Endothelial Cell Response in Kawasaki Disease and Multisystem Inflammatory Syndrome in Children
Abstract
:1. Introduction
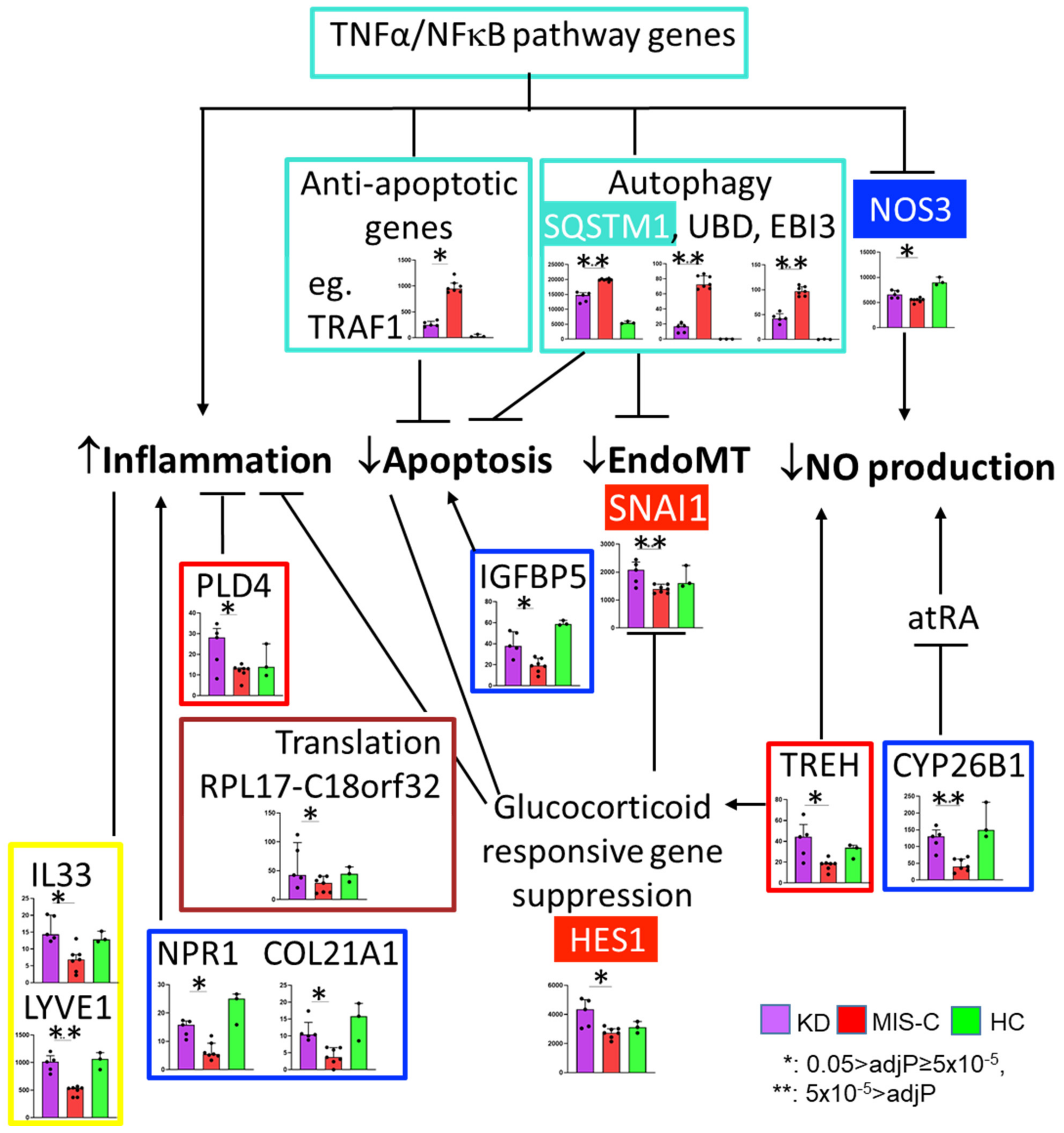
2. Results
2.1. RNA-seq and Differential Expression Analysis on Cultured ECs Incubated with Sera from the Patients with KD and MIS-C
2.2. Weighted Gene Co-Expression Analysis (WGCNA)
2.3. Differential Expression Analysis with Rigorous Filter
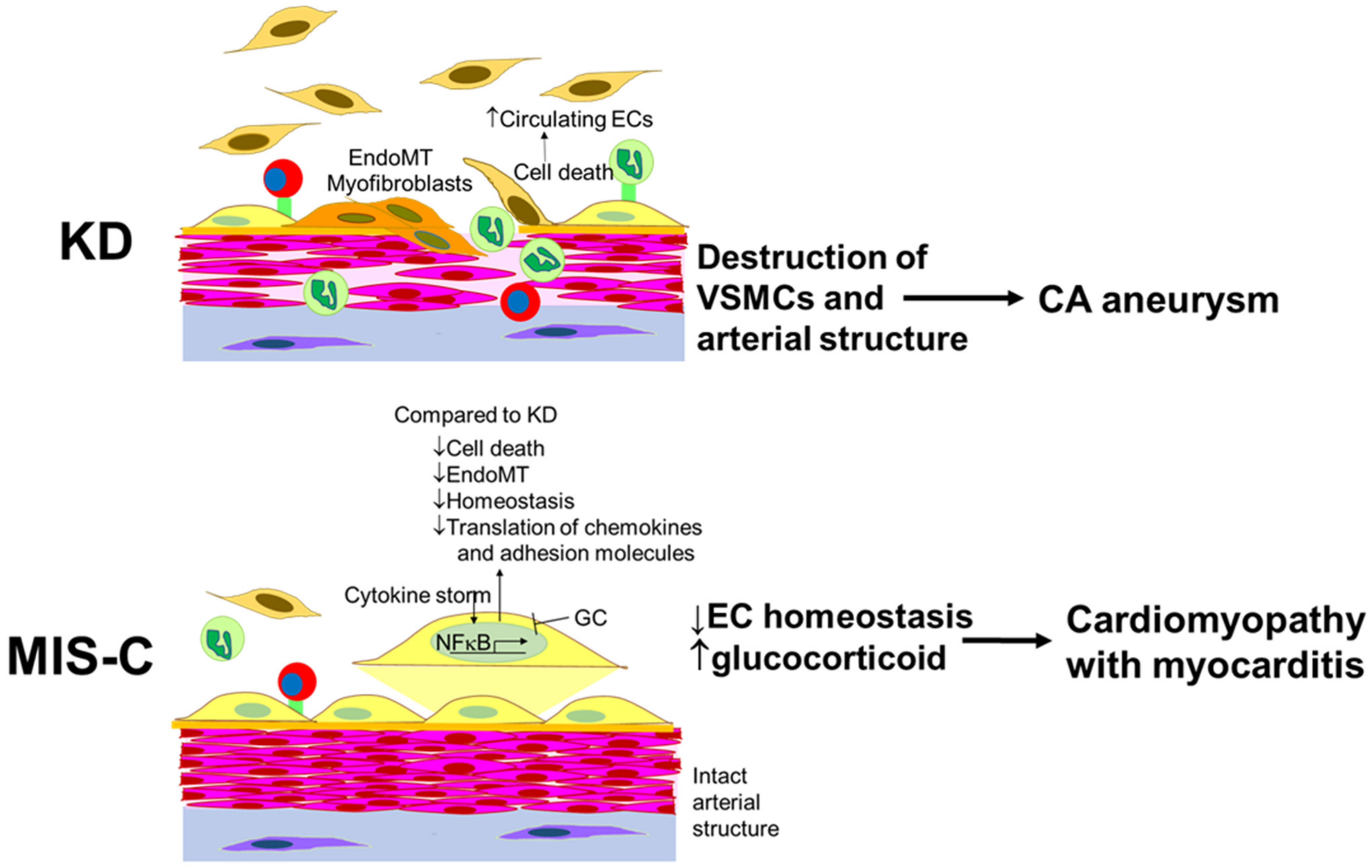
3. Discussion
4. Limitations
5. Materials and Methods
5.1. Patients and Samples
5.2. Cell Culture
5.3. RNA Extraction, RNA Sequencing, and RT-PCR
5.4. Differential Expression Analysis
5.5. Weighted Gene Correlation Network Analysis (WGCNA) Analysis
5.6. Statistical Analysis
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whittaker, E.; Bamford, A.; Kenny, J.; Kaforou, M.; Jones, C.E.; Shah, P.; Ramnarayan, P.; Fraisse, A.; Miller, O.; Davies, P.; et al. Clinical Characteristics of 58 Children with a Pediatric Inflammatory Multisystem Syndrome Temporally Associated with SARS-CoV-2. JAMA 2020, 324, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.Y.; Shimizu, C.; Tremoulet, A.H.; Bainto, E.; Roberts, S.C.; Sivilay, N.; Gardiner, M.A.; Kanegaye, J.T.; Hogan, A.H.; Salazar, J.C.; et al. A machine-learning algorithm for diagnosis of multisystem inflammatory syndrome in children and Kawasaki disease in the USA: A retrospective model development and validation study. Lancet Digit. Health 2022, 4, e717–e726. [Google Scholar] [CrossRef] [PubMed]
- Skochko, S.M.; Jain, S.; Sun, X.; Sivilay, N.; Kanegaye, J.T.; Pancheri, J.; Shimizu, C.; Sheets, R.; Tremoulet, A.H.; Burns, J.C. Kawasaki Disease Outcomes and Response to Therapy in a Multiethnic Community: A 10-Year Experience. J. Pediatr. 2018, 203, 408–415.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newburger, J.W.; Takahashi, M.; Beiser, A.S.; Burns, J.C.; Bastian, J.; Chung, K.J.; Colan, S.D.; Duffy, C.E.; Fulton, D.R.; Glode, M.P.; et al. A single intravenous infusion of gamma globulin as compared with four infusions in the treatment of acute Kawasaki syndrome. N. Engl. J. Med. 1991, 324, 1633–1639. [Google Scholar] [CrossRef]
- Son, M.B.F.; Murray, N.; Friedman, K.; Young, C.C.; Newhams, M.M.; Feldstein, L.R.; Loftis, L.L.; Tarquinio, K.M.; Singh, A.R.; Heidemann, S.M.; et al. Multisystem Inflammatory Syndrome in Children—Initial Therapy and Outcomes. N. Engl. J. Med. 2021, 385, 23–34. [Google Scholar] [CrossRef]
- Alsaied, T.; Tremoulet, A.H.; Burns, J.C.; Saidi, A.; Dionne, A.; Lang, S.M.; Newburger, J.W.; de Ferranti, S.; Friedman, K.G. Review of Cardiac Involvement in Multisystem Inflammatory Syndrome in Children. Circulation 2021, 143, 78–88. [Google Scholar] [CrossRef]
- Mayordomo-Colunga, J.; Vivanco-Allende, A.; Lopez-Alonso, I.; Lopez-Martinez, C.; Fernandez-Vega, I.; Gil-Pena, H.; Rey, C. SARS-CoV-2 Spike Protein in Intestinal Cells of a Patient with Coronavirus Disease 2019 Multisystem Inflammatory Syndrome. J. Pediatr. 2022, 243, 214–218.e5. [Google Scholar] [CrossRef]
- Duarte-Neto, A.N.; Caldini, E.G.; Gomes-Gouvea, M.S.; Kanamura, C.T.; de Almeida Monteiro, R.A.; Ferranti, J.F.; Ventura, A.M.C.; Regalio, F.A.; Fiorenzano, D.M.; Gibelli, M.; et al. An autopsy study of the spectrum of severe COVID-19 in children: From SARS to different phenotypes of MIS-C. EClinicalMedicine 2021, 35, 100850. [Google Scholar] [CrossRef]
- Ghosh, P.; Katkar, G.D.; Shimizu, C.; Kim, J.; Khandelwal, S.; Tremoulet, A.H.; Kanegaye, J.T.; Pediatric Emergency Medicine Kawasaki Disease Research Group; Bocchini, J.; Das, S.; et al. An Artificial Intelligence-guided signature reveals the shared host immune response in MIS-C and Kawasaki disease. Nat. Commun. 2022, 13, 2687. [Google Scholar] [CrossRef]
- He, M.; Chen, Z.; Martin, M.; Zhang, J.; Sangwung, P.; Woo, B.; Tremoulet, A.H.; Shimizu, C.; Jain, M.K.; Burns, J.C.; et al. miR-483 Targeting of CTGF Suppresses Endothelial-to-Mesenchymal Transition: Therapeutic Implications in Kawasaki Disease. Circ. Res. 2017, 120, 354–365. [Google Scholar] [CrossRef]
- Qian, X.X.; Mata-Greenwood, E.; Liao, W.X.; Zhang, H.; Zheng, J.; Chen, D.B. Transcriptional regulation of endothelial nitric oxide synthase expression in uterine artery endothelial cells by c-Jun/AP-1. Mol. Cell. Endocrinol. 2007, 279, 39–51. [Google Scholar] [CrossRef] [Green Version]
- Wallerath, T.; Witte, K.; Schafer, S.C.; Schwarz, P.M.; Prellwitz, W.; Wohlfart, P.; Kleinert, H.; Lehr, H.A.; Lemmer, B.; Forstermann, U. Down-regulation of the expression of endothelial NO synthase is likely to contribute to glucocorticoid-mediated hypertension. Proc. Natl. Acad. Sci. USA 1999, 96, 13357–13362. [Google Scholar] [CrossRef]
- Ahmad, S.; Hewett, P.W.; Wang, P.; Al-Ani, B.; Cudmore, M.; Fujisawa, T.; Haigh, J.J.; le Noble, F.; Wang, L.; Mukhopadhyay, D.; et al. Direct evidence for endothelial vascular endothelial growth factor receptor-1 function in nitric oxide-mediated angiogenesis. Circ. Res. 2006, 99, 715–722. [Google Scholar] [CrossRef]
- Park-Windhol, C.; Ng, Y.S.; Yang, J.; Primo, V.; Saint-Geniez, M.; D’Amore, P.A. Endomucin inhibits VEGF-induced endothelial cell migration, growth, and morphogenesis by modulating VEGFR2 signaling. Sci. Rep. 2017, 7, 17138. [Google Scholar] [CrossRef] [Green Version]
- Liao, K.H.; Chang, S.J.; Chang, H.C.; Chien, C.L.; Huang, T.S.; Feng, T.C.; Lin, W.W.; Shih, C.C.; Yang, M.H.; Yang, S.H.; et al. Endothelial angiogenesis is directed by RUNX1T1-regulated VEGFA, BMP4 and TGF-beta2 expression. PLoS ONE 2017, 12, e0179758. [Google Scholar] [CrossRef] [Green Version]
- Baby, N.; Li, Y.; Ling, E.A.; Lu, J.; Dheen, S.T. Runx1t1 (Runt-related transcription factor 1; translocated to, 1) epigenetically regulates the proliferation and nitric oxide production of microglia. PLoS ONE 2014, 9, e89326. [Google Scholar] [CrossRef]
- Lin, C.Y.; Hung, S.Y.; Chen, H.T.; Tsou, H.K.; Fong, Y.C.; Wang, S.W.; Tang, C.H. Brain-derived neurotrophic factor increases vascular endothelial growth factor expression and enhances angiogenesis in human chondrosarcoma cells. Biochem. Pharmacol. 2014, 91, 522–533. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Y.; Zhou, Z.; Shi, H.; Qiu, X.; Xiong, J.; Chen, Y. BDNF regulates the expression and secretion of VEGF from osteoblasts via the TrkB/ERK1/2 signaling pathway during fracture healing. Mol. Med. Rep. 2017, 15, 1362–1367. [Google Scholar] [CrossRef] [Green Version]
- Revollo, J.R.; Oakley, R.H.; Lu, N.Z.; Kadmiel, M.; Gandhavadi, M.; Cidlowski, J.A. HES1 is a master regulator of glucocorticoid receptor-dependent gene expression. Sci. Signal 2013, 6, ra103. [Google Scholar] [CrossRef] [Green Version]
- Leclerc, N.; Luppen, C.A.; Ho, V.V.; Nagpal, S.; Hacia, J.G.; Smith, E.; Frenkel, B. Gene expression profiling of glucocorticoid-inhibited osteoblasts. J. Mol. Endocrinol. 2004, 33, 175–193. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.W.; Lee, S.A.; Shin, J.M.; Park, I.H.; Lee, H.M. Glucocorticoids ameliorate TGF-beta1-mediated epithelial-to-mesenchymal transition of airway epithelium through MAPK and Snail/Slug signaling pathways. Sci. Rep. 2017, 7, 3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostafa, M.M.; Rider, C.F.; Shah, S.; Traves, S.L.; Gordon, P.M.K.; Miller-Larsson, A.; Leigh, R.; Newton, R. Glucocorticoid-driven transcriptomes in human airway epithelial cells: Commonalities, differences and functional insight from cell lines and primary cells. BMC Med. Genom. 2019, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Guo, F.; Li, Y.; Saaoud, F.; Kimmis, B.D.; Sandhu, J.; Fan, M.; Maulik, D.; Lessner, S.; Papasian, C.J.; et al. Adiporedoxin suppresses endothelial activation via inhibiting MAPK and NF-kappaB signaling. Sci. Rep. 2016, 6, 38975. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yoshida, T.; Wu, L.; Maiti, D.; Cebotaru, L.; Duh, E.J. Transcription factor MEF2C suppresses endothelial cell inflammation via regulation of NF-kappaB and KLF2. J. Cell Physiol. 2015, 230, 1310–1320. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Banerji, S.; Lawrance, W.; Gileadi, U.; Prota, G.; Holder, K.A.; Roshorm, Y.M.; Hanke, T.; Cerundolo, V.; Gale, N.W.; et al. Dendritic cells enter lymph vessels by hyaluronan-mediated docking to the endothelial receptor LYVE-1. Nat. Immunol. 2017, 18, 762–770. [Google Scholar] [CrossRef] [PubMed]
- Nikitenko, L.L.; Smith, D.M.; Bicknell, R.; Rees, M.C. Transcriptional regulation of the CRLR gene in human microvascular endothelial cells by hypoxia. FASEB J. 2003, 17, 1499–1501. [Google Scholar] [CrossRef] [Green Version]
- Speksnijder, N.; Christensen, K.V.; Didriksen, M.; De Kloet, E.R.; Datson, N.A. Glucocorticoid receptor and myocyte enhancer factor 2 cooperate to regulate the expression of c-JUN in a neuronal context. J. Mol. Neurosci. 2012, 48, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes. Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Amarachintha, S.; Wilson, A.; Pang, Q. The immune receptor Trem1 cooperates with diminished DNA damage response to induce preleukemic stem cell expansion. Leukemia 2017, 31, 423–433. [Google Scholar] [CrossRef]
- Camara-Quilez, M.; Barreiro-Alonso, A.; Vizoso-Vazquez, A.; Rodriguez-Belmonte, E.; Quindos-Varela, M.; Lamas-Maceiras, M.; Cerdan, M.E. The HMGB1-2 Ovarian Cancer Interactome. The Role of HMGB Proteins and Their Interacting Partners MIEN1 and NOP53 in Ovary Cancer and Drug-Response. Cancers 2020, 12, 2435. [Google Scholar] [CrossRef]
- Tan, C.T.; Zhou, Q.L.; Su, Y.C.; Fu, N.Y.; Chang, H.C.; Tao, R.N.; Sukumaran, S.K.; Baksh, S.; Tan, Y.J.; Sabapathy, K.; et al. MOAP-1 Mediates Fas-Induced Apoptosis in Liver by Facilitating tBid Recruitment to Mitochondria. Cell Rep. 2016, 16, 174–185. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, A.; Mizoguchi, I.; Hasegawa, H.; Katahira, Y.; Inoue, S.; Sakamoto, E.; Furusaka, Y.; Sekine, A.; Miyakawa, S.; Murakami, F.; et al. A Chaperone-Like Role for EBI3 in Collaboration with Calnexin under Inflammatory Conditions. Front. Immunol. 2021, 12, 757669. [Google Scholar] [CrossRef]
- Aichem, A.; Kalveram, B.; Spinnenhirn, V.; Kluge, K.; Catone, N.; Johansen, T.; Groettrup, M. The proteomic analysis of endogenous FAT10 substrates identifies p62/SQSTM1 as a substrate of FAT10ylation. J. Cell Sci. 2012, 125 Pt 19, 4576–4585. [Google Scholar] [CrossRef] [Green Version]
- Grassi, G.; Di Caprio, G.; Santangelo, L.; Fimia, G.M.; Cozzolino, A.M.; Komatsu, M.; Ippolito, G.; Tripodi, M.; Alonzi, T. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting Snail degradation. Cell Death Dis. 2015, 6, e1880. [Google Scholar] [CrossRef] [Green Version]
- Achan, V.; Tran, C.T.; Arrigoni, F.; Whitley, G.S.; Leiper, J.M.; Vallance, P. All-trans-Retinoic acid increases nitric oxide synthesis by endothelial cells: A role for the induction of dimethylarginine dimethylaminohydrolase. Circ. Res. 2002, 90, 764–769. [Google Scholar] [CrossRef]
- LaRocca, T.J.; Henson, G.D.; Thorburn, A.; Sindler, A.L.; Pierce, G.L.; Seals, D.R. Translational evidence that impaired autophagy contributes to arterial ageing. J. Physiol. 2012, 590, 3305–3316. [Google Scholar] [CrossRef]
- Khan, S.H.; Arnott, J.A.; Kumar, R. Naturally occurring osmolyte, trehalose induces functional conformation in an intrinsically disordered activation domain of glucocorticoid receptor. PLoS ONE 2011, 6, e19689. [Google Scholar] [CrossRef]
- Qiao, D.; Xu, J.; Le, C.; Huang, E.; Liu, C.; Qiu, P.; Lin, Z.; Xie, W.B.; Wang, H. Insulin-like growth factor binding protein 5 (IGFBP5) mediates methamphetamine-induced dopaminergic neuron apoptosis. Toxicol. Lett. 2014, 230, 444–453. [Google Scholar] [CrossRef]
- Gavin, A.L.; Huang, D.; Blane, T.R.; Thinnes, T.C.; Murakami, Y.; Fukui, R.; Miyake, K.; Nemazee, D. Cleavage of DNA and RNA by PLD3 and PLD4 limits autoinflammatory triggering by multiple sensors. Nat. Commun. 2021, 12, 5874. [Google Scholar] [CrossRef]
- Gogulamudi, V.R.; Mani, I.; Subramanian, U.; Pandey, K.N. Genetic disruption of Npr1 depletes regulatory T cells and provokes high levels of proinflammatory cytokines and fibrosis in the kidneys of female mutant mice. Am. J. Physiol. Renal Physiol. 2019, 316, F1254–F1272. [Google Scholar] [CrossRef]
- Zhang, K.; Tao, C.; Xu, J.; Ruan, J.; Xia, J.; Zhu, W.; Xin, L.; Ye, H.; Xie, N.; Xia, B.; et al. CD8(+) T Cells Involved in Metabolic Inflammation in Visceral Adipose Tissue and Liver of Transgenic Pigs. Front. Immunol. 2021, 12, 690069. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.M. Role of IL-33 in inflammation and disease. J. Inflamm. 2011, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- Panganiban, R.P.; Vonakis, B.M.; Ishmael, F.T.; Stellato, C. Coordinated post-transcriptional regulation of the chemokine system: Messages from CCL2. J. Interferon Cytokine Res. 2014, 34, 255–266. [Google Scholar] [CrossRef]
- Zielinska, K.A.; Van Moortel, L.; Opdenakker, G.; De Bosscher, K.; Van den Steen, P.E. Endothelial Response to Glucocorticoids in Inflammatory Diseases. Front. Immunol. 2016, 7, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruver-Yates, A.L.; Cidlowski, J.A. Tissue-specific actions of glucocorticoids on apoptosis: A double-edged sword. Cells 2013, 2, 202–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, O.J.; Iniguez-Lluhi, J.A.; Romanelli, A.; Kimball, S.R.; Jefferson, L.S. The activated glucocorticoid receptor modulates presumptive autoregulation of ribosomal protein S6 protein kinase, p70 S6K. J. Biol. Chem. 2002, 277, 2525–2533. [Google Scholar] [CrossRef] [Green Version]
- Shah, O.J.; Kimball, S.R.; Jefferson, L.S. Acute attenuation of translation initiation and protein synthesis by glucocorticoids in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E76–E82. [Google Scholar] [CrossRef]
- Meyuhas, O.; Baldin, V.; Bouche, G.; Amalric, F. Glucocorticoids repress ribosome biosynthesis in lymphosarcoma cells by affecting gene expression at the level of transcription, posttranscription and translation. Biochim. Biophys. Acta 1990, 1049, 38–44. [Google Scholar] [CrossRef]
- Munzel, T.; Templin, C.; Cammann, V.L.; Hahad, O. Takotsubo Syndrome: Impact of endothelial dysfunction and oxidative stress. Free Radic. Biol. Med. 2021, 169, 216–223. [Google Scholar] [CrossRef]
- Fabi, M.; Petrovic, B.; Andreozzi, L.; Corinaldesi, E.; Filice, E.; Biagi, C.; Rizzello, A.; Mattesini, B.E.; Bugani, S.; Lanari, M. Circulating Endothelial Cells: A New Possible Marker of Endothelial Damage in Kawasaki Disease, Multisystem Inflammatory Syndrome in Children and Acute SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 10106. [Google Scholar] [CrossRef]
- Tao, R.; Wang, Z.F.; Qiu, W.; He, Y.F.; Yan, W.Q.; Sun, W.Y.; Li, H.J. Role of S100A3 in human hepatocellular carcinoma and the anticancer effect of sodium cantharidinate. Exp. Ther. Med. 2017, 13, 2812–2818. [Google Scholar] [CrossRef] [Green Version]
- Rogers, P.R.; Song, J.; Gramaglia, I.; Killeen, N.; Croft, M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity 2001, 15, 445–455. [Google Scholar] [CrossRef] [Green Version]
- Frazzi, R. BIRC3 and BIRC5: Multi-faceted inhibitors in cancer. Cell Biosci. 2021, 11, 8. [Google Scholar] [CrossRef]
- Hoh, J.F.; Hughes, S. Myogenic and neurogenic regulation of myosin gene expression in cat jaw-closing muscles regenerating in fast and slow limb muscle beds. J. Muscle Res. Cell Motil. 1988, 9, 59–72. [Google Scholar]
- Hu, Y.W.; Wu, S.G.; Zhao, J.J.; Ma, X.; Lu, J.B.; Xiu, J.C.; Zhang, Y.; Huang, C.; Qiu, Y.R.; Sha, Y.H.; et al. VNN1 promotes atherosclerosis progression in apoE-/- mice fed a high-fat/high-cholesterol diet. J. Lipid Res. 2016, 57, 1398–1411. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.Y.; Liu, Y.H.; Chen, Y.J.; Yeh, Y.Y.; Huang, H.M. CD69 partially inhibits apoptosis and erythroid differentiation via CD24, and their knockdown increase imatinib sensitivity in BCR-ABL-positive cells. J. Cell Physiol. 2018, 233, 7467–7479. [Google Scholar] [CrossRef]
- Barcellos-de-Souza, P.; Canetti, C.; Barja-Fidalgo, C.; Arruda, M.A. Leukotriene B(4) inhibits neutrophil apoptosis via NADPH oxidase activity: Redox control of NF-kappaB pathway and mitochondrial stability. Biochim. Biophys. Acta 2012, 1823, 1990–1997. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Y.; Mayo, M.W.; Korneluk, R.G.; Goeddel, D.V.; Baldwin, A.S., Jr. NF-kappaB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998, 281, 1680–1683. [Google Scholar] [CrossRef]
- Shiozaki, A.; Marunaka, Y.; Otsuji, E. Roles of Ion and Water Channels in the Cell Death and Survival of Upper Gastrointestinal Tract Cancers. Front. Cell Dev. Biol. 2021, 9, 616933. [Google Scholar] [CrossRef]
- Duan, C.; Allard, J.B. Insulin-Like Growth Factor Binding Protein-5 in Physiology and Disease. Front. Endocrinol. 2020, 11, 100. [Google Scholar] [CrossRef] [Green Version]
- Islam, M.A.; Sooro, M.A.; Zhang, P. Autophagic Regulation of p62 is Critical for Cancer Therapy. Int. J. Mol. Sci. 2018, 19, 1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gugnoni, M.; Sancisi, V.; Manzotti, G.; Gandolfi, G.; Ciarrocchi, A. Autophagy and epithelial-mesenchymal transition: An intricate interplay in cancer. Cell Death Dis. 2016, 7, e2520. [Google Scholar] [CrossRef] [PubMed]
- Orenstein, J.M.; Shulman, S.T.; Fox, L.M.; Baker, S.C.; Takahashi, M.; Bhatti, T.R.; Russo, P.A.; Mierau, G.W.; de Chadarevian, J.P.; Perlman, E.J.; et al. Three linked vasculopathic processes characterize kawasaki disease: A light and transmission electron microscopic study. PLoS ONE 2012, 7, e38998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, C.; Oharaseki, T.; Takahashi, K.; Kottek, A.; Franco, A.; Burns, J.C. The role of TGF-beta and myofibroblasts in the arteritis of Kawasaki disease. Hum. Pathol. 2013, 44, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Bolli, R.; Marban, E. Molecular and cellular mechanisms of myocardial stunning. Physiol. Rev. 1999, 79, 609–634. [Google Scholar] [CrossRef] [Green Version]
- Myers, V.D.; McClung, J.M.; Wang, J.; Tahrir, F.G.; Gupta, M.K.; Gordon, J.; Kontos, C.H.; Khalili, K.; Cheung, J.Y.; Feldman, A.M. The Multifunctional Protein BAG3: A Novel Therapeutic Target in Cardiovascular Disease. JACC Basic. Transl. Sci. 2018, 3, 122–131. [Google Scholar] [CrossRef]
- Shokr, M.; Rashed, A.; Lata, K.; Kondur, A. Dexamethasone Associated ST Elevation Myocardial Infarction Four Days after an Unremarkable Coronary Angiogram-Another Reason for Cautious Use of Steroids: A Case Report and Review of the Literature. Case Rep. Cardiol. 2016, 2016, 4970858. [Google Scholar] [CrossRef] [Green Version]
- Rogers, K.M.; Bonar, C.A.; Estrella, J.L.; Yang, S. Inhibitory effect of glucocorticoid on coronary artery endothelial function. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H1922–H1928. [Google Scholar] [CrossRef]
- Jun, S.S.; Chen, Z.; Pace, M.C.; Shaul, P.W. Glucocorticoids downregulate cyclooxygenase-1 gene expression and prostacyclin synthesis in fetal pulmonary artery endothelium. Circ. Res. 1999, 84, 193–200. [Google Scholar] [CrossRef]
- Feldstein, L.R.; Tenforde, M.W.; Friedman, K.G.; Newhams, M.; Rose, E.B.; Dapul, H.; Soma, V.L.; Maddux, A.B.; Mourani, P.M.; Bowens, C.; et al. Characteristics and Outcomes of US Children and Adolescents with Multisystem Inflammatory Syndrome in Children (MIS-C) Compared With Severe Acute COVID-19. JAMA 2021, 325, 1074–1087. [Google Scholar] [CrossRef]
- Valverde, I.; Singh, Y.; Sanchez-de-Toledo, J.; Theocharis, P.; Chikermane, A.; Di Filippo, S.; Kucinska, B.; Mannarino, S.; Tamariz-Martel, A.; Gutierrez-Larraya, F.; et al. Acute Cardiovascular Manifestations in 286 Children with Multisystem Inflammatory Syndrome Associated With COVID-19 Infection in Europe. Circulation 2021, 143, 21–32. [Google Scholar] [CrossRef]
- Bermejo, I.A.; Bautista-Rodriguez, C.; Fraisse, A.; Voges, I.; Gatehouse, P.; Kang, H.; Piccinelli, E.; Rowlinson, G.; Lane, M.; Semple, T.; et al. Short-Term sequelae of Multisystem Inflammatory Syndrome in Children Assessed by CMR. JACC Cardiovasc. Imaging 2021, 14, 1666–1667. [Google Scholar] [CrossRef]
- Penner, J.; Abdel-Mannan, O.; Grant, K.; Maillard, S.; Kucera, F.; Hassell, J.; Eyre, M.; Berger, Z.; Hacohen, Y.; Moshal, K.; et al. 6-month multidisciplinary follow-up and outcomes of patients with paediatric inflammatory multisystem syndrome (PIMS-TS) at a UK tertiary paediatric hospital: A retrospective cohort study. Lancet Child. Adolesc. Health 2021, 5, 473–482. [Google Scholar]
- Ciftel, M.; Ates, N.; Yilmaz, O. Investigation of endothelial dysfunction and arterial stiffness in multisystem inflammatory syndrome in children. Eur. J. Pediatr. 2022, 181, 91–97. [Google Scholar] [CrossRef]
- Anderson, P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010, 10, 24–35. [Google Scholar] [CrossRef]
- Holz, M.K.; Ballif, B.A.; Gygi, S.P.; Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 2005, 123, 569–580. [Google Scholar] [CrossRef] [Green Version]
- Maurya, M.R.; Gupta, S.; Li, J.Y.; Ajami, N.E.; Chen, Z.B.; Shyy, J.Y.; Chien, S.; Subramaniam, S. Longitudinal shear stress response in human endothelial cells to atheroprone and atheroprotective conditions. Proc. Natl. Acad. Sci. USA 2021, 118, e2023236118. [Google Scholar] [CrossRef]
- McCrindle, B.W.; Rowley, A.H.; Newburger, J.W.; Burns, J.C.; Bolger, A.F.; Gewitz, M.; Baker, A.L.; Jackson, M.A.; Takahashi, M.; Shah, P.B.; et al. Diagnosis, Treatment, and Long-Term Management of Kawasaki Disease: A Scientific Statement for Health Professionals from the American Heart Association. Circulation 2017, 135, e927–e999. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Multisystem Inflammatory Syndrome in Children (MIS-C) Associated with Coronavirus Disease 2019 (COVID-19). Emergency Preparedness and Response. 2021. Available online: https://emergency.cdc.gov/han/2020/han00432.asp (accessed on 28 July 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| MIS-C | KD | HC *4 | p *5 | ||
|---|---|---|---|---|---|
| n = 7 | n = 5 | n = 3 | |||
| Age, yrs *1 | 10.5 (8.8–12.2) | 1.9 (1.6–3.8) | 5.5 (4.2–7.1) | 0.005 | |
| Male, n (%) | 6 (86) | 3 (60) | 2 (67) | NS | |
| Ethnicity, n (%) | Asian | 0 | 1 (20) | 0 | NS |
| AA | 2 (29) | 0 | 0 | ||
| White | 0 | 1 (20) | 2 (67) | ||
| Hispanic | 4 (57) | 2 (40) | 1 (33) | ||
| >2 races | 1 (14) | 1 (20) | 0 | ||
| Illness day of serum collection *2 | 3 (3–4.5) | 5 (4–5) | 418 (416–704) | NS | |
| Coronary artery Zmax *3 | 2.1 (1.6–2.7) | 3.2 (1.7–3.3) | NA | NS | |
| EF min, % | 58 (46–62) | 61 (58–66) | NA | NS | |
| Laboratory data | WBC, 103/uL | 6.5 (5.0–11.1) | 18.4 (12.3–20.7) | NA | 0.048 |
| PLT, 103/mm3 | 140 (93–221) | 358 (181–361) | NA | NS | |
| ESR, mm/h | 44 (36.5–53.5) | 48 (42–58) | NA | NS | |
| CRP, mg/dL | 21.3 (20.3–26.5) | 7.0 (4.8–8.8) | NA | 0.048 | |
| Troponin max, ng/mL | 0.050 (0.015–0.225) | ND | NA | NA | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Shimizu, C.; He, M.; Wang, H.; Hoffman, H.M.; Tremoulet, A.H.; Shyy, J.Y.-J.; Burns, J.C. Endothelial Cell Response in Kawasaki Disease and Multisystem Inflammatory Syndrome in Children. Int. J. Mol. Sci. 2023, 24, 12318. https://doi.org/10.3390/ijms241512318
Kim J, Shimizu C, He M, Wang H, Hoffman HM, Tremoulet AH, Shyy JY-J, Burns JC. Endothelial Cell Response in Kawasaki Disease and Multisystem Inflammatory Syndrome in Children. International Journal of Molecular Sciences. 2023; 24(15):12318. https://doi.org/10.3390/ijms241512318
Chicago/Turabian StyleKim, Jihoon, Chisato Shimizu, Ming He, Hao Wang, Hal M. Hoffman, Adriana H. Tremoulet, John Y.-J. Shyy, and Jane C. Burns. 2023. "Endothelial Cell Response in Kawasaki Disease and Multisystem Inflammatory Syndrome in Children" International Journal of Molecular Sciences 24, no. 15: 12318. https://doi.org/10.3390/ijms241512318
APA StyleKim, J., Shimizu, C., He, M., Wang, H., Hoffman, H. M., Tremoulet, A. H., Shyy, J. Y.-J., & Burns, J. C. (2023). Endothelial Cell Response in Kawasaki Disease and Multisystem Inflammatory Syndrome in Children. International Journal of Molecular Sciences, 24(15), 12318. https://doi.org/10.3390/ijms241512318






