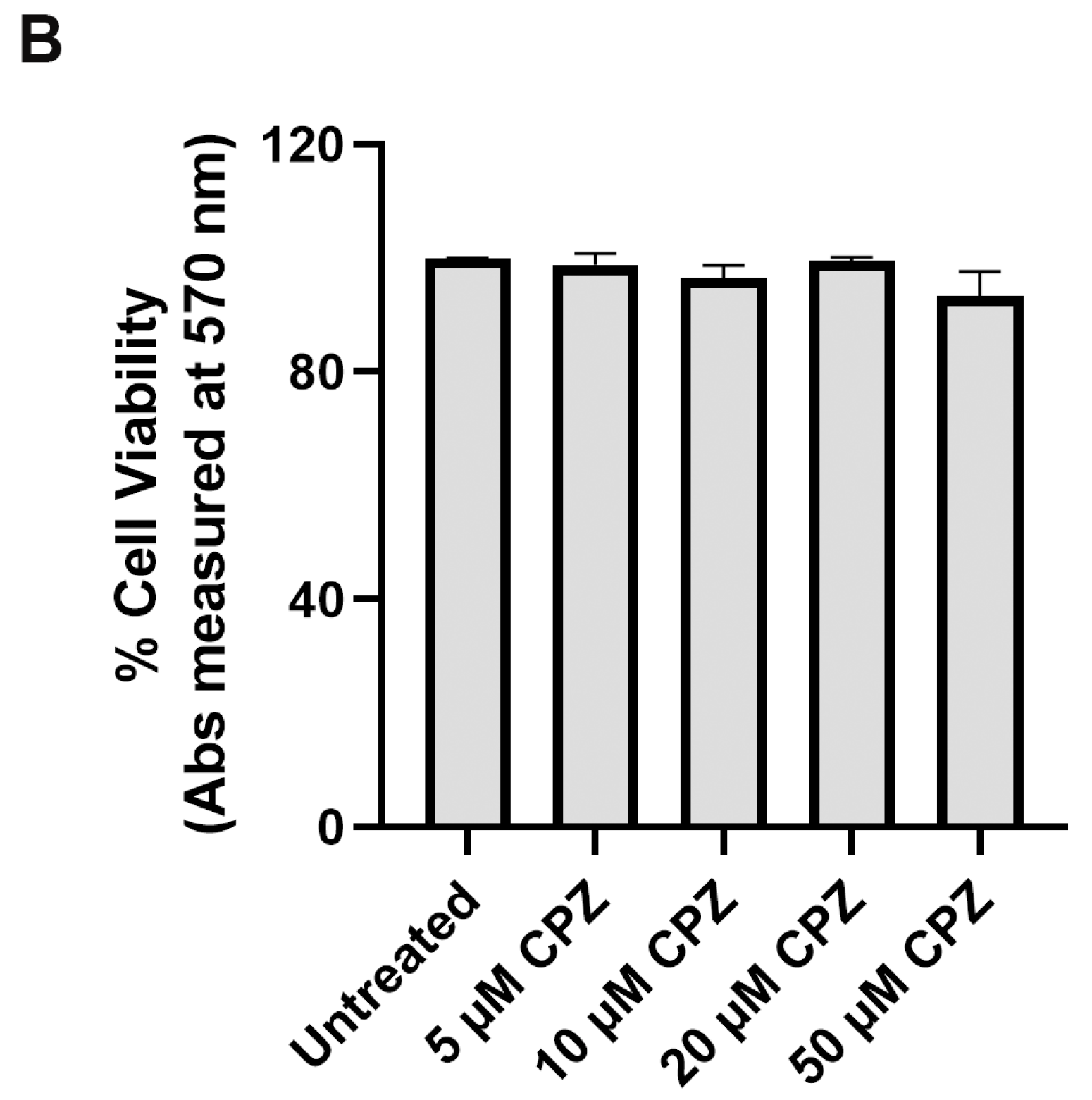
4.1. Antibodies, Inhibitors, and Reagents
4.2. Cells and Culture
4.3. Immunoblot Analysis
4.4. Production of SARS-CoV-2 Spike Protein Pseudovirions
4.4.1. Measurement of Physical and Infectious Viral Titer
4.4.2. Pseudovirus Infection Assay
4.5. Cell Viability Assay
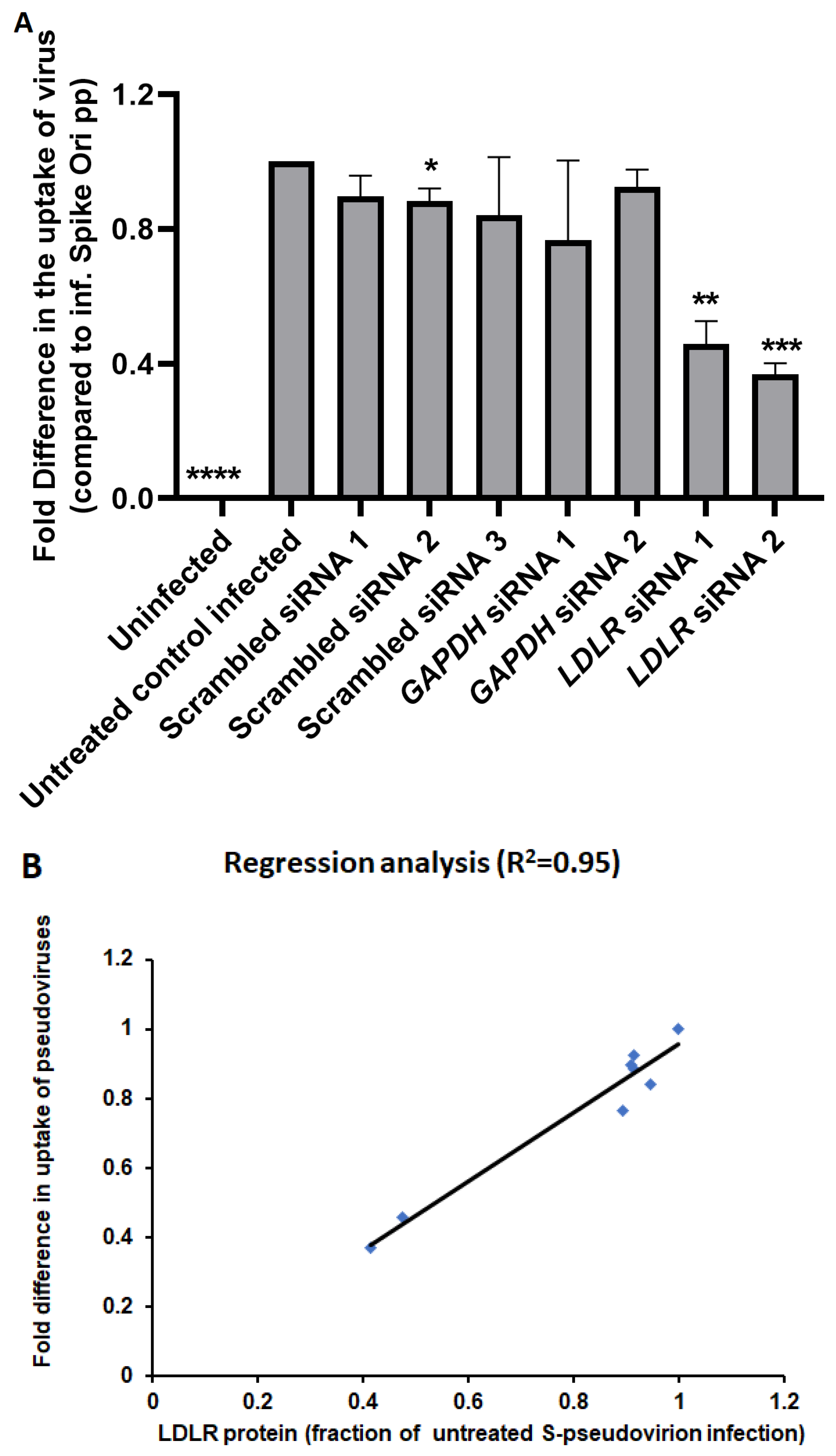
4.6. siRNA Treatment and Pseudovirion Infection of ARPE-19 Cells
4.7. Multiple Alignments Analyses
4.8. Statistical Analysis of Data
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904.e899. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef] [PubMed]
- Eriksen, A.Z.; Moller, R.; Makovoz, B.; Uhl, S.A.; tenOever, B.R.; Blenkinsop, T.A. SARS-CoV-2 infects human adult donor eyes and hESC-derived ocular epithelium. Cell Stem Cell 2021, 28, 1205–1220.e1207. [Google Scholar] [CrossRef]
- Martinez-Colon, G.J.; Ratnasiri, K.; Chen, H.; Jiang, S.; Zanley, E.; Rustagi, A.; Verma, R.; Chen, H.; Andrews, J.R.; Mertz, K.D.; et al. SARS-CoV-2 infection drives an inflammatory response in human adipose tissue through infection of adipocytes and macrophages. Sci. Transl. Med. 2022, 14, eabm9151. [Google Scholar] [CrossRef]
- Partridge, L.J.; Urwin, L.; Nicklin, M.J.H.; James, D.C.; Green, L.R.; Monk, P.N. ACE2-Independent Interaction of SARS-CoV-2 Spike Protein with Human Epithelial Cells Is Inhibited by Unfractionated Heparin. Cells 2021, 10, 1419. [Google Scholar] [CrossRef]
- Shen, X.R.; Geng, R.; Li, Q.; Chen, Y.; Li, S.F.; Wang, Q.; Min, J.; Yang, Y.; Li, B.; Jiang, R.D.; et al. ACE2-independent infection of T lymphocytes by SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 83. [Google Scholar] [CrossRef]
- Borsa, M.; Mazet, J.M. Attacking the defence: SARS-CoV-2 can infect immune cells. Nat. Rev. Immunol. 2020, 20, 592. [Google Scholar] [CrossRef]
- Davanzo, G.G.; Codo, A.C.; Brunetti, N.S.; Boldrini, V.; Knittel, T.L.; Monterio, L.B.; de Moraes, D.; Ferrari, A.J.R.; de Souza, G.F.; Muraro, S.P.; et al. SARS-CoV-2 Uses CD4 to Infect T Helper Lymphocytes. medRxiv 2020. [Google Scholar] [CrossRef]
- Karthika, T.; Joseph, J.; Das, V.R.A.; Nair, N.; Charulekha, P.; Roji, M.D.; Raj, V.S. SARS-CoV-2 Cellular Entry Is Independent of the ACE2 Cytoplasmic Domain Signaling. Cells 2021, 10, 1814. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wan, L.; Yan, Q.; Wang, X.; Zhang, J.; Yang, X.; Zhang, Y.; Fan, C.; Li, D.; Deng, Y.; et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020, 2, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.L.; Teng, J.L.L.; Jia, L.; Zhang, C.; Huang, C.; Cai, J.P.; Zhou, R.; Chan, K.H.; Zhao, H.; Zhu, L.; et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 2021, 184, 2212–2228.e2212. [Google Scholar] [CrossRef]
- Pia, L.; Rowland-Jones, S. Omicron entry route. Nat. Rev. Immunol. 2022, 22, 144. [Google Scholar] [CrossRef]
- Filippini, A.; D’Alessio, A. Caveolae and Lipid Rafts in Endothelium: Valuable Organelles for Multiple Functions. Biomolecules 2020, 10, 1218. [Google Scholar] [CrossRef]
- Sorice, M.; Misasi, R.; Riitano, G.; Manganelli, V.; Martellucci, S.; Longo, A.; Garofalo, T.; Mattei, V. Targeting Lipid Rafts as a Strategy Against Coronavirus. Front. Cell Dev. Biol. 2020, 8, 618296. [Google Scholar] [CrossRef]
- Yang, N.; Shen, H.M. Targeting the Endocytic Pathway and Autophagy Process as a Novel Therapeutic Strategy in COVID-19. Int. J. Biol. Sci. 2020, 16, 1724–1731. [Google Scholar] [CrossRef]
- Davis, G.; Li, K.; Thankam, F.G.; Wilson, D.R.; Agrawal, D.K. Ocular transmissibility of COVID-19: Possibilities and perspectives. Mol. Cell. Biochem. 2022, 477, 849–864. [Google Scholar] [CrossRef]
- Petronio Petronio, G.; Di Marco, R.; Costagliola, C. Do Ocular Fluids Represent a Transmission Route of SARS-CoV-2 Infection? Front. Med. 2020, 7, 620412. [Google Scholar] [CrossRef]
- Yamasoba, D.; Uriu, K.; Plianchaisuk, A.; Kosugi, Y.; Pan, L.; Zahradnik, J.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 Omicron XBB.1.16 variant. Lancet Infect. Dis. 2023, 23, 655–656. [Google Scholar] [CrossRef]
- Postnikova, O.A.; Uppal, S.; Huang, W.; Kane, M.A.; Villasmil, R.; Rogozin, I.B.; Poliakov, E.; Redmond, T.M. The Functional Consequences of the Novel Ribosomal Pausing Site in SARS-CoV-2 Spike Glycoprotein RNA. Int. J. Mol. Sci. 2021, 22, 6490. [Google Scholar] [CrossRef]
- Wang, H.; Yang, P.; Liu, K.; Guo, F.; Zhang, Y.; Zhang, G.; Jiang, C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008, 18, 290–301. [Google Scholar] [CrossRef]
- Amraei, R.; Xia, C.; Olejnik, J.; White, M.R.; Napoleon, M.A.; Lotfollahzadeh, S.; Hauser, B.M.; Schmidt, A.G.; Chitalia, V.; Muhlberger, E.; et al. Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2113874119. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Sanders, D.W.; Jumper, C.C.; Ackerman, P.J.; Bracha, D.; Donlic, A.; Kim, H.; Kenney, D.; Castello-Serrano, I.; Suzuki, S.; Tamura, T.; et al. SARS-CoV-2 requires cholesterol for viral entry and pathological syncytia formation. Elife 2021, 10, e65962. [Google Scholar] [CrossRef]
- Amar, M.J.; Kaler, M.; Courville, A.B.; Shamburek, R.; Sampson, M.; Remaley, A.T. Randomized double blind clinical trial on the effect of oral alpha-cyclodextrin on serum lipids. Lipids Health Dis. 2016, 15, 115. [Google Scholar] [CrossRef]
- Coisne, C.; Tilloy, S.; Monflier, E.; Wils, D.; Fenart, L.; Gosselet, F. Cyclodextrins as Emerging Therapeutic Tools in the Treatment of Cholesterol-Associated Vascular and Neurodegenerative Diseases. Molecules 2016, 21, 1748. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E.; Karten, B. Niemann-Pick C disease and mobilization of lysosomal cholesterol by cyclodextrin. J. Lipid Res. 2014, 55, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, Y.; Irie, T.; Uekama, K.; Fukunaga, K.; Pitha, J. Differential effects of alpha-, beta- and gamma-cyclodextrins on human erythrocytes. Eur. J. Biochem. 1989, 186, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Yancey, P.G.; Rodrigueza, W.V.; Kilsdonk, E.P.; Stoudt, G.W.; Johnson, W.J.; Phillips, M.C.; Rothblat, G.H. Cellular cholesterol efflux mediated by cyclodextrins. Demonstration Of kinetic pools and mechanism of efflux. J. Biol. Chem. 1996, 271, 16026–16034. [Google Scholar] [CrossRef] [PubMed]
- Wittkowski, K.M.; Dadurian, C.; Seybold, M.P.; Kim, H.S.; Hoshino, A.; Lyden, D. Complex polymorphisms in endocytosis genes suggest alpha-cyclodextrin as a treatment for breast cancer. PLoS ONE 2018, 13, e0199012. [Google Scholar] [CrossRef]
- El-Darzi, N.; Mast, N.; Petrov, A.M.; Pikuleva, I.A. 2-Hydroxypropyl-beta-cyclodextrin reduces retinal cholesterol in wild-type and Cyp27a1−/− Cyp46a1−/− mice with deficiency in the oxysterol production. Br. J. Pharmacol. 2021, 178, 3220–3234. [Google Scholar] [CrossRef]
- Civra, A.; Francese, R.; Gamba, P.; Testa, G.; Cagno, V.; Poli, G.; Lembo, D. 25-Hydroxycholesterol and 27-hydroxycholesterol inhibit human rotavirus infection by sequestering viral particles into late endosomes. Redox Biol. 2018, 19, 318–330. [Google Scholar] [CrossRef]
- Shang, C.; Zhuang, X.; Zhang, H.; Li, Y.; Zhu, Y.; Lu, J.; Ge, C.; Cong, J.; Li, T.; Tian, M.; et al. Inhibitors of endosomal acidification suppress SARS-CoV-2 replication and relieve viral pneumonia in hACE2 transgenic mice. Virol. J. 2021, 18, 46. [Google Scholar] [CrossRef]
- Blanc, M.; Hsieh, W.Y.; Robertson, K.A.; Kropp, K.A.; Forster, T.; Shui, G.; Lacaze, P.; Watterson, S.; Griffiths, S.J.; Spann, N.J.; et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 2013, 38, 106–118. [Google Scholar] [CrossRef]
- Liu, S.Y.; Sanchez, D.J.; Aliyari, R.; Lu, S.; Cheng, G. Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl. Acad. Sci. USA 2012, 109, 4239–4244. [Google Scholar] [CrossRef]
- Moog, C.; Aubertin, A.M.; Kirn, A.; Luu, B. Oxysterols, but not cholesterol, inhibit human immunodeficiency virus replication in vitro. Antivir. Chem. Chemother. 1998, 9, 491–496. [Google Scholar] [CrossRef]
- Shibata, N.; Carlin, A.F.; Spann, N.J.; Saijo, K.; Morello, C.S.; McDonald, J.G.; Romanoski, C.E.; Maurya, M.R.; Kaikkonen, M.U.; Lam, M.T.; et al. 25-Hydroxycholesterol activates the integrated stress response to reprogram transcription and translation in macrophages. J. Biol. Chem. 2013, 288, 35812–35823. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, J.; Li, M.; Chen, M.; Sun, C. Multifaceted Functions of CH25H and 25HC to Modulate the Lipid Metabolism, Immune Responses, and Broadly Antiviral Activities. Viruses 2020, 12, 727. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Scott, A.L. Cholesterol 25-hydroxylase production by dendritic cells and macrophages is regulated by type I interferons. J. Leukoc. Biol. 2010, 88, 1081–1087. [Google Scholar] [CrossRef]
- Zu, S.; Deng, Y.Q.; Zhou, C.; Li, J.; Li, L.; Chen, Q.; Li, X.F.; Zhao, H.; Gold, S.; He, J.; et al. 25-Hydroxycholesterol is a potent SARS-CoV-2 inhibitor. Cell Res. 2020, 30, 1043–1045. [Google Scholar] [CrossRef]
- Leussink, S.; Aranda-Pardos, I.; A-Gonzalez, N. Lipid metabolism as a mechanism of immunomodulation in macrophages: The role of liver X receptors. Curr. Opin. Pharmacol. 2020, 53, 18–26. [Google Scholar] [CrossRef]
- Zhang, L.; Reue, K.; Fong, L.G.; Young, S.G.; Tontonoz, P. Feedback regulation of cholesterol uptake by the LXR-IDOL-LDLR axis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2541–2546. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, V.; Nelson, J.K.; Maspero, E.; Marques, A.R.A.; Scheer, L.; Polo, S.; Zelcer, N. The LXR-IDOL axis defines a clathrin-, caveolae-, and dynamin-independent endocytic route for LDLR internalization and lysosomal degradation. J. Lipid Res. 2013, 54, 2174–2184. [Google Scholar] [CrossRef]
- Glebov, O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020, 287, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Chau, N.; Abdel-Hamid, M.K.; Hu, L.; von Kleist, L.; Whiting, A.; Krishnan, S.; Maamary, P.; Joseph, S.R.; Simpson, F.; et al. Phenothiazine-derived antipsychotic drugs inhibit dynamin and clathrin-mediated endocytosis. Traffic 2015, 16, 635–654. [Google Scholar] [CrossRef]
- Preta, G.; Cronin, J.G.; Sheldon, I.M. Dynasore—not just a dynamin inhibitor. Cell Commun. Signal. 2015, 13, 24. [Google Scholar] [CrossRef] [PubMed]
- McGuinn, K.P.; Mahoney, M.G. Lipid rafts and detergent-resistant membranes in epithelial keratinocytes. Methods Mol. Biol. 2014, 1195, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, C.M.; Brumell, J.H.; Finlay, B.B. Microbial pathogenesis: Lipid rafts as pathogen portals. Curr. Biol. 2000, 10, R823–R825. [Google Scholar] [CrossRef]
- Palacios-Rapalo, S.N.; De Jesus-Gonzalez, L.A.; Cordero-Rivera, C.D.; Farfan-Morales, C.N.; Osuna-Ramos, J.F.; Martinez-Mier, G.; Quistian-Galvan, J.; Munoz-Perez, A.; Bernal-Dolores, V.; Del Angel, R.M.; et al. Cholesterol-Rich Lipid Rafts as Platforms for SARS-CoV-2 Entry. Front. Immunol. 2021, 12, 796855. [Google Scholar] [CrossRef] [PubMed]
- Fecchi, K.; Anticoli, S.; Peruzzu, D.; Iessi, E.; Gagliardi, M.C.; Matarrese, P.; Ruggieri, A. Coronavirus Interplay with Lipid Rafts and Autophagy Unveils Promising Therapeutic Targets. Front. Microbiol. 2020, 11, 1821. [Google Scholar] [CrossRef]
- Bai, X.L.; Yang, X.Y.; Li, J.Y.; Ye, L.; Jia, X.; Xiong, Z.F.; Wang, Y.M.; Jin, S. Cavin-1 regulates caveolae-mediated LDL transcytosis: Crosstalk in an AMPK/eNOS/NF-kappaB/Sp1 loop. Oncotarget 2017, 8, 103985–103995. [Google Scholar] [CrossRef]
- Vishnyakova, T.G.; Bocharov, A.V.; Baranova, I.N.; Kurlander, R.; Drake, S.K.; Chen, Z.; Amar, M.; Sviridov, D.; Vaisman, B.; Poliakov, E.; et al. SR-BI mediates neutral lipid sorting from LDL to lipid droplets and facilitates their formation. PLoS ONE 2020, 15, e0240659. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, H.; Han, G.Z. Host-Virus Arms Races Drive Elevated Adaptive Evolution in Viral Receptors. J. Virol. 2020, 94, e00684-20. [Google Scholar] [CrossRef]
- Nielsen, R.; Hellmann, I.; Hubisz, M.; Bustamante, C.; Clark, A.G. Recent and ongoing selection in the human genome. Nat. Rev. Genet. 2007, 8, 857–868. [Google Scholar] [CrossRef]
- Kosakovsky Pond, S.L.; Frost, S.D. Not so different after all: A comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Wertheim, J.O.; Moola, S.; Weighill, T.; Scheffler, K.; Kosakovsky Pond, S.L. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012, 8, e1002764. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A receptor-mediated pathway for cholesterol homeostasis. Science 1986, 232, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Dunn, K.W.; Pytowski, B.; McGraw, T.E. Endosome acidification and receptor trafficking: Bafilomycin A1 slows receptor externalization by a mechanism involving the receptor’s internalization motif. Mol. Biol. Cell 1993, 4, 1251–1266. [Google Scholar] [CrossRef] [PubMed]
- Cyr, Y.; Bissonnette, S.; Lamantia, V.; Wassef, H.; Loizon, E.; Ngo Sock, E.T.; Vidal, H.; Mayer, G.; Chretien, M.; Faraj, M. White Adipose Tissue Surface Expression of LDLR and CD36 is Associated with Risk Factors for Type 2 Diabetes in Adults with Obesity. Obesity 2020, 28, 2357–2367. [Google Scholar] [CrossRef]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996, 62, 155–169. [Google Scholar] [CrossRef]
- Samuel, W.; Jaworski, C.; Postnikova, O.A.; Kutty, R.K.; Duncan, T.; Tan, L.X.; Poliakov, E.; Lakkaraju, A.; Redmond, T.M. Appropriately differentiated ARPE-19 cells regain phenotype and gene expression profiles similar to those of native RPE cells. Mol. Vis. 2017, 23, 60–89. [Google Scholar]
- Matsui, M.; Sakurai, F.; Elbashir, S.; Foster, D.J.; Manoharan, M.; Corey, D.R. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem. Biol. 2010, 17, 1344–1355. [Google Scholar] [CrossRef]






| Antibody | Catalog No. | Company Name |
|---|---|---|
| Normal Mouse IgG (1 mg/mL) | 12-371 | Millipore Sigma (Burlington, MA, USA) |
| Normal Rabbit IgG (1 mg/mL) | 12-370 | Millipore Sigma (Burlington, MA, USA) |
| Mouse anti-EGFR monoclonal clone LA22 | 05-104 | Millipore Sigma (Burlington, MA, USA) |
| ACE-2 mouse monoclonal IgG2A clone 171606 | MAB933 | R&D Systems (Minneapolis, MN, USA) |
| ACE-2 mouse monoclonal IgG2A clone 535919 | MAB9332-100 | R&D Systems (Minneapolis, MN, USA) |
| ACE-2 goat polyclonal IgG (0.2 mg/mL) | AF933 | R&D Systems (Minneapolis, MN, USA) |
| Mouse anti-human monoclonal Neuropilin-1 (0.34 mg/mL) | MAB3870 | R&D Systems (Minneapolis, MN, USA) |
| Mouse monoclonal Ultra-Leaf™ Purified anti-human CD147 (2 mg/mL) | 306221 | Biolegend® (San Diego, CA, USA) |
| Axl rabbit polyclonal IgG (0.34 mg/mL) | PA5-34658 | Thermo Fisher Scientific (Waltham, MA, USA) |
| Caveolin-1 rabbit polyclonal IgG (1 mg/mL) | PA1-064 | Thermo Fisher Scientific (Waltham, MA, USA) |
| Vimentin (V9) mouse monoclonal (0.5 mg/mL) | sc-6260 | Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) |
| Vimentin chicken polyclonal IgY | NB-300-223 | Novus Biologicals (Centennial, CO, USA) |
| Dynamin I/II rabbit polyclonal IgG | 2342 | Cell Signaling Technology® (Danvers, MA, USA) |
| Flotilin-1 rabbit polyclonal IgG (0.34 mg/mL) | 3253 | Cell Signaling Technology® (Danvers, MA, USA) |
| Clathrin HC (C-20) goat polyclonal IgG (200 µg/mL) | sc-6579 | Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) |
| LDL Receptor Mouse monoclonal [1B10H10] antibody (1 mg/mL) | ab204941 | Abcam (Waltham, MA, USA) |
| LDL Receptor Mouse monoclonal clone C7 | MABS26 | Millipore Sigma (Burlington, MA, USA) |
| SR-B1 rabbit polyclonal IgG | NB400-113 | Novus Biologicals (Centennial, CO, USA) |
| Secondary IRDye 800CW Goat Anti-Rabbit | 925-32211 | LI-COR Biosciences, Lincoln, NE, USA |
| Secondary IRDye 680RD Goat Anti-Mouse | 926-68070 | LI-COR Biosciences, Lincoln, NE, USA |
| Secondary IRDye 680RD Donkey Anti-Goat | 925-68074 | LI-COR Biosciences, Lincoln, NE, USA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uppal, S.; Postnikova, O.; Villasmil, R.; Rogozin, I.B.; Bocharov, A.V.; Eggerman, T.L.; Poliakov, E.; Redmond, T.M. Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells. Int. J. Mol. Sci. 2023, 24, 11860. https://doi.org/10.3390/ijms241411860
Uppal S, Postnikova O, Villasmil R, Rogozin IB, Bocharov AV, Eggerman TL, Poliakov E, Redmond TM. Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells. International Journal of Molecular Sciences. 2023; 24(14):11860. https://doi.org/10.3390/ijms241411860
Chicago/Turabian StyleUppal, Sheetal, Olga Postnikova, Rafael Villasmil, Igor B. Rogozin, Alexander V. Bocharov, Thomas L. Eggerman, Eugenia Poliakov, and T. Michael Redmond. 2023. "Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells" International Journal of Molecular Sciences 24, no. 14: 11860. https://doi.org/10.3390/ijms241411860
APA StyleUppal, S., Postnikova, O., Villasmil, R., Rogozin, I. B., Bocharov, A. V., Eggerman, T. L., Poliakov, E., & Redmond, T. M. (2023). Low-Density Lipoprotein Receptor (LDLR) Is Involved in Internalization of Lentiviral Particles Pseudotyped with SARS-CoV-2 Spike Protein in Ocular Cells. International Journal of Molecular Sciences, 24(14), 11860. https://doi.org/10.3390/ijms241411860









