Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep
Abstract
:1. Introduction
2. Results
2.1. Cell Culture and Identification
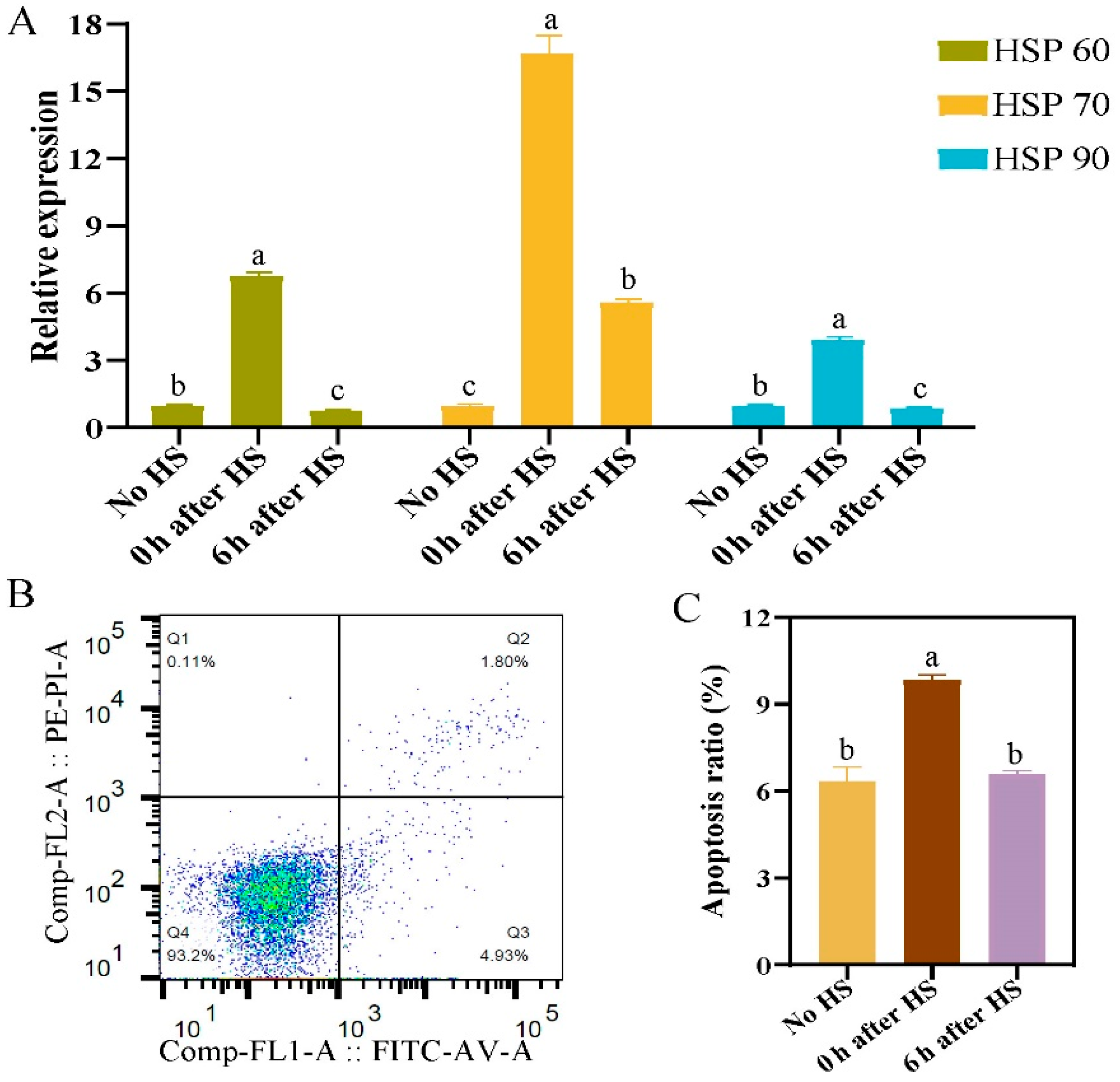
2.2. Establishment of an In Vitro Heat Stress Model in Primary Hepatocytes and Adipocytes
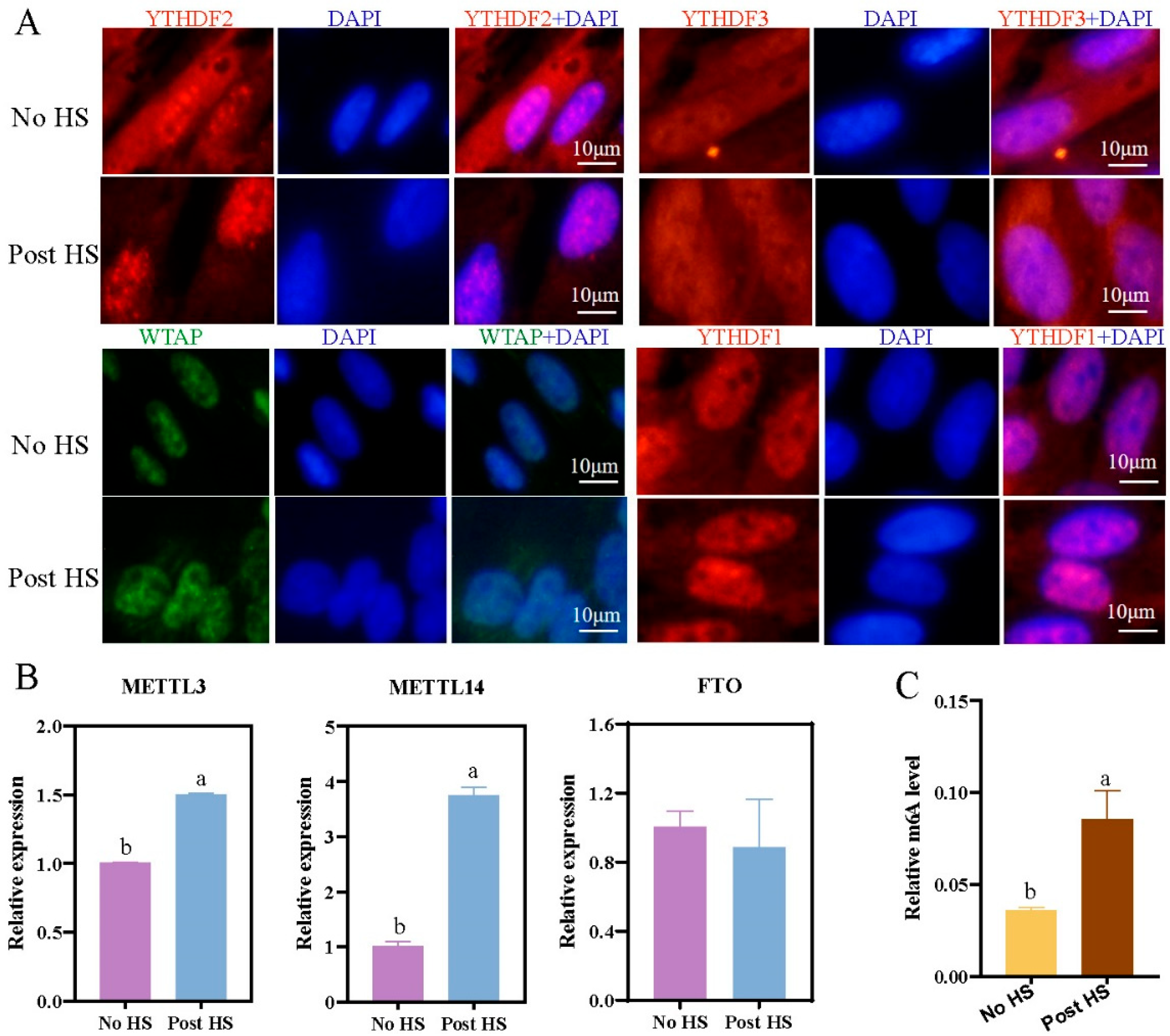
2.3. Effect of Heat Stress on Expression of Methylation-Related Factors
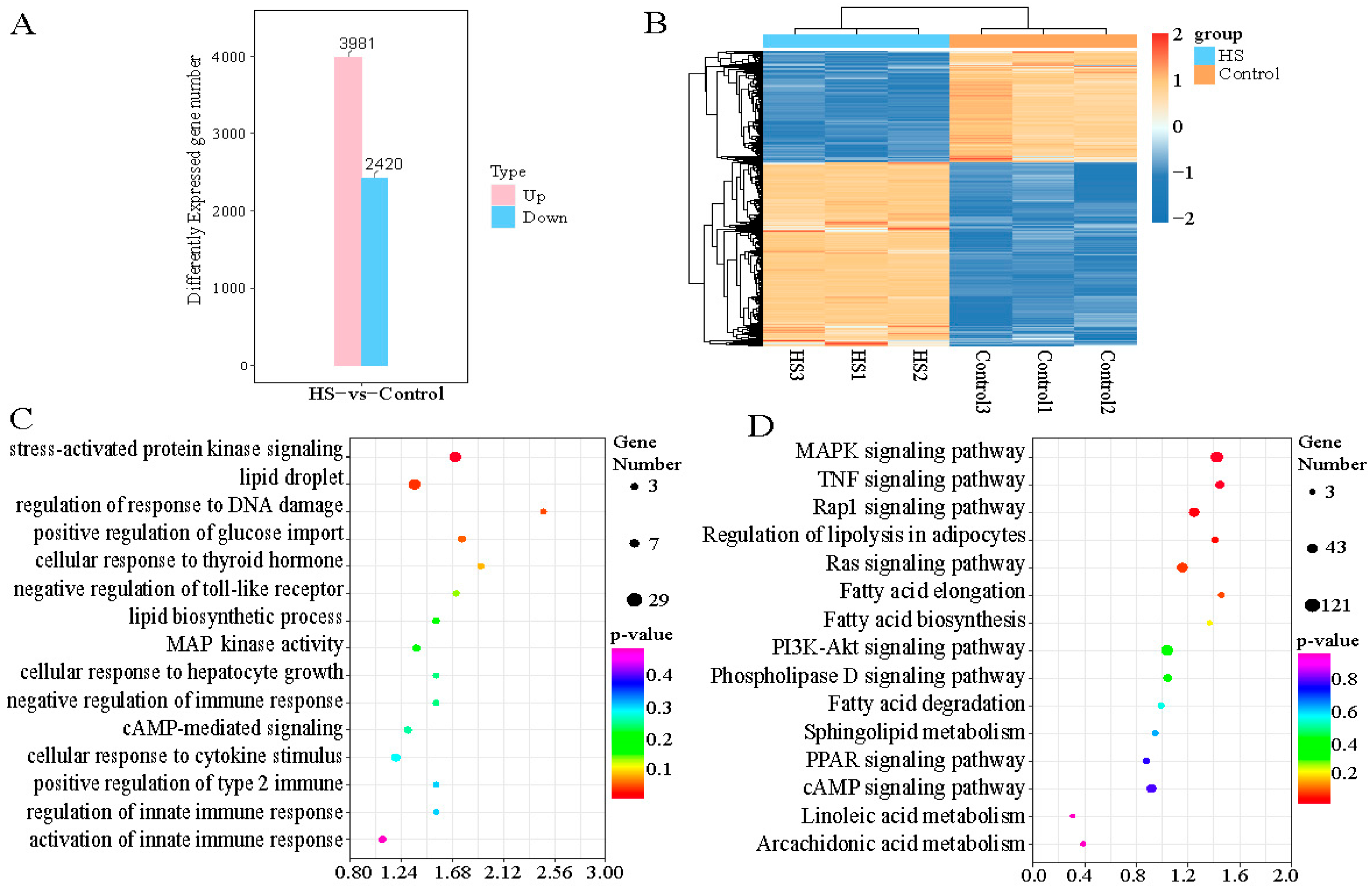
2.4. Identification of Heat-Stress-Related Differentially Expressed Genes (DEGs) and Functional Enrichment Analysis of Primary Hepatocytes
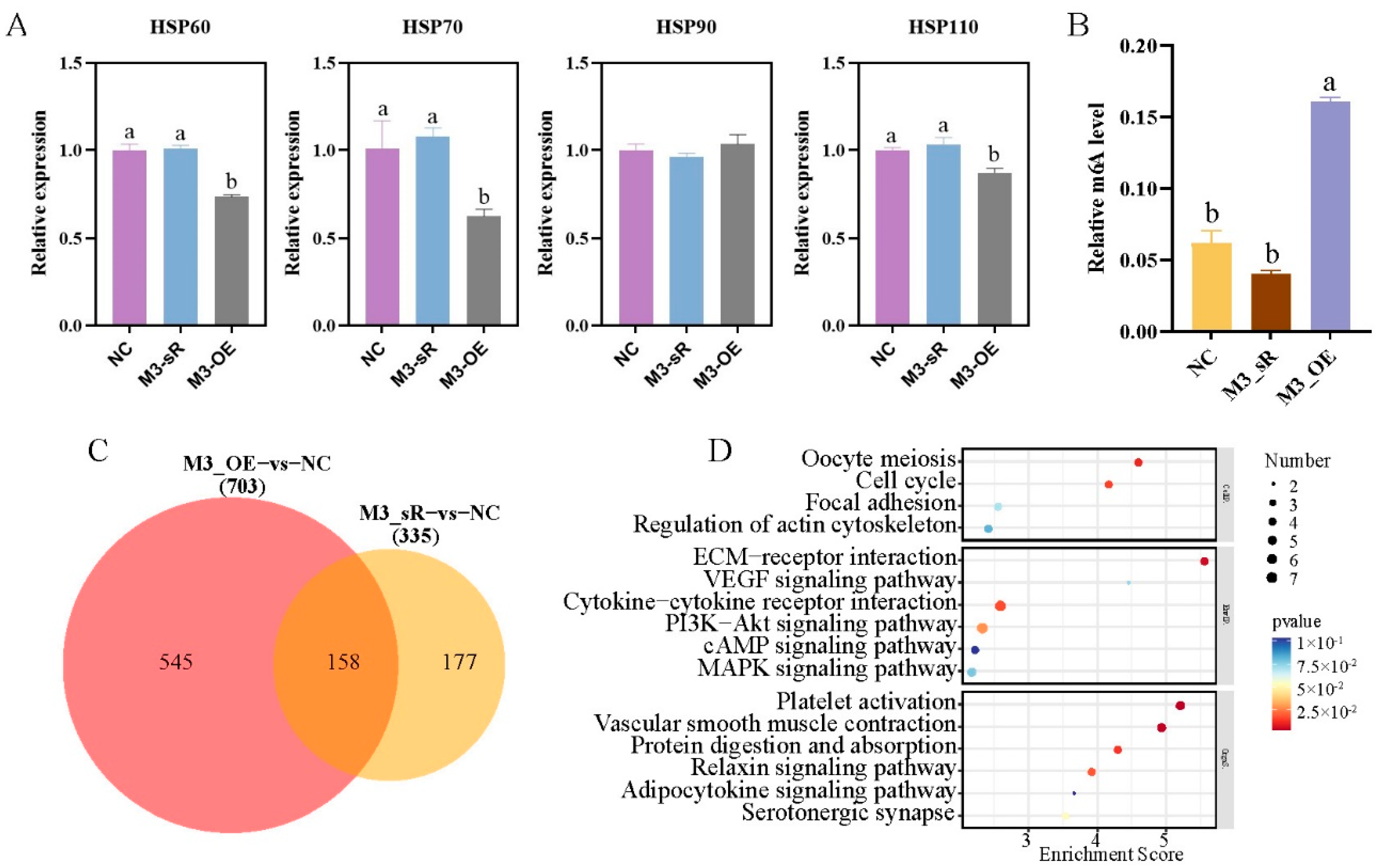
2.5. Molecular Mechanism of METTL3 in the Regulation of Heat Stress
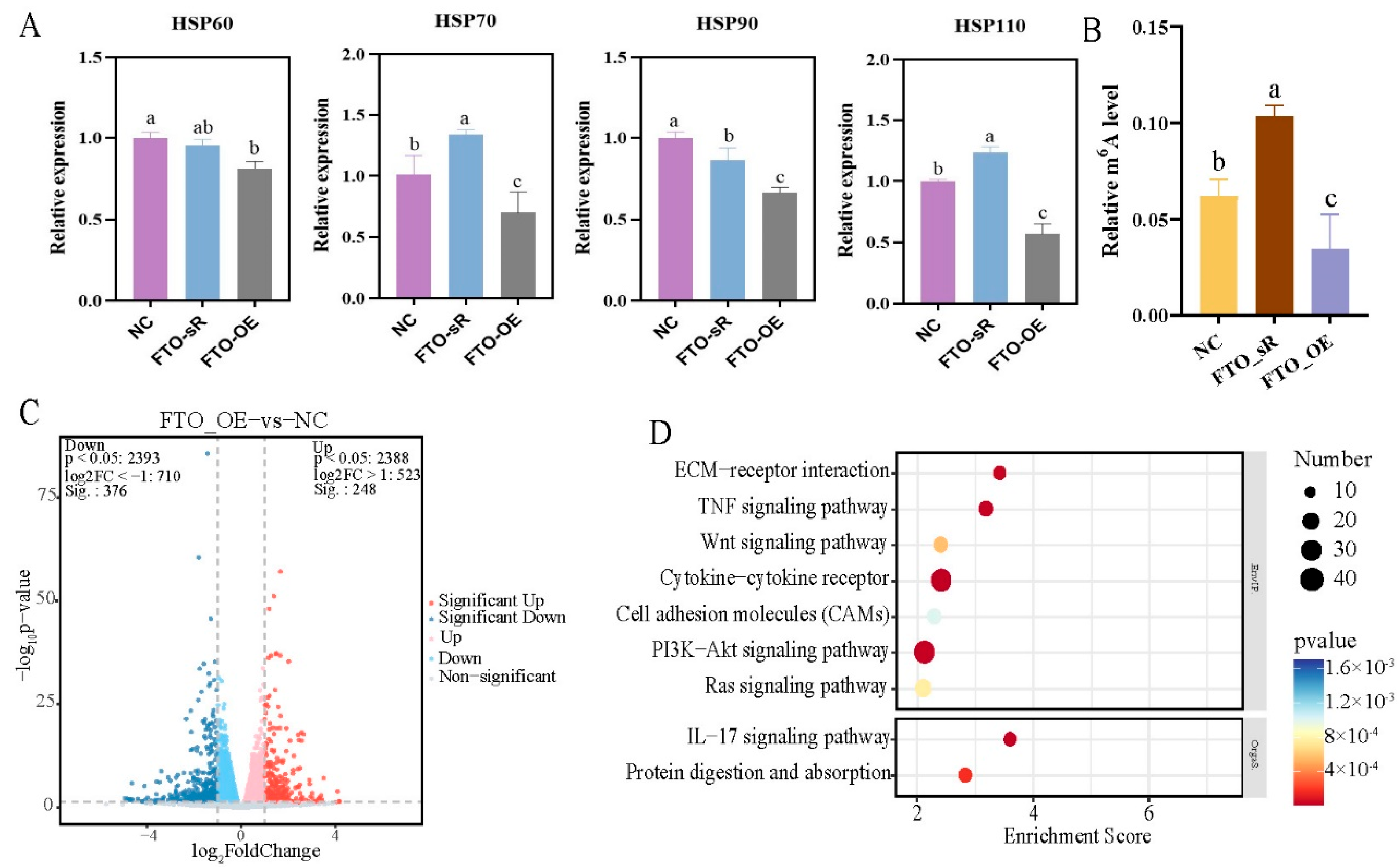
2.6. Molecular Mechanism of FTO in the Regulation of Heat Stress
3. Discussion
4. Materials and Methods
4.1. Cell Isolation and Culture Procedures
4.2. Cell Proliferation and Apoptosis Detection
4.3. Oil Red O Staining and Immunofluorescence Assay
4.4. Heat Stress Induction
4.5. m6A RNA Methylation Quantification
4.6. Lentiviral Overexpression and RNAi Constructs and Infection of Cells
4.7. RNA-Seq Data Analysis
4.8. RNA Extraction, Reverse Transcription, and qRT-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFP | Alpha-fetoprotein |
| ALKBK5 | Alk B homologue 5 |
| CCK-8 | Cell Counting Kit-8 |
| DEGs | Differentially expressed genes |
| FTO | Fat mass and obesity associated protein |
| GO | Gene Ontology |
| HS | Heat stress |
| HSP | Heat stress protein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| m6A | N6-methyladenosine |
| METTL3 | Methyltransferase-like protein 3 |
| METTL14 | Methyltransferase-like protein 14 |
| qRT-PCR | Quantitative reverse transcriptase-PCR |
| RNAi | RNA interference |
| WTAP | Wilms tumor 1 associated protein |
| YTHDF1 | YTH domain family proteins 1 |
| YTHDF2 | YTH domain family proteins 2 |
| YTHDC1 | YTH domain containing proteins 1 |
| YTHDC2 | YTH domain containing proteins 2 |
References
- Zhao, H.; Wang, Z.; Li, X.; Chu, Z.; Zhao, C.; Zhao, F. Research on the evolution characteristics of future climate change in West Liao River Basin. Environ. Sci. Pollut. Res. Int. 2022, 29, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Sejian, V.; Srivastava, R.S. Effects of Melatonin on Adrenal Cortical Functions of Indian Goats under Thermal Stress. Vet. Med. Int. 2010, 2010, 348919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Xie, T.; Li, S.; Wang, W.; Wang, Y.; Cao, Z.; Yang, H. Effects of Selenium as a Dietary Source on Performance, Inflammation, Cell Damage, and Reproduction of Livestock Induced by Heat Stress: A Review. Front. Immunol. 2021, 12, 820853. [Google Scholar] [CrossRef]
- Chang-Fung-Martel, J.; Harrison, M.T.; Brown, J.N.; Rawnsley, R.; Smith, A.P.; Meinke, H. Negative relationship between dry matter intake and the temperature-humidity index with increasing heat stress in cattle: A global meta-analysis. Int. J. Biometeorol. 2021, 65, 2099–2109. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ezernieks, V.; Wang, J.; Arachchillage, N.; Garner, J.; Wales, W.; Cocks, B.; Rochfort, S. Heat Stress in Dairy Cattle Alters Lipid Composition of Milk. Sci. Rep. 2017, 7, 961. [Google Scholar] [CrossRef] [Green Version]
- Chelmicki, T.; Roger, E.; Teissandier, A.; Dura, M.; Bonneville, L.; Rucli, S.; Dossin, F.; Fouassier, C.; Lameiras, S.; Bourc’his, D. m6A RNA methylation regulates the fate of endogenous retroviruses. Nature 2021, 591, 312–316. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Huai, Q.L.; Sun, S.J.; Guo, L.; Xue, X.M.; Song, P.; Ying, J.M.; Gao, Y.B.; Gao, S.G.; He, J. Development and validation of m6A RNA methylation regulators-based signature in lung adenocarcinoma. Chin. Med. J. 2021, 134, 3. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, H.; Yu, B.; Tao, D.; Xu, X.; Xu, Y.; Wang, J.; Jiao, Y.; Wang, L. The role of m6A methylation in therapy resistance in cancer. Mol. Cancer 2023, 22, 91. [Google Scholar] [CrossRef]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Rep. 2018, 28, 9. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.; Patil, D.; Zhou, J.; Zinoviev, A.; Jaffrey, S. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Ma, Y.; Li, Q.; Liu, E.; Jin, M.; Zhang, L.; Wei, C. The role of N6-methyladenosine RNA methylation in the heat stress response of sheep (Ovis aries). Cell Stress Chaperones 2019, 24, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Chu, M.; Li, Q.; Jin, M.; Fei, X.; Ma, L.; Zhang, L.; Wei, C. Transcriptomic Analysis Provides Novel Insights into Heat Stress Responses in Sheep. Animals 2019, 9, 387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.; Dou, X.; Zhang, D.; Liu, T.; Yang, B.; Lu, Z. Development of an Improved Method for the Isolation and Culture of Newborn Sheep Primary Hepatocytes. Curr. Issues Mol. Biol. 2022, 44, 3621–3631. [Google Scholar] [CrossRef]
- Yu, J.; Li, Y.; Wang, T.; Zhong, X. Modification of N6-methyladenosine RNA methylation on heat shock protein expression. PLoS ONE 2018, 13, e0198604. [Google Scholar] [CrossRef]
- Zuo, L.; Christofi, F.L.; Wright, V.P.; Liu, C.Y.; Merola, A.J.; Berliner, L.J.; Clanton, T.L. Intra- and extracellular measurement of reactive oxygen species produced during heat stress in diaphragm muscle. Am. J. Physiol. Cell Physiol. 2000, 279, C1058–C1066. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Xie, H.; Pan, P.; Qu, Q.; Xia, Q.; Gao, X.; Zhang, S.; Jiang, Q. Heat stress promotes lipid accumulation by inhibiting the AMPK-PGC-1α signaling pathway in 3T3-L1 preadipocytes. Cell Stress Chaperones 2021, 26, 563–574. [Google Scholar] [CrossRef]
- Qu, H.; Donkin, S.S.; Ajuwon, K.M. Heat stress enhances adipogenic differentiation of subcutaneous fat depot-derived porcine stromovascular cells. J. Anim. Sci. 2015, 93, 3832–3842. [Google Scholar] [CrossRef] [Green Version]
- Barnes, T.L.; Cadaret, C.N.; Beede, K.A.; Schmidt, T.B.; Petersen, J.L.; Yates, D.T. Hypertrophic muscle growth and metabolic efficiency were impaired by chronic heat stress, improved by zilpaterol supplementation, and not affected by ractopamine supplementation in feedlot lambs1. J. Anim. Sci. 2019, 97, 4101–4113. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, D.D.; Klauck, V.; Campigotto, G.; Alba, D.F.; Dos Reis, J.H.; Gebert, R.R.; Souza, C.F.; Baldissera, M.D.; Schogor, A.L.B.; Santos, I.D.; et al. Benefits of the inclusion of açai oil in the diet of dairy sheep in heat stress on health and milk production and quality. J. Therm. Biol. 2019, 84, 250–258. [Google Scholar]
- Liu, H.; Li, K.; Mingbin, L.; Zhao, J.; Xiong, B. Effects of chestnut tannins on the meat quality, welfare, and antioxidant status of heat-stressed lambs. Meat Sci. 2016, 116, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Ringseis, R.; Eder, K. Heat stress in pigs and broilers: Role of gut dysbiosis in the impairment of the gut-liver axis and restoration of these effects by probiotics, prebiotics and synbiotics. J. Anim. Sci. Biotechnol. 2022, 13, 126. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [Green Version]
- Danesh Mesgaran, M.; Kargar, H.; Janssen, R.; Danesh Mesgaran, S.; Ghesmati, A.; Vatankhah, A. Rumen-protected zinc-methionine dietary inclusion alters dairy cow performances, and oxidative and inflammatory status under long-term environmental heat stress. Front. Vet. Sci. 2022, 9, 935939. [Google Scholar] [CrossRef]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular Mechanisms Driving mRNA Degradation by m6A Modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Horodyska, J.; Wimmers, K.; Reyer, H.; Trakooljul, N.; Mullen, A.M.; Lawlor, P.G.; Hamill, R.M. RNA-seq of muscle from pigs divergent in feed efficiency and product quality identifies differences in immune response, growth, and macronutrient and connective tissue metabolism. BMC Genom. 2018, 19, 791. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Wu, S.-B.; Roberts, J. RNA-sequencing analysis of shell gland shows differences in gene expression profile at two time-points of eggshell formation in laying chickens. BMC Genom. 2019, 20, 89. [Google Scholar] [CrossRef] [Green Version]
- Salleh, S.M.; Mazzoni, G.; Løvendahl, P.; Kadarmideenco, H.N. Gene expression networks from RNA sequencing of dairy cattle identifies genes and pathways affecting feed efficiency. BMC Bioinform. 2018, 19, 513. [Google Scholar] [CrossRef]
- Borrie, S.C.; Brems, H.; Legius, E.; Bagni, C. Cognitive Dysfunctions in Intellectual Disabilities: The Contributions of the Ras-MAPK and PI3K-AKT-mTOR Pathways. Annu. Rev. Genomics Hum. Genet. 2017, 18, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Frische, E.W.; Zwartkruis, F.J.T. Rap1, a mercenary among the Ras-like GTPases. Dev. Biol. 2010, 340, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kam, C.Y.; Dubash, A.D.; Magistrati, E.; Polo, S.; Satchell, K.J.F.; Sheikh, F.; Lampe, P.D.; Green, K.J. Desmoplakin maintains gap junctions by inhibiting Ras/MAPK and lysosomal degradation of connexin-43. J. Cell Biol. 2018, 217, 3219–3235. [Google Scholar] [CrossRef]
- Lee, H.-W.; Rhee, D.-K.; Kim, B.-O.; Pyo, S. Inhibitory effect of sinigrin on adipocyte differentiation in 3T3-L1 cells: Involvement of AMPK and MAPK pathways. Biomed. Pharmacother. 2018, 102, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, D.; Zeng, R.; Xian, T.; Lu, Y.; Zeng, G.; Sun, Z.; Huang, B.; Huang, Q. Epigallocatechin-3-gallate inhibits adipogenesis through down-regulation of PPARγ and FAS expression mediated by PI3K-AKT signaling in 3T3-L1 cells. Eur. J. Pharmacol. 2017, 795, 134–142. [Google Scholar] [CrossRef]
- Shevtsov, M.; Multhoff, G. Heat Shock Protein-Peptide and HSP-Based Immunotherapies for the Treatment of Cancer. Front. Immunol. 2016, 7, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Choi, R.-Y.; Lee, M.-K. Polygonum multiflorum Thunb. Hot Water Extract Reverses High-Fat Diet-Induced Lipid Metabolism of White and Brown Adipose Tissues in Obese Mice. Plants 2021, 10, 1509. [Google Scholar] [CrossRef]
- Siddiqui, S.H.; Khan, M.; Park, J.; Lee, J.; Choe, H.; Shim, K.; Kang, D. COPA3 peptide supplementation alleviates the heat stress of chicken fibroblasts. Front. Vet. Sci. 2023, 10, 985040. [Google Scholar] [CrossRef]
- Yao, Y.; Bi, Z.; Wu, R.; Zhao, Y.; Liu, Y.; Liu, Q.; Wang, Y.; Wang, X. METTL3 inhibits BMSC adipogenic differentiation by targeting the JAK1/STAT5/C/EBPβ pathway via an m6A-YTHDF2-dependent manner. FASEB J. 2019, 33, 7529–7544. [Google Scholar] [CrossRef]
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y.; et al. Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cell Rep. 2018, 25, 1816–1828. [Google Scholar] [CrossRef] [Green Version]
- Song, T.; Yang, Y.; Jiang, S.; Peng, J. Novel Insights into Adipogenesis from the Perspective of Transcriptional and RNA N6-Methyladenosine-Mediated Post-Transcriptional Regulation. Adv. Sci. 2020, 7, 2001563. [Google Scholar] [CrossRef]
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235. [Google Scholar] [CrossRef]
- Chauhan, S.S.; Rashamol, V.P.; Bagath, M.; Sejian, V.; Dunshea, F.R. Impacts of heat stress on immune responses and oxidative stress in farm animals and nutritional strategies for amelioration. Int. J. Biometeorol. 2021, 65, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhao, Z.; Shi, Y.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. CircEZH2 Regulates Milk Fat Metabolism through miR-378b Sponge Activity. Animals 2022, 12, 718. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Tang, S.; Liu, L.; Cao, A.; Xie, J.; Zhang, H. Effects of Bile Acids on Growth Performance and Lipid Metabolism during Chronic Heat Stress in Broiler Chickens. Animals 2021, 11, 630. [Google Scholar] [CrossRef]
- Li, A.; Gu, Y.; Zhang, X.; Yu, H.; Liu, D.; Pang, Q. Betaine Regulates the Production of Reactive Oxygen Species through Wnt10b Signaling in the Liver of Zebrafish. Front. Physiol. 2022, 13, 877178. [Google Scholar] [CrossRef]
- Dunshea, F.R.; Oluboyede, K.; DiGiacomo, K.; Leury, B.J.; Cottrell, J.J. Betaine Improves Milk Yield in Grazing Dairy Cows Supplemented with Concentrates at High Temperatures. Animals 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Huang, W.; Wang, Z.; Nie, Q. m6A demethylase FTO regulate CTNNB1 to promote adipogenesis of chicken preadipocyte. J. Anim. Sci. Biotechnol. 2022, 13, 147. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, J.; Chen, J.; Wu, W.; Wang, X.; Wang, Y. The beneficial effects of betaine on dysfunctional adipose tissue and N6-methyladenosine mRNA methylation requires the AMP-activated protein kinase α1 subunit. J. Nutr. Biochem. 2015, 26, 1678–1684. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Yuan, C.; Guo, T.; Liu, J.; Yang, B.; Lu, Z. Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep. Int. J. Mol. Sci. 2023, 24, 11926. https://doi.org/10.3390/ijms241511926
Chen B, Yuan C, Guo T, Liu J, Yang B, Lu Z. Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep. International Journal of Molecular Sciences. 2023; 24(15):11926. https://doi.org/10.3390/ijms241511926
Chicago/Turabian StyleChen, Bowen, Chao Yuan, Tingting Guo, Jianbin Liu, Bohui Yang, and Zengkui Lu. 2023. "Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep" International Journal of Molecular Sciences 24, no. 15: 11926. https://doi.org/10.3390/ijms241511926
APA StyleChen, B., Yuan, C., Guo, T., Liu, J., Yang, B., & Lu, Z. (2023). Molecular Mechanism of m6A Methylation Modification Genes METTL3 and FTO in Regulating Heat Stress in Sheep. International Journal of Molecular Sciences, 24(15), 11926. https://doi.org/10.3390/ijms241511926





