Biochemical and Functional Characterization of the Three Zebrafish Transglutaminases 2
Abstract
1. Introduction
2. Results
2.1. Evolutionary History of Zebrafish Transglutaminase 2
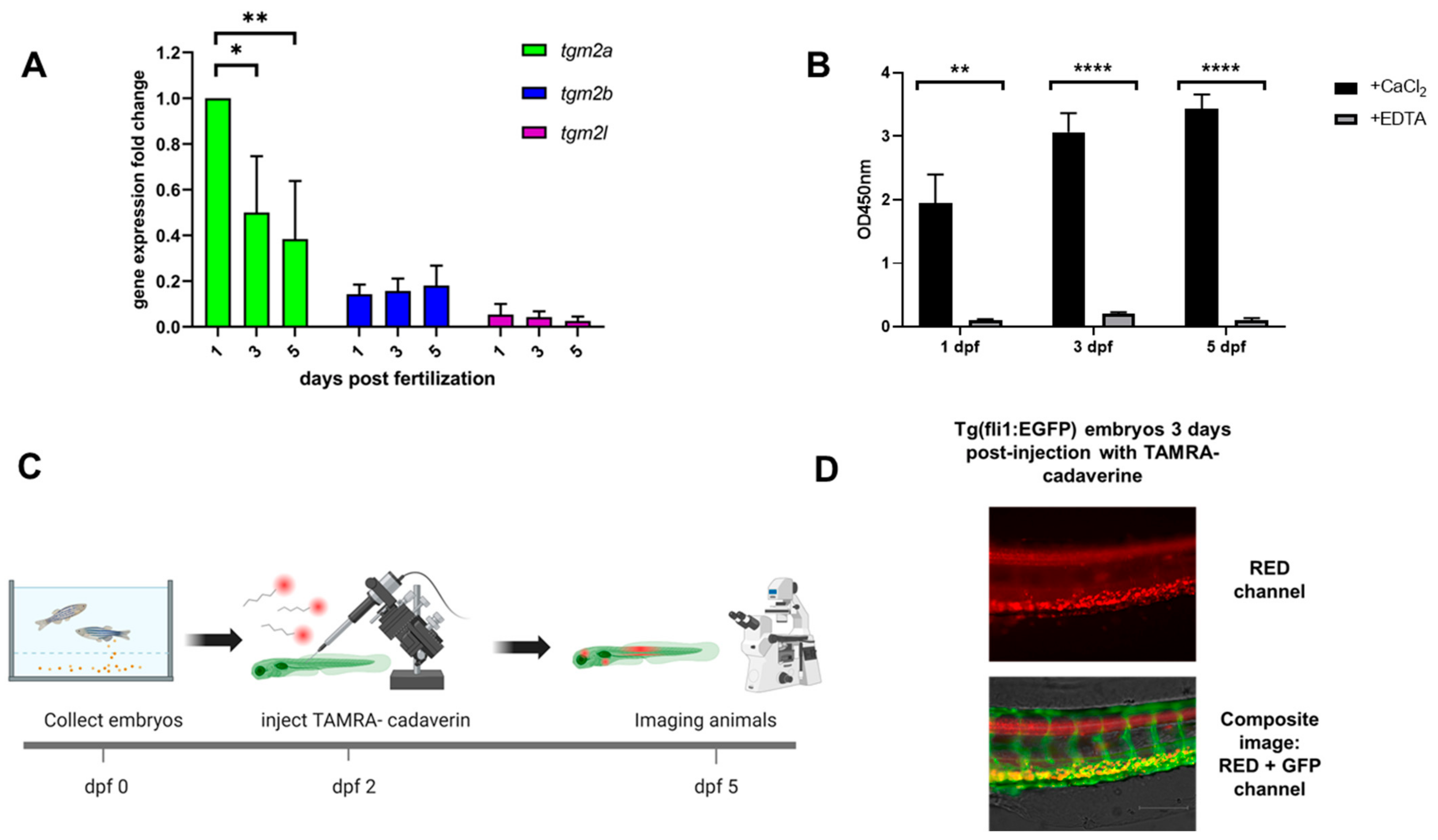
2.2. Gene Expression Analysis of Zebrafish Transglutaminase 2 Genes
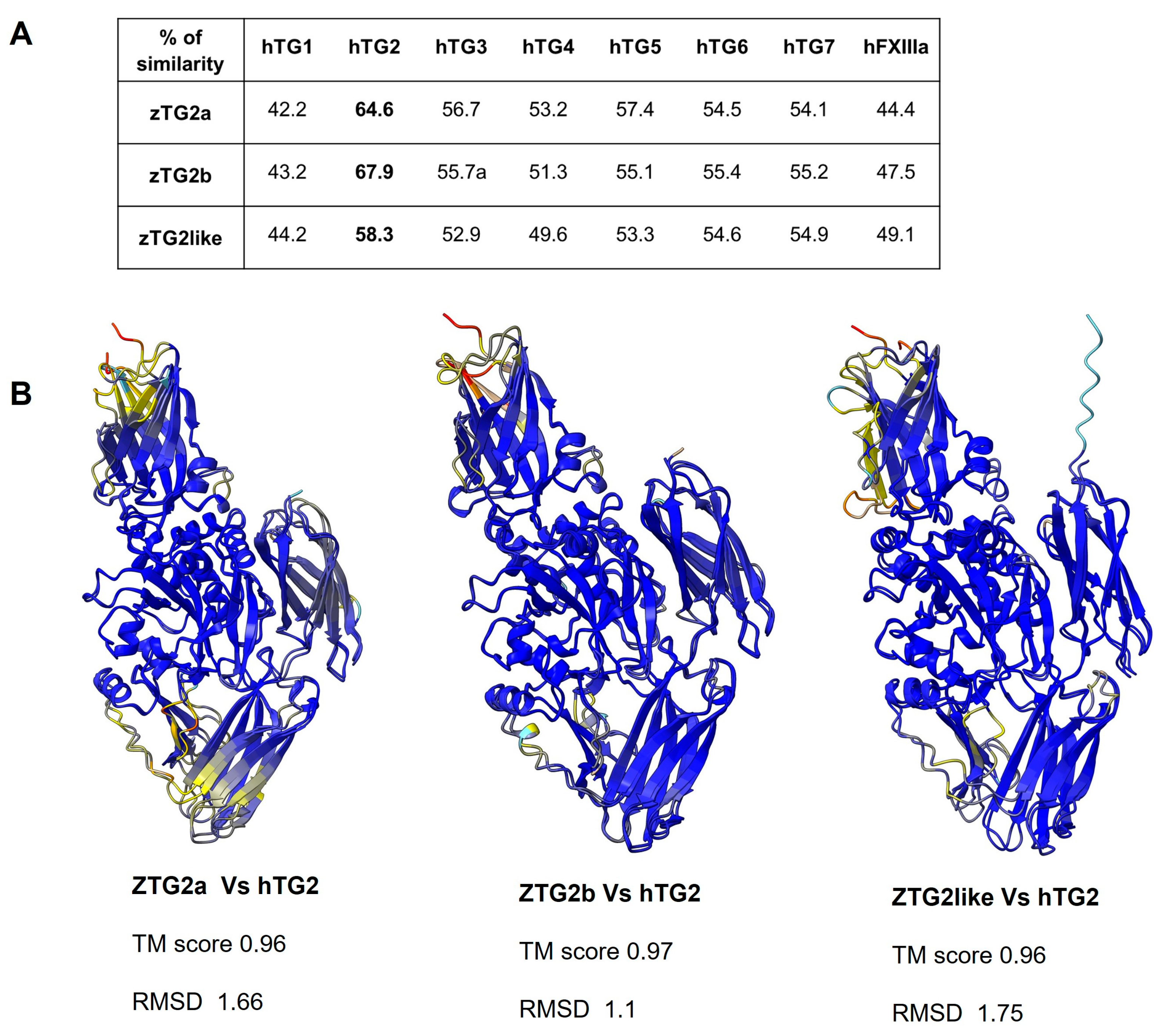
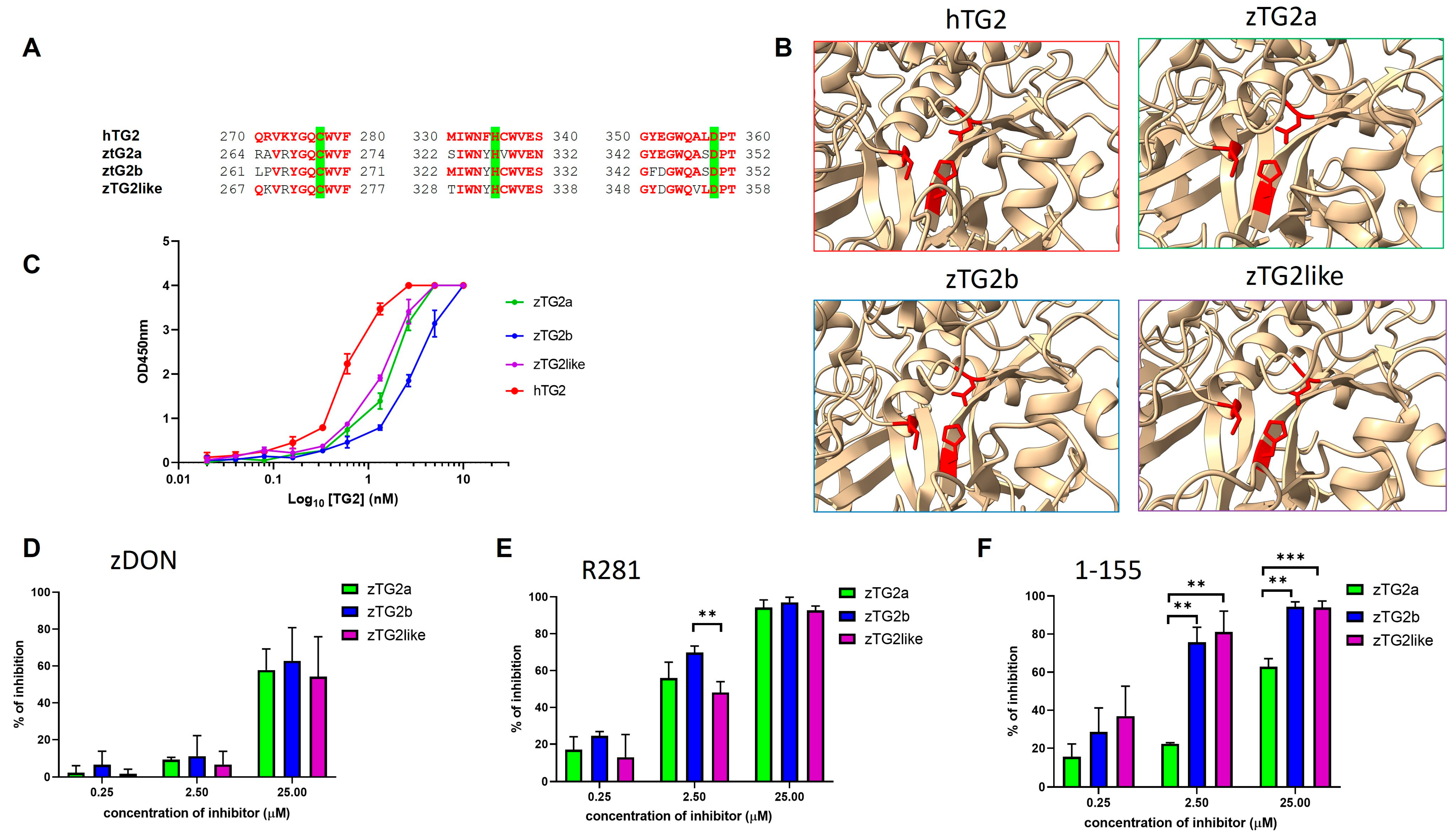
2.3. Zebrafish TGs2 Proteins Sequence and Structure Analysis
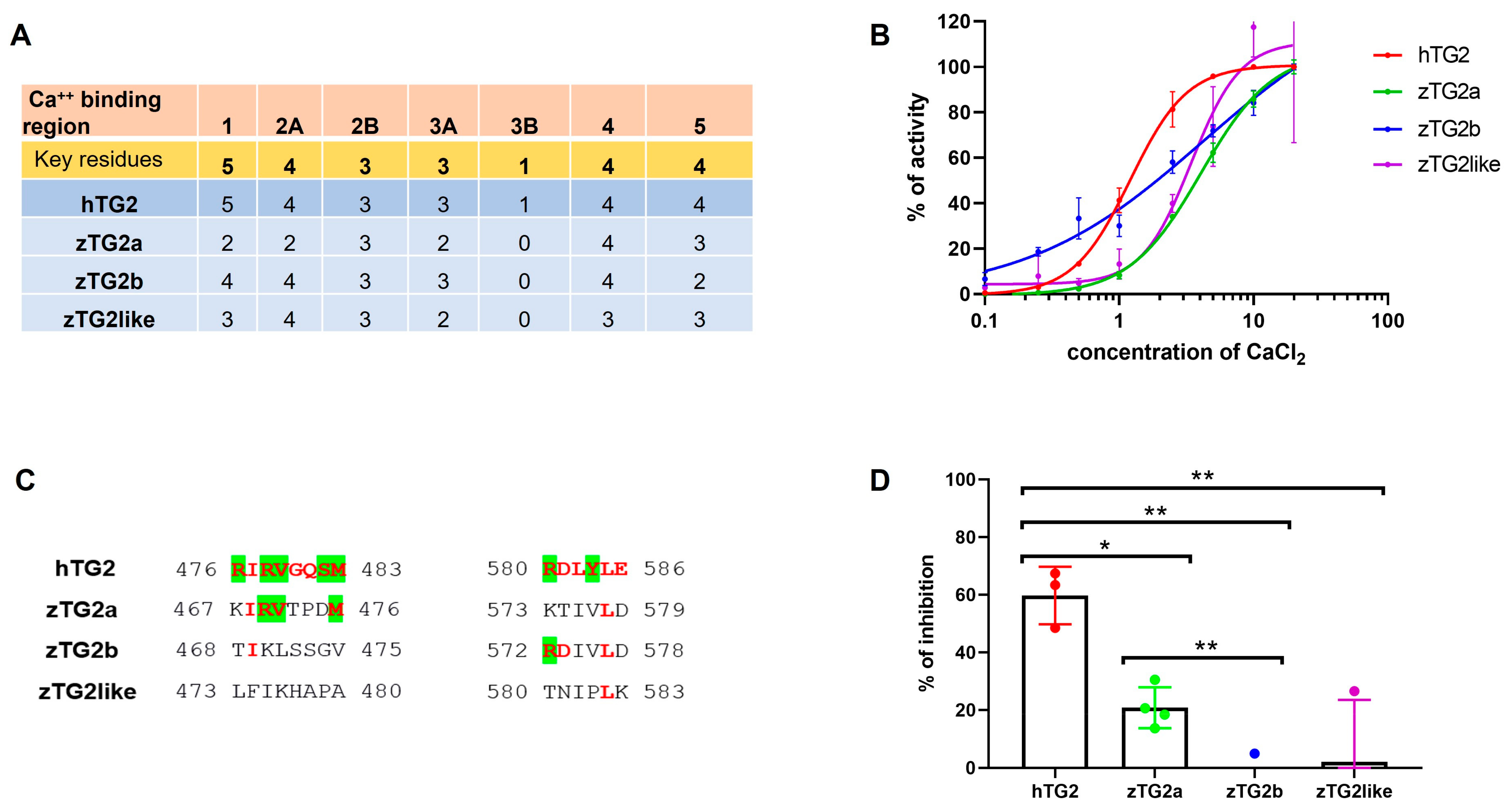
2.4. Recombinant zTGs2 Proteins Have a Ca2+-Dependent Activity
2.5. The zTGs2 Proteins Have a Ca2+-Dependent Activity but Are Not Inhibited by GTP
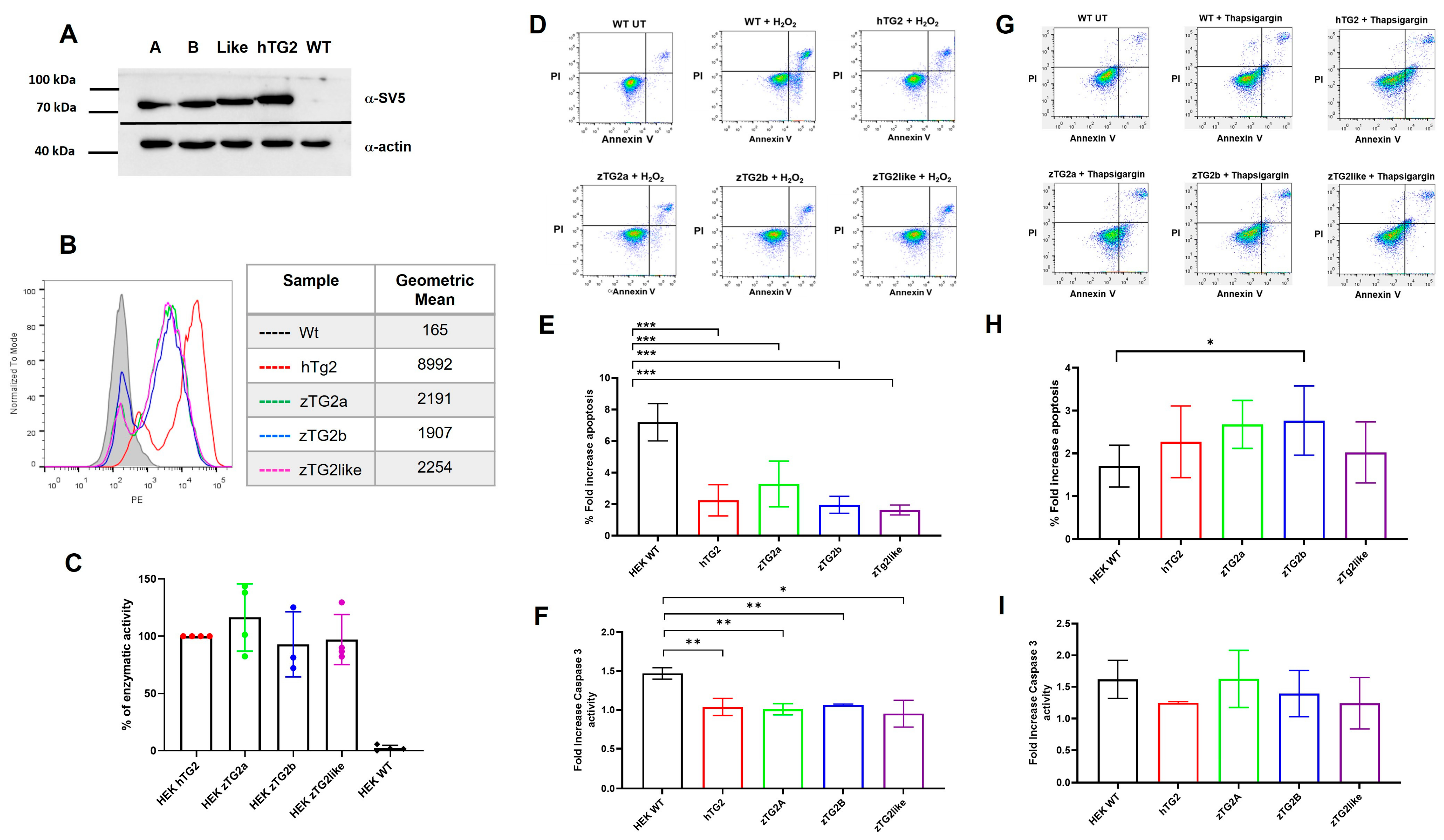
2.6. Intracellular zTGs2 Proteins in HEK293 Cells Mediate Different Effects against Cell Death in a Stimulus-Dependent Manner
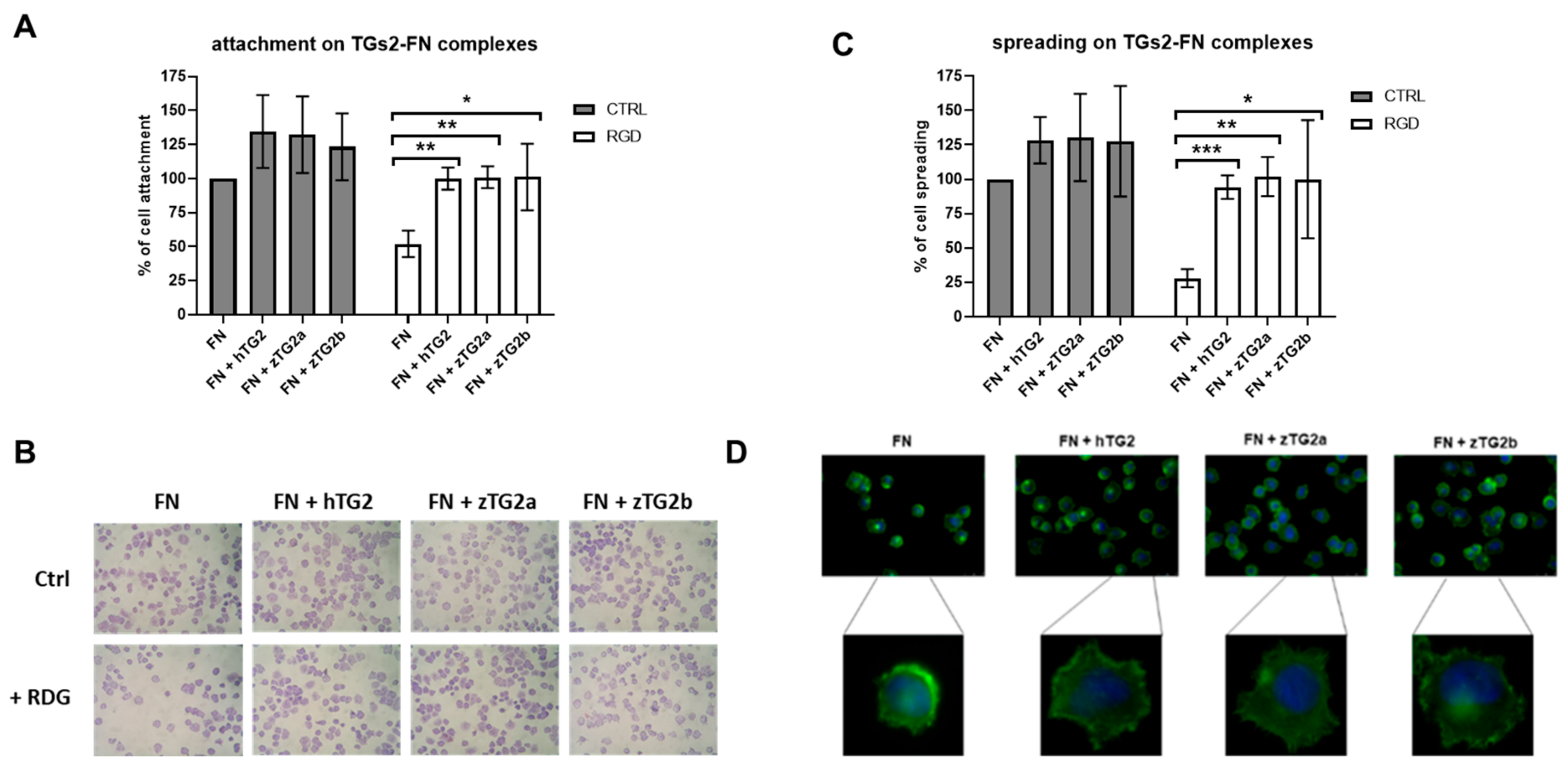
2.7. The zTGs2 Proteins Mediate Can Mediate RGD-Independent Cell Adhesion and Spreading
3. Discussion
4. Materials and Methods
4.1. Genetic Analysis of TG2 Orthologues in the Zebrafish Genome
4.2. Zebrafish Embryos Handling and Maintenance
4.3. Gene Expression Study of the Three Zebrafish TG2 Genes during the Embryonic Development
4.4. Zebrafish Embryos Lysates Transamidation Activity Assay
4.5. Fluorescent Cadaverine Incorporation in Live Zebrafish Embryos
4.6. Molecular Cloning of Zebrafish Transglutaminase 2 Sequence into Bacterial Expression Vector
4.7. Recombinant Protein Production and Purification
4.8. zTGs2 Transamidation Activity Curve and Inhibition Assays
4.9. RGD-Independent Cell Adhesion Assay
4.10. Actin Cytoskeletal Staining
4.11. Molecular Cloning of Zebrafish Transglutaminase 2 Sequence into Eukaryotic Expression Vector
4.12. Transient Cell Transfection and Selection of Stable TG2-Expressing HEK293 Cells
4.13. HEK293 Anti-SV5 Tag Western Blot
4.14. HEK293 Cell Lysates Transamidation Activity Assay
4.15. HEK293 Cells Flow Cytometry Analysis
4.16. Apoptosis Assays with Hydrogen Peroxide and Thapsigargin
4.17. Caspase3 Activity Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Beninati, S.; Piacentini, M. The Transglutaminase Family: An Overview. Minireview Article. Amino Acids 2004, 26, 367–372. [Google Scholar]
- Bergamini, C.M.; Collighan, R.J.; Wang, Z.; Griffin, M. Structure and Regulation of Type 2 Transglutaminase in Relation to Its Physiological Functions and Pathological Roles. In Advances in Enzymology: And Related Areas of Molecular Biology; John Wiley & Sons: Hoboken, NJ, USA, 2011; Volume 78, pp. 1–46. [Google Scholar] [CrossRef]
- Pinkas, D.M.; Strop, P.; Brunger, A.T.; Khosla, C. Transglutaminase 2 Undergoes a Large Conformational Change upon Activation. PLoS Biol. 2007, 5, 327. [Google Scholar] [CrossRef]
- Bergamini, C.M. GTP Modulates Calcium Binding and Cation-Induced Conformational Changes in Erythrocyte Transglutaminase. FEBS Lett. 1988, 239, 255–258. [Google Scholar] [CrossRef]
- Jeong, E.M.; Lee, K.B.; Kim, G.E.; Kim, C.M.; Lee, J.-H.; Kim, H.-J.; Shin, J.-W.; Kwon, M.-A.; Ho Park, H.; Kim, I.-G. Competitive Binding of Magnesium to Calcium Binding Sites Reciprocally Regulates Transamidase and GTP Hydrolysis Activity of Transglutaminase 2. Int. J. Mol. Sci. 2020, 21, 791. [Google Scholar] [CrossRef]
- Baek, K.J.; Kang, S.K.; Damron, D.S.; Im, M.J. Phospholipase Cδ1 Is a Guanine Nucleotide Exchanging Factor for Transglutaminase II (Gαh) and Promotes α 1B-Adrenoreceptor-Mediated GTP Binding and Intracellular Calcium Release. J. Biol. Chem. 2001, 276, 5591–5597. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, S.E.; Begg, G.E.; Graham, R.M. Graham Cross-Linking Transglutaminases with G Protein-Coupled Receptor Signaling. Sci. STKE 2006, 2006, pe34. [Google Scholar] [CrossRef][Green Version]
- Eckert, R.L.; Kaartinen, M.T.; Nurminskaya, M.; Belkin, A.M.; Colak, G.; Johnson, G.V.W.; Mehta, K. Transglutaminase Regulation of Cell Function. Physiol. Rev. 2014, 94, 383–417. [Google Scholar] [CrossRef]
- Odii, B.O.; Coussons, P. Biological Functionalities of Transglutaminase 2 and the Possibility of Its Compensation by Other Members of the Transglutaminase Family. Sci. World J. 2014, 2014, 714561. [Google Scholar] [CrossRef]
- Hasegawa, G.; Suwa, M.; Ichikawa, Y.; Ohtsuka, T.; Kumagai, S.; Kikuchi, M.; Sato, Y.; Saito, Y. A Novel Function of Tissue-Type Transglutaminase: Protein Disulphide Isomerase. Biochem. J. 2003, 373, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.-F.; Tatsukawa, H.; Kojima, S. New Insights into the Functions and Localization of Nuclear Transglutaminase 2. FEBS J. 2011, 278, 4756–4767. [Google Scholar] [CrossRef] [PubMed]
- Tatsukawa, H.; Furutani, Y.; Hitomi, K.; Kojima, S. Transglutaminase 2 Has Opposing Roles in the Regulation of Cellular Functions as Well as Cell Growth and Death. Cell Death. Dis. 2016, 7, e2244. [Google Scholar] [CrossRef]
- Belkin, A.M. Extracellular TG2: Emerging Functions and Regulation. FEBS J. 2011, 278, 4704–4716. [Google Scholar] [CrossRef]
- Wang, Z.; Griffin, M. TG2, a Novel Extracellular Protein with Multiple Functions. Amino Acids 2012, 42, 939–949. [Google Scholar]
- Wang, Z.; Collighan, R.J.; Gross, S.R.; Danen, E.H.J.; Orend, G.; Telci, D.; Griffin, M. RGD-Independent Cell Adhesion via a Tissue Transglutaminase-Fibronectin Matrix Promotes Fibronectin Fibril Deposition and Requires Syndecan-4/2 and A5β1 Integrin Co-Signaling. J. Biol. Chem. 2010, 285, 40212–40229. [Google Scholar] [CrossRef]
- Telci, D.; Wang, Z.; Li, X.; Verderio, E.A.M.; Humphries, M.J.; Baccarini, M.; Basaga, H.; Griffin, M. Fibronectin-Tissue Transglutaminase Matrix Rescues RGD-Impaired Cell Adhesion through Syndecan-4 and Β1 Integrin Co-Signaling. J. Biol. Chem. 2008, 283, 20937–20947. [Google Scholar] [CrossRef]
- Lai, T.-S.; Lin, C.-J.; Wu, Y.-T.; Wu, C.-J. Tissue Transglutaminase (TG2) and Mitochondrial Function and Dysfunction. Front. Biosci. 2017, 22, 1114–1137. [Google Scholar]
- D’Eletto, M.; Rossin, F.; Occhigrossi, L.; Farrace, M.G.; Faccenda, D.; Desai, R.; Marchi, S.; Refolo, G.; Falasca, L.; Antonioli, M.; et al. Transglutaminase Type 2 Regulates ER-Mitochondria Contact Sites by Interacting with GRP75. Cell Rep. 2018, 25, 3573–3581.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Telci, D.; Griffin, M. Importance of Syndecan-4 and Syndecan -2 in Osteoblast Cell Adhesion and Survival Mediated by a Tissue Transglutaminase-Fibronectin Complex. Exp. Cell Res. 2011, 317, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Perez, M.; Caja, S.; Melino, G.; Johnson, T.S.; Lindfors, K.; Griffin, M. A Novel Extracellular Role for Tissue Transglutaminase in Matrix-Bound VEGF-Mediated Angiogenesis. Cell Death Dis. 2013, 4, e808. [Google Scholar] [CrossRef]
- Zonca, S.; Pinton, G.; Wang, Z.; Soluri, M.F.; Tavian, D.; Griffin, M.; Sblattero, D.; Moro, L. Tissue Transglutaminase (TG2) Enables Survival of Human Malignant Pleural Mesothelioma Cells in Hypoxia. Cell Death Dis. 2017, 8, e2592. [Google Scholar] [CrossRef]
- Meyers, J.R. Zebrafish: Development of a Vertebrate Model Organism. Curr. Protoc. Essent. Lab. Tech. 2018, 16, e19. [Google Scholar] [CrossRef]
- Zang, L.; Torraca, V.; Shimada, Y.; Nishimura, N. Editorial: Zebrafish Models for Human Disease Studies. Front. Cell Dev. Biol. 2022, 10, 861941. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of Embryonic Development of the Zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Deasey, S.; Grichenko, O.; Du, S.; Nurminskaya, M. Characterization of the Transglutaminase Gene Family in Zebrafish and in Vivo Analysis of Transglutaminase-Dependent Bone Mineralization. Amino Acids 2012, 42, 1065–1075. [Google Scholar] [CrossRef]
- Rossin, F.; Costa, R.; Bordi, M.; D’Eletto, M.; Occhigrossi, L.; Farrace, M.G.; Barlev, N.; Ciccosanti, F.; Muccioli, S.; Chieregato, L.; et al. Transglutaminase Type 2 Regulates the Wnt/β-Catenin Pathway in Vertebrates. Cell Death Dis. 2021, 12, 249. [Google Scholar] [CrossRef]
- Watanabe, Y.; Okuya, K.; Takada, Y.; Kinoshita, M.; Yokoi, S.; Chisada, S.; Kamei, Y.; Tatsukawa, H.; Yamamoto, N.; Abe, H.; et al. Gene Disruption of Medaka (Oryzias latipes) Orthologue for Mammalian Tissue-Type Transglutaminase (TG2) Causes Movement Retardation. J. Biochem. 2020, 168, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Watanabe, Y.; Okuya, K.; Tatsukawa, H.; Hashimoto, H.; Hitomi, K. Biochemical Characterization of the Medaka (Oryzias latipes) Orthologue for Mammalian Tissue-Type Transglutaminase (TG2). Biosci. Biotechnol. Biochem. 2017, 81, 469–474. [Google Scholar] [CrossRef][Green Version]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic. Acids. Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J.; et al. The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-Teleost Comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Griffin, M.; Mongeot, A.; Collighan, R.; Saint, R.E.; Jones, R.A.; Coutts, I.G.C.; Rathbone, D.L. Synthesis of Potent Water-Soluble Tissue Transglutaminase Inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 5559–5562. [Google Scholar] [CrossRef]
- Badarau, E.; Wang, Z.; Rathbone, D.L.; Costanzi, A.; Thibault, T.; Murdoch, C.E.; El Alaoui, S.; Bartkeviciute, M.; Griffin, M. Development of Potent and Selective Tissue Transglutaminase Inhibitors: Their Effect on TG2 Function and Application in Pathological Conditions. Chem. Biol. 2015, 22, 1347–1361. [Google Scholar] [CrossRef]
- Király, R.; Cssz, É.; Kurtán, T.; Antus, S.; Szigeti, K.; Simon-Vecsei, Z.; Korponay-Szabó, I.R.; Keresztessy, Z.; Fésüs, L. Functional Significance of Five Noncanonical Ca2+-Binding Sites of Human Transglutaminase 2 Characterized by Site-Directed Mutagenesis. FEBS J. 2009, 276, 7083–7096. [Google Scholar] [CrossRef]
- Jones, R.A.; Kotsakis, P.; Johnson, T.S.; Chau, D.Y.S.; Ali, S.; Melino, G.; Griffin, M. Matrix Changes Induced by Transglutaminase 2 Lead to Inhibition of Angiogenesis and Tumor Growth. Cell Death Differ. 2006, 13, 1442–1453. [Google Scholar] [CrossRef]
- Wang, Z.; Perez, M.; Lee, E.S.; Kojima, S.; Griffin, M. The Functional Relationship between Transglutaminase 2 and Transforming Growth Factor Β1 in the Regulation of Angiogenesis and Endothelial–Mesenchymal Transition. Cell Death Dis. 2017, 8, e3032. [Google Scholar] [CrossRef]
- Kalliokoski, S.; Sulic, A.M.; Korponay-Szabó, I.R.; Szondy, Z.; Frias, R.; Perez, M.A.; Martucciello, S.; Roivainen, A.; Pelliniemi, L.J.; Esposito, C.; et al. Celiac Disease-Specific TG2-Targeted Autoantibodies Inhibit Angiogenesis Ex Vivo and In Vivo in Mice by Interfering with Endothelial Cell Dynamics. PLoS ONE 2013, 8, e65887. [Google Scholar] [CrossRef]
- Tempest, R.; Guarnerio, S.; Maani, R.; Cooper, J.; Peake, N. The Biological and Biomechanical Role of Transglutaminase-2 in the Tumour Microenvironment. Cancers 2021, 13, 2788. [Google Scholar] [CrossRef]
- Ayinde, O.; Wang, Z.; Griffin, M. Tissue Transglutaminase Induces Epithelial-Mesenchymal-Transition and the Acquisition of Stem Cell like Characteristics in Colorectal Cancer Cells. Oncotarget 2017, 8, 20025–20041. [Google Scholar] [CrossRef] [PubMed]
- Ivashkin, E.; Melnikova, V.; Kurtova, A.; Brun, N.R.; Obukhova, A.; Khabarova, M.Y.; Yakusheff, A.; Adameyko, I.; Gribble, K.E.; Voronezhskaya, E.E.; et al. Transglutaminase Activity Determines Nuclear Localization of Serotonin Immunoreactivity in the Early Embryos of Invertebrates and Vertebrates. ACS Chem. Neurosci. 2019, 10, 3888–3899. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein Domains Identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Han, B.G.; Cho, J.W.; Cho, Y.D.; Jeong, K.C.; Kim, S.Y.; Lee, B. Il Crystal Structure of Human Transglutaminase 2 in Complex with Adenosine Triphosphate. Int. J. Biol. Macromol. 2010, 47, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Milakovic, T.; Tucholski, J.; McCoy, E.; Johnson, G.V.W. Intracellular Localization and Activity State of Tissue Transglutaminase Differentially Impacts Cell Death. J. Biol. Chem. 2004, 279, 8715–8722. [Google Scholar] [CrossRef]
- Fesus, L.; Thomazy, V.; Falus, A. Induction and Activation of Tissue Transglutaminase during Programmed Cell Death. FEBS Lett. 1987, 224, 104–108. [Google Scholar] [CrossRef]
- Tatsukawa, H.; Hitomi, K.; Edwards, V.; Kaartinen, M.T.; Van Dam, A.-M. Role of Transglutaminase 2 in Cell Death, Survival, and Fibrosis. Cells 2021, 10, 1842. [Google Scholar] [CrossRef]
- Fok, J.Y.; Kapil Mehta, A.E. Tissue Transglutaminase Induces the Release of Apoptosis Inducing Factor and Results in Apoptotic Death of Pancreatic Cancer Cells. Apoptosis 2007, 12, 1455–1463. [Google Scholar] [CrossRef]
- Cho, S.-Y.; Lee, J.-H.; Bae, H.-D.; Jeong, E.M.; Jang, G.-Y.; Kim, C.-W.; Shin, D.-M.; Jeon, J.-H.; Kim, I.-G. Transglutaminase 2 Inhibits Apoptosis Induced by Calcium-Overload through down-Regulation of Bax. Exp. Mol. Med. 2010, 42, 639–650. [Google Scholar] [CrossRef]
- Ku, B.M.; Kim, D.S.; Kim, K.H.; Yoo, B.C.; Kim, S.H.; Gong, Y.D.; Kim, S.Y. Transglutaminase 2 Inhibition Found to Induce P53 Mediated Apoptosis in Renal Cell Carcinoma. FASEB J. 2013, 27, 3487–3495. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast Approximation for Phylogenetic Bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An Improved General Amino Acid Replacement Matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic. Acids. Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Xie, S.; Yao, J. Identification of Novel Reference Genes Suitable for QRT-PCR Normalization with Respect to the Zebrafish Developmental Stage. PLoS ONE 2016, 11, e0149277. [Google Scholar] [CrossRef]
- Tang, R.; Dodd, A.; Lai, D.; McNabb, W.C.; Love, D.R. Validation of Zebrafish (Danio Rerio) Reference Genes for Quantitative Real-Time RT-PCR Normalization. Acta Biochim. Biophys. Sin. 2007, 39, 384–390. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Sequence 5′-3′ |
|---|---|
| zTG2a Forward | CCAAAGCAGTGGGTCGAGAT |
| zTG2a Reverse | GTGCGTAAAACATCACCCGG |
| zTG2b Forward | GATGGGTCGTTCCGATCTCC |
| zTG2b Reverse | GCTGATCTTCTGGCCCACTT |
| zTG2like Forward | CACCTGCAGCCCTGAAAAAC |
| zTG2like Reverse | TCTGGTTGGGATGCCAAGAC |
| Beta actin Forward | CGAGCTGTCTTCCCATCCA |
| Beta actin Reverse | TCACCAACGTAGCTGTCTTTCTG |
| Lsm12b Forward | AGTTGTCCCAAGCCTATGCAATCAG |
| Lsm12b Reverse | CCACTCAGGAGGATAAAGACGAGTC |
| Hsp70 Forward | TCAAGCGCAACACAACCATC |
| Hsp70 Reverse | ATTTGCCCAGCAGGTTGTTG |
| Primer Name | Sequnce 5′-3′ |
|---|---|
| zTG2a sense KpnI | AGATCTGGGTACCATGGAGAGAGTGGTGGAG |
| zTG2a anti HindIII | TCCTCGAGAAGCTTTTATTCATAGATAACCAG |
| zTG2b sense KpnI | AGATCTGGGTACCATGGCTCTGGACATCGGC |
| zTG2b anti HindIII | TCCTCGAGAAGCTTTCATTTCCCGATGATGAC |
| zTG2like sense KpnI | AGATCTGGGTACCATGGCCAGCTATAATGCC |
| zTG2like anti HindIII | TCCTCGAGAAGCTTCTAAATTTCTGGGACAGC |
| Primer Name | Sequence 5′-3′ |
|---|---|
| hTG2-HYGRO-Sense_NheI | GCTGGCTAGCTGCCACCATGGCCGAGGAGCTGGTC |
| hTG2-HYGRO-anti_SV5 | GATTGGTTTGCCACTAGTGGCGGGGCCAATGATGAC |
| zTG2a-HYGRO-Sense_Xba | AGCTGTCTAGATGCCACCATGGAGAGAGTGGTGG |
| zTG2a-HYGRO-anti_SV5 | GATTGGTTTGCCACTAGTTTCATAGATAACCAGATTC |
| zTG2b-HYGRO-Sense_NheI | GCTGGCTAGCTGCCACCATGGCTCTGGACATCGGC |
| zTG2b-HYGRO-anti_SV5 | GATTGGTTTGCCACTAGTTTTCCCGATGATGACGTTC |
| zTG2like-HYGRO-Sense_Xba | GCTGTCTAGATGCCACCATGGCCAGCTATAATGCC |
| zTG2like-HYGRO-anti_SV5 | GATTGGTTTGCCACTAGTAATTTCTGGGACAGCATTTAC |
| SV5_HindIII_anti | AGCTAAGCTTTTAAGTACTATCCAGGCCCAGCAG TGGGTTTGGGATTGGTTTGCCACTAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisetto, M.; Fattorini, M.; Lanza, A.; Gerdol, M.; Griffin, M.; Wang, Z.; Ferrara, F.; Sblattero, D. Biochemical and Functional Characterization of the Three Zebrafish Transglutaminases 2. Int. J. Mol. Sci. 2023, 24, 12041. https://doi.org/10.3390/ijms241512041
Lisetto M, Fattorini M, Lanza A, Gerdol M, Griffin M, Wang Z, Ferrara F, Sblattero D. Biochemical and Functional Characterization of the Three Zebrafish Transglutaminases 2. International Journal of Molecular Sciences. 2023; 24(15):12041. https://doi.org/10.3390/ijms241512041
Chicago/Turabian StyleLisetto, Manuel, Mariagiulia Fattorini, Andrea Lanza, Marco Gerdol, Martin Griffin, Zhuo Wang, Fortunato Ferrara, and Daniele Sblattero. 2023. "Biochemical and Functional Characterization of the Three Zebrafish Transglutaminases 2" International Journal of Molecular Sciences 24, no. 15: 12041. https://doi.org/10.3390/ijms241512041
APA StyleLisetto, M., Fattorini, M., Lanza, A., Gerdol, M., Griffin, M., Wang, Z., Ferrara, F., & Sblattero, D. (2023). Biochemical and Functional Characterization of the Three Zebrafish Transglutaminases 2. International Journal of Molecular Sciences, 24(15), 12041. https://doi.org/10.3390/ijms241512041






