Triangular Silver Nanoplates as a Bioanalytical Tool: Potential COVID-19 Detection
Abstract
1. Introduction
2. Results
2.1. Detection of Native Fn Using Anti-Fn Antibody-Functionalized PEGAuTSNP
2.2. Spike Protein Optimal Functionalization Volume Determination
2.3. Spike-PEGAuTSNP Detection Limit Determination
2.4. Specificity Analysis of the Spike–Anti-Spike System
2.5. Spike Protein Detection-Platform Testing within a Complex System
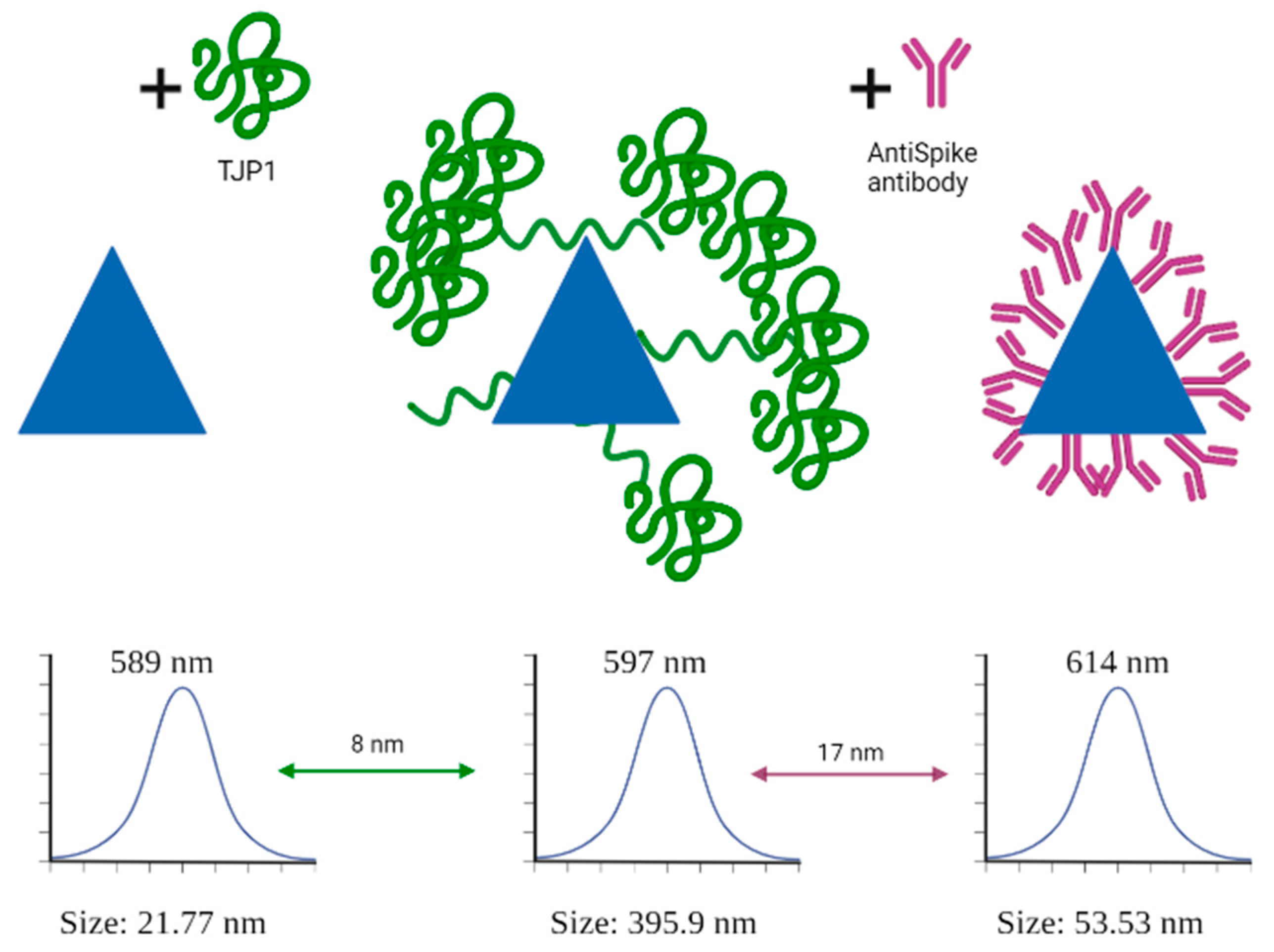
2.6. Dinamic Light Scattering (DLS) Size Measurements
2.7. Spike–Anti-Spike Binding PEGAuTSNP-Based Immunoassay within Horse Serum
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Anti-Fn-PEGAuTSNP Preparation
4.3. Extracellular Matrix Isolation Protocol
4.4. Detection of Native Fibronectin with Anti-Fn-PEGAuTSNP
4.5. Spike Protein Optimal Functionalization Volume Determination
4.6. Nanoplate-Based Immunoassay with Changing Concentrations of Anti-Spike Antibody
4.7. Spike–Anti-Spike Binding PEGAuTSNP-Based Immunoassay
4.8. Spike–Anti-Spike Binding PEGAuTSNP-Based Immunoassay within Horse Serum
4.9. Dynamic Light Scattering (DLS) Size Measurements
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vashist, S.K.; Luong, J.H.T. Immunoassays: An Overview; Elsevier Inc.: Amsterdam, The Netherlands, 2018; ISBN 9780128117620. [Google Scholar]
- Darwish, I.A. Immunoassay Methods and Their Applications in Pharmaceutical Analysis: Basic Methodology and Recent Advances. Int. J. Biomed. Sci. 2006, 2, 217. [Google Scholar]
- Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Gholami, A.; Omidifar, N.; Babapoor, A.; Vijayakameswara Rao, N.; Chiang, W.H. Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors 2022, 12, 286. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ning, J.; Peng, D.; Chen, T.; Ahmad, I.; Ali, A.; Lei, Z.; Abu bakr Shabbir, M.; Cheng, G.; Yuan, Z. Current Advances in Immunoassays for the Detection of Antibiotics Residues: A Review. Food Agric. Immunol. 2020, 31, 268–290. [Google Scholar] [CrossRef]
- Cox, K.L.; Devanarayan, V.; Kriauciunas, A.; Manetta, J.; Montrose, C.; Sittampalam, S. Immunoassay Methods. In Assay Guidance Manual; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2019. [Google Scholar]
- Gan, S.D.; Patel, K.R. Enzyme Immunoassay and Enzyme-Linked Immunosorbent Assay. J. Investig. Dermatol. 2013, 133, e12. [Google Scholar] [CrossRef]
- Lajtha, A.; Baker, G.; Dunn, S.; Holt, A. Handbook of Neurochemistry and Molecular Neurobiology: Practical Neurochemistry Methods; Springer Science: New York, NY, USA, 2007; ISBN 978-0-387-30359-8. [Google Scholar]
- Farka, Z.; Juřík, T.; Kovář, D.; Trnková, L.; Skládal, P. Nanoparticle-Based Immunochemical Biosensors and Assays: Recent Advances and Challenges. Chem. Rev. 2017, 117, 9973–10042. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Barroso, L.G.; Azaman, F.A.; Pogue, R.; Devine, D.; Fournet, M.B. Monitoring In Vitro Extracellular Matrix Protein Conformations in the Presence of Biomimetic Bone-Regeneration Scaffolds Using Functionalized Gold-Edge-Coated Triangular Silver Nanoparticles. Nanomaterials 2023, 13, 57. [Google Scholar] [CrossRef]
- Mercadal, P.A.; Fraire, J.C.; Motrich, R.D.; Coronado, E.A. Enzyme-Free Immunoassay Using Silver Nanoparticles for Detection of Gliadin at Ultralow Concentrations. ACS Omega 2018, 3, 2340–2350. [Google Scholar] [CrossRef]
- Mallah, S.I.; Ghorab, O.K.; Al-Salmi, S.; Abdellatif, O.S.; Tharmaratnam, T.; Iskandar, M.A.; Sefen, J.A.N.; Sidhu, P.; Atallah, B.; El-Lababidi, R.; et al. COVID-19: Breaking down a Global Health Crisis. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 1–36. [Google Scholar] [CrossRef]
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19); StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Valdivia, A.; Torres, I.; Latorre, V.; Francés-Gómez, C.; Albert, E.; Gozalbo-Rovira, R.; Alcaraz, M.J.; Buesa, J.; Rodríguez-Díaz, J.; Geller, R.; et al. Inference of SARS-CoV-2 Spike-Binding Neutralizing Antibody Titers in Sera from Hospitalized COVID-19 Patients by Using Commercial Enzyme and Chemiluminescent Immunoassays. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 485–494. [Google Scholar] [CrossRef]
- Yatsunyk, L.A.; Mendoza, O.; Mergny, J.-L. “Nano-Oddities”: Unusual Nucleic Acid Assemblies for DNA-Based Nanostructures and Nanodevices. Acc. Chem. Res. 2014, 47, 1836–1844. [Google Scholar] [CrossRef]
- Zhang, Y.; Charles, D.E.; Ledwith, D.M.; Aherne, D.; Cunningham, S.; Voisin, M.; Blau, W.J.; Gun’Ko, Y.K.; Kelly, J.M.; Brennan-Fournet, M.E. Wash-Free Highly Sensitive Detection of C-Reactive Protein Using Gold Derivatised Triangular Silver Nanoplates. RSC Adv. 2014, 4, 29022–29031. [Google Scholar] [CrossRef]
- Jia, H.; Xu, W.; An, J.; Li, D.; Zhao, B. A Simple Method to Synthesize Triangular Silver Nanoparticles by Light Irradiation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2006, 64, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Askari, H.; Sanadgol, N.; Azarnezhad, A.; Tajbakhsh, A.; Rafiei, H.; Safarpour, A.R.; Gheibihayat, S.M.; Raeis-Abdollahi, E.; Savardashtaki, A.; Ghanbariasad, A.; et al. Kidney Diseases and COVID-19 Infection: Causes and Effect, Supportive Therapeutics and Nutritional Perspectives. Heliyon 2021, 7, e06008. [Google Scholar] [CrossRef]
- Ren, X.; Meng, X.; Chen, D.; Tang, F.; Jiao, J. Using Silver Nanoparticle to Enhance Current Response of Biosensor. Biosens. Bioelectron. 2005, 21, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Rad, A.S.; Mirabi, A.; Binaian, E.; Tayebi, H. A Review on Glucose and Hydrogen Peroxide Biosensor Based on Modified Electrode Included Silver Nanoparticles. Int. J. Electrochem. Sci. 2011, 6, 3671–3683. [Google Scholar] [CrossRef]
- Chen, Z.P.; Peng, Z.F.; Luo, Y.; Qu, B.; Jiang, J.H.; Zhang, X.B.; Shen, G.L.; Yu, R.Q. Successively Amplified Electrochemical Immunoassay Based on Biocatalytic Deposition of Silver Nanoparticles and Silver Enhancement. Biosens. Bioelectron. 2007, 23, 485–491. [Google Scholar] [CrossRef]
- Khristunova, E.; Barek, J.; Kratochvil, B.; Korotkova, E.; Dorozhko, E.; Vyskocil, V. Electrochemical Immunoassay for the Detection of Antibodies to Tick-Borne Encephalitis Virus by Using Various Types of Bioconjugates Based on Silver Nanoparticles. Bioelectrochemistry 2020, 135, 107576. [Google Scholar] [CrossRef]
- Rodriguez Barroso, L.G.; Garcia, E.L.; Mojicevic, M.; Huerta, M.; Pogue, R.; Devine, D.M.; Brennan-fournet, M. Triangular Silver Nanoparticles Synthesis: Investigating Potential Application in Materials and Biosensing. Appl. Sci. 2023, 13, 8100. [Google Scholar] [CrossRef]
- Choi, H.K.; Lee, M.J.; Lee, S.N.; Kim, T.H.; Oh, B.K. Noble Metal Nanomaterial-Based Biosensors for Electrochemical and Optical Detection of Viruses Causing Respiratory Illnesses. Front. Chem. 2021, 9, 672739. [Google Scholar] [CrossRef]
- Macdonald, P.J.; Ruan, Q.; Grieshaber, J.L.; Swift, K.M.; Taylor, R.E.; Prostko, J.C.; Tetin, S.Y. Affinity of Anti-Spike Antibodies in SARS-CoV-2 Patient Plasma and Its Effect on COVID-19 Antibody Assays. eBioMedicine 2022, 75, 103796. [Google Scholar] [CrossRef]
- Brennan-Fournet, M.E.; Huerta, M.; Zhang, Y.; Malliaras, G.; Owens, R.M. Detection of Fibronectin Conformational Changes in the Extracellular Matrix of Live Cells Using Plasmonic Nanoplates. J. Mater. Chem. B 2015, 3, 9140–9147. [Google Scholar] [CrossRef] [PubMed]
- Castelletti, F.; Donadelli, R.; Banterla, F.; Hildebrandt, F.; Zipfel, P.F.; Bresin, E.; Otto, E.; Skerka, C.; Renieri, A.; Todeschini, M.; et al. Mutations in FN1 Cause Glomerulopathy with Fibronectin Deposits. Proc. Natl. Acad. Sci. USA 2008, 105, 2538–2543. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Hielscher, A. Fibronectin: How Its Aberrant Expression in Tumors May Improve Therapeutic Targeting. J. Cancer 2017, 8, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Dobrynin, D.; Polischuk, I.; Pokroy, B. A Comparison Study of the Detection Limit of Omicron SARS-CoV-2 Nucleocapsid by Various Rapid Antigen Tests. Biosensors 2022, 12, 1083. [Google Scholar] [CrossRef] [PubMed]
- Sediq, A.S.; Van Duijvenvoorde, R.B.; Jiskoot, W.; Nejadnik, M.R. No Touching! Abrasion of Adsorbed Protein Is the Root Cause of Subvisible Particle Formation during Stirring. J. Pharm. Sci. 2016, 105, 519–529. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, B.A.S.; Alev, C.; Mechael, R.; Salci, K.R.; Lee, J.B.; Fiebig-Comyn, A.; Guezguez, B.; Wu, Y.; Sheng, G.; Bhatia, M. Expansive Generation of Functional Airway Epithelium From Human Embryonic Stem Cells. Stem Cells Transl. Med. 2014, 3, 7–17. [Google Scholar] [CrossRef]
- Riond, B.; Wenger-Riggenbach, B.; Hofmann-Lehmann, R.; Lutz, H. Serum Protein Concentrations from Clinically Healthy Horses Determined by Agarose Gel Electrophoresis. Vet. Clin. Pathol. 2009, 38, 73–77. [Google Scholar] [CrossRef]
- Vasile, E.; Rusen, E.; Mocanu, A.; Patrascu, M.; Calinescu, I. Polymer Colloids and Silver Nanoparticles Hybrid Materials. Colloid Polym. Sci. 2012, 290, 193–201. [Google Scholar] [CrossRef]
- Schintke, S.; Frau, E. Modulated 3d Cross-Correlation Dynamic Light Scattering Applications for Optical Biosensing and Time-Dependent Monitoring of Nanoparticle-Biofluid Interactions. Appl. Sci. 2020, 10, 8969. [Google Scholar] [CrossRef]
- Hellewell, A.L.; Rosini, S.; Adams, J.C. A Rapid, Scalable Method for the Isolation, Functional Study, and Analysis of Cell-Derived Extracellular Matrix. J. Vis. Exp. 2017, 2017, 55051. [Google Scholar] [CrossRef]
- Stetefeld, J.; Mckenna, S.A.; Patel, T.R. Dynamic Light Scattering: A Practical Guide and Applications in Biomedical Sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
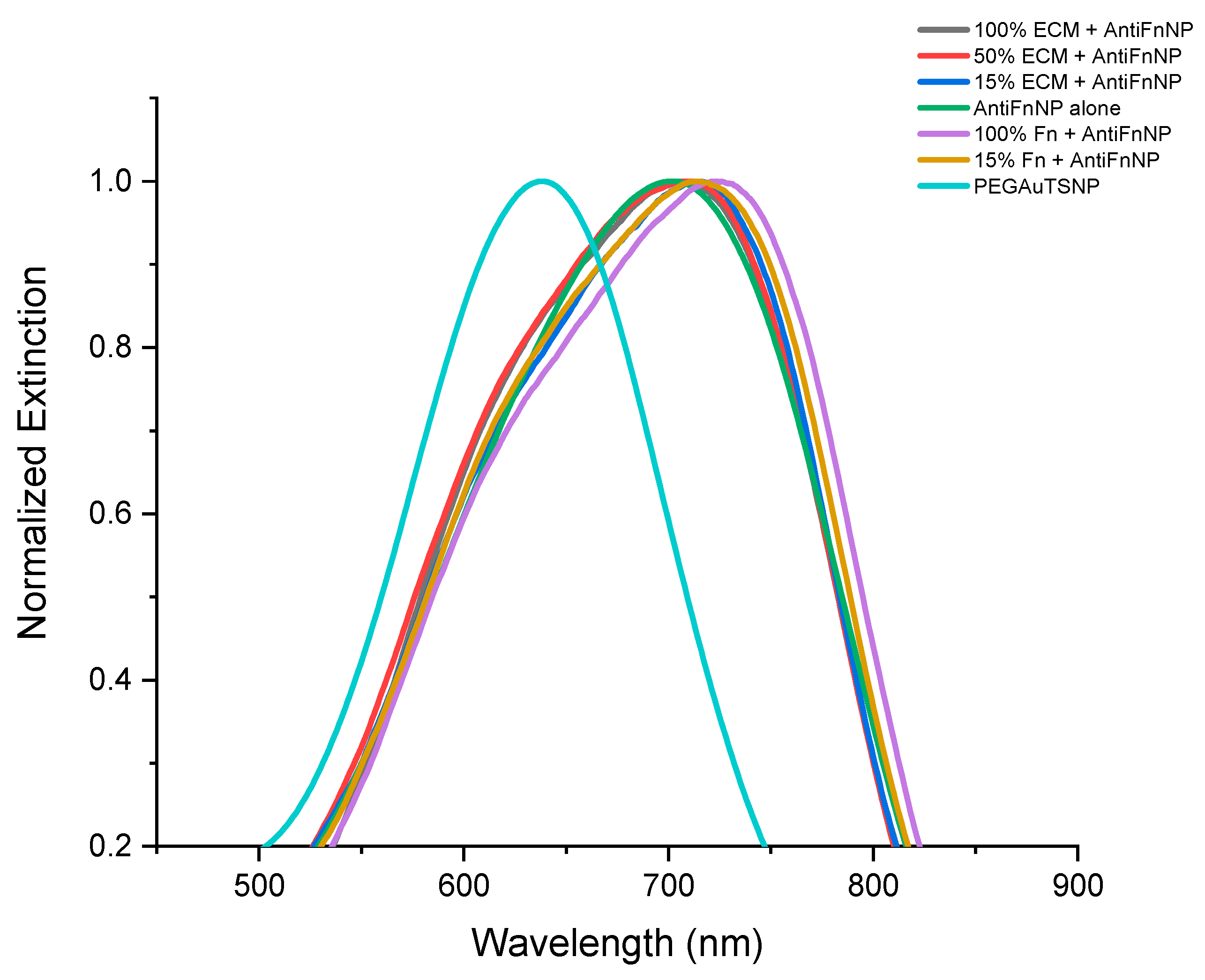

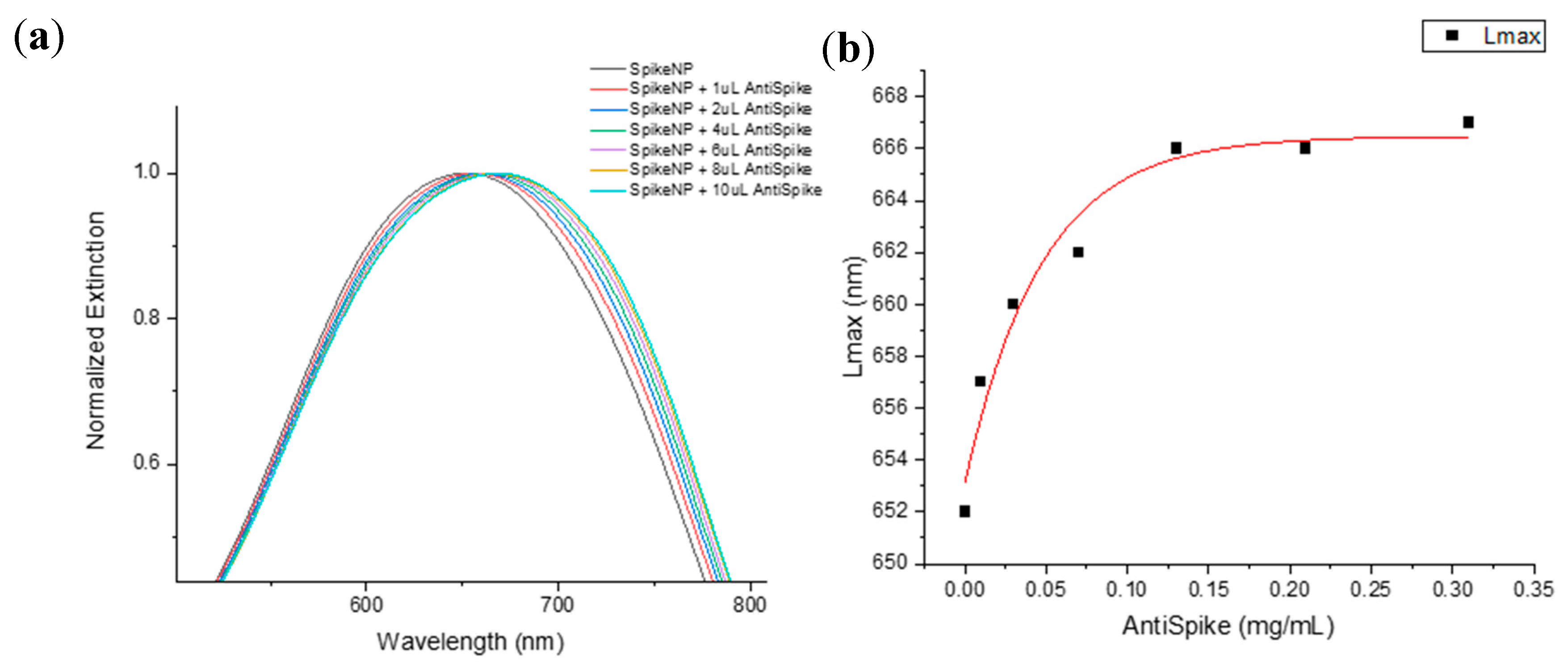

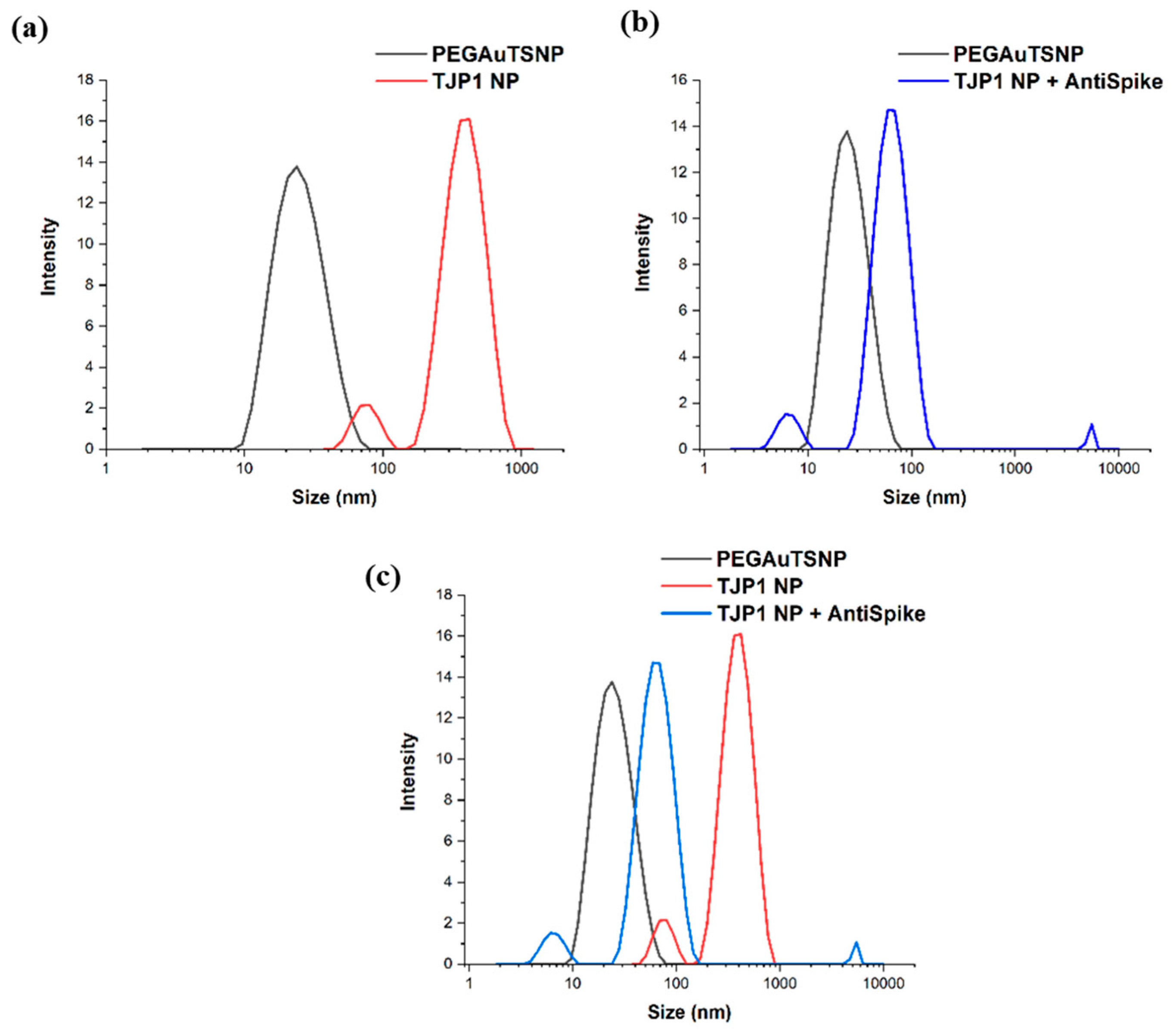
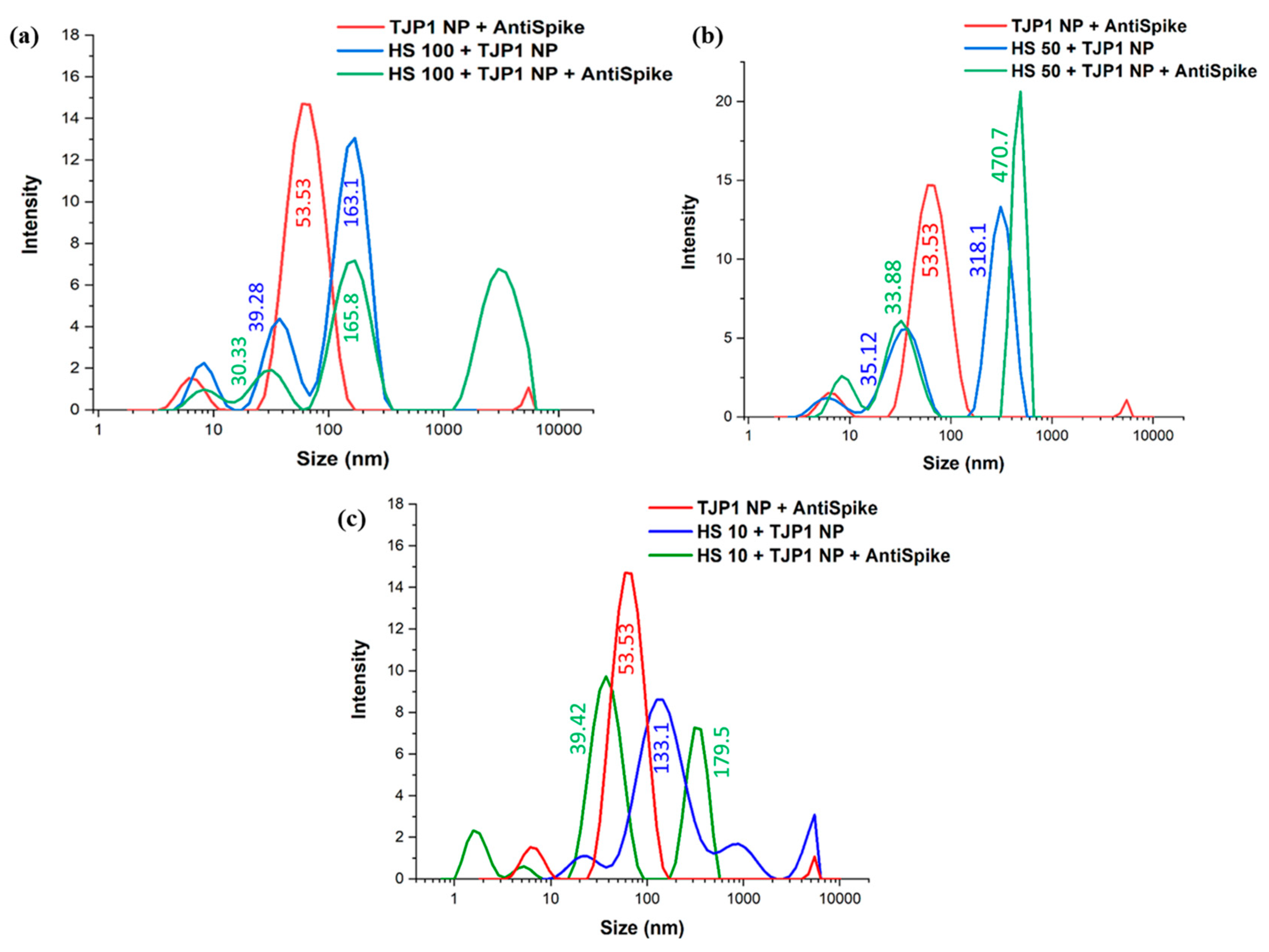
| Treatment | λmax |
|---|---|
| PEGAuTSNP | 638 nm |
| Anti-Fn-PEGAuTSNP | 703 nm |
| 100% ECM + Anti-Fn-PEGAuTSNP | 707 nm |
| 50% ECM + Anti-Fn-PEGAuTSNP | 711 nm |
| 15% ECM + Anti-Fn-PEGAuTSNP | 715 nm |
| 100% Fn + Anti-Fn-PEGAuTSNP | 723 nm |
| 15% Fn + Anti-Fn-PEGAuTSNP | 714 nm |
| Treatment | λmax |
|---|---|
| PEGAuTSNP | 638 nm |
| Anti-TJP1-PEGAuTSNP | 675 nm |
| 100% ECM + Anti-TJP1-PEGAuTSNP | 677 nm |
| 50% ECM + Anti-TJP1-PEGAuTSNP | 677 nm |
| 15% ECM + Anti-TJP1-PEGAuTSNP | 671 nm |
| 100% Fn + Anti-TJP1-PEGAuTSNP | 678 nm |
| Spike volume | 1 µg | 3 µg | 5 µg | 7 µg | 10 µg |
| λmax before anti-Spike (nm) | 603 | 647 | 641 | 632 | 632 |
| λmax after anti-Spike (nm) | 615 | 651 | 647 | 639 | 638 |
| Δλ (nm) | 12 | 4 | 6 | 7 | 6 |
| λmax Spike-PEGAuTSNP (nm) | λmax TJP1-PEGAuTSNP (nm) | |
|---|---|---|
| Before anti-Spike | 639 | 600 |
| After anti-Spike | 646 | 601 |
| Δλ | 7 | 1 |
| Spike NP | HS 100% + Spike | HS 50% + Spike | HS 10% + Spike | TJP1 NP | HS 100% + TJP1 NP | HS 50% + TJP1 NP | HS 10% + TJP1 NP | |
|---|---|---|---|---|---|---|---|---|
| λmax before anti-Spike (nm) | 590 | 609 | 612 | 607 | 596 | 612 | 617 | 604 |
| λmax after anti-Spike (nm) | 603 | 617 | 619 | 609 | 597 | 620 | 620 | 606 |
| Δλ (nm) | 13 | 8 | 7 | 2 | 1 | 8 | 3 | 2 |
| PEG AuTSNP | Spike NP | TJP1 NP | HS 100% +TJP1 NP | HS 50% +TJP1 NP | HS 10% +TJP1 NP | |
|---|---|---|---|---|---|---|
| λmax before anti-Spike (nm) | 589 | 604 | 597 | 628 | 627 | 618 |
| λmax after anti-Spike (nm) | 614 | 619 | 614 | 633 | 632 | 620 |
| Δλ | 25 | 15 | 17 | 5 | 5 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez Barroso, L.G.; Lanzagorta Garcia, E.; Mojicevic, M.; Alkan Tas, B.; Huerta, M.; Pogue, R.; Devine, D.M.; Brennan-Fournet, M. Triangular Silver Nanoplates as a Bioanalytical Tool: Potential COVID-19 Detection. Int. J. Mol. Sci. 2023, 24, 11974. https://doi.org/10.3390/ijms241511974
Rodriguez Barroso LG, Lanzagorta Garcia E, Mojicevic M, Alkan Tas B, Huerta M, Pogue R, Devine DM, Brennan-Fournet M. Triangular Silver Nanoplates as a Bioanalytical Tool: Potential COVID-19 Detection. International Journal of Molecular Sciences. 2023; 24(15):11974. https://doi.org/10.3390/ijms241511974
Chicago/Turabian StyleRodriguez Barroso, Laura G., Eduardo Lanzagorta Garcia, Marija Mojicevic, Buket Alkan Tas, Miriam Huerta, Robert Pogue, Declan M. Devine, and Margaret Brennan-Fournet. 2023. "Triangular Silver Nanoplates as a Bioanalytical Tool: Potential COVID-19 Detection" International Journal of Molecular Sciences 24, no. 15: 11974. https://doi.org/10.3390/ijms241511974
APA StyleRodriguez Barroso, L. G., Lanzagorta Garcia, E., Mojicevic, M., Alkan Tas, B., Huerta, M., Pogue, R., Devine, D. M., & Brennan-Fournet, M. (2023). Triangular Silver Nanoplates as a Bioanalytical Tool: Potential COVID-19 Detection. International Journal of Molecular Sciences, 24(15), 11974. https://doi.org/10.3390/ijms241511974








