Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis
Abstract
1. Introduction
2. Connection between Radioprotective Natural Products and miRNAs
2.1. Function of Radioprotective Natural Products in Non-Cancer Tissue Studies
2.2. Functions of Radioprotective Natural Products in Anticancer Studies
2.3. Function of Radioprotective Natural Products in Non-Cancer Tissue and Anticancer Studies
2.4. Other Radioprotective Natural Products Needing Further Investigation
3. Connection between Natural-Product-Regulated miRNAs and Radiation-Modulating Effects
3.1. Function of Natural-Product-Regulated miRNAs in Non-Cancer Radiation Studies
3.1.1. Some Natural-Product-Regulated miRNAs Are Highly Expressed in Non-Cancer Radiation Studies
3.1.2. Some Natural-Product-Regulated miRNAs Can Function as Radioprotectors in Non-Cancer Tissues
3.2. Function of Natural-Product-Regulated miRNAs in Cancer Radiation Studies
3.2.1. Some Natural-Product-Regulated miRNAs Can Function as Radiosensitizers in Cancer Cells
3.2.2. Some Natural-Product-Regulated miRNAs Can Have a Radioresistance Function in Cancer Cells
3.3. Function of Natural-Product-Regulated miRNAs in Both Non-Cancer and Cancer Radiation Studies
3.4. Other Natural-Product-Regulated miRNA Candidates May Have Radiomodulating Effects
4. Connection between Natural-Product-Regulated miRNAs and Exosome Biogenesis-Modulating Effects
4.1. Function of Natural-Product-Regulated miRNAs in Non-Cancer Exosome Studies
4.2. Function of Natural-Product-Regulated miRNAs in Cancer Exosome Studies
4.3. Other Natural-Product-Regulated miRNAs May Have Exosome Biogenesis-Modulating Effects
5. Relationship between Radiation and Exosome Biogenesis Modulation by Natural-Product-Regulated miRNAs
6. Overview of Natural Products That Regulate miRNAs to Modulate Radiation Responses
7. Overview of Natural Products That Regulate Exosomal miRNAs Modulating Exosome Biogenesis
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Citrin, D.E. Radiation Modifiers. Hematol. Oncol. Clin. N. Am. 2019, 33, 1041–1055. [Google Scholar] [CrossRef] [PubMed]
- Mun, G.I.; Kim, S.; Choi, E.; Kim, C.S.; Lee, Y.S. Pharmacology of natural radioprotectors. Arch. Pharm. Res. 2018, 41, 1033–1050. [Google Scholar] [CrossRef] [PubMed]
- Kuruba, V.; Gollapalli, P. Natural radioprotectors and their impact on cancer drug discovery. Radiat. Oncol. J. 2018, 36, 265–275. [Google Scholar] [CrossRef]
- Yi, J.; Zhu, J.; Zhao, C.; Kang, Q.; Zhang, X.; Suo, K.; Cao, N.; Hao, L.; Lu, J. Potential of natural products as radioprotectors and radiosensitizers: Opportunities and challenges. Food Funct. 2021, 12, 5204–5218. [Google Scholar] [CrossRef]
- Fischer, N.; Seo, E.J.; Efferth, T. Prevention from radiation damage by natural products. Phytomedicine 2018, 47, 192–200. [Google Scholar] [CrossRef]
- Jit, B.P.; Pradhan, B.; Dash, R.; Bhuyan, P.P.; Behera, C.; Behera, R.K.; Sharma, A.; Alcaraz, M.; Jena, M. Phytochemicals: Potential therapeutic modulators of radiation induced signaling pathways. Antioxidants 2021, 11, 49. [Google Scholar] [CrossRef]
- Kang, Q.; Zhang, X.; Cao, N.; Chen, C.; Yi, J.; Hao, L.; Ji, Y.; Liu, X.; Lu, J. EGCG enhances cancer cells sensitivity under (60)Cogamma radiation based on miR-34a/Sirt1/p53. Food Chem. Toxicol. 2019, 133, 110807. [Google Scholar] [CrossRef]
- Parihar, V.K.; Dhawan, J.; Kumar, S.; Manjula, S.N.; Subramanian, G.; Unnikrishnan, M.K.; Rao, C.M. Free radical scavenging and radioprotective activity of dehydrozingerone against whole body gamma irradiation in Swiss albino mice. Chem. Biol. Interact. 2007, 170, 49–58. [Google Scholar] [CrossRef]
- Li, G.H.; Wang, D.L.; Hu, Y.D.; Pu, P.; Li, D.Z.; Wang, W.D.; Zhu, B.; Hao, P.; Wang, J.; Xu, X.Q.; et al. Berberine inhibits acute radiation intestinal syndrome in human with abdomen radiotherapy. Med. Oncol. 2010, 27, 919–925. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, H.; Zhang, C.; Cheng, Y.; Hu, L.; Meng, X.; Zhao, Y. Protective effects of berberine on radiation-induced lung injury via intercellular adhesion molecular-1 and transforming growth factor-beta-1 in patients with lung cancer. Eur. J. Cancer 2008, 44, 2425–2432. [Google Scholar] [CrossRef]
- Jagetia, G.C.; Baliga, M.S. Radioprotection by mangiferin in DBAxC57BL mice: A preliminary study. Phytomedicine 2005, 12, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ahn, K.S.; Alharbi, S.A.; Shair, O.H.; Arfuso, F.; Sethi, G.; Chinnathambi, A.; Tang, F.R. Celastrol alleviates gamma irradiation-induced damage by modulating diverse inflammatory mediators. Int. J. Mol. Sci. 2020, 21, 1084. [Google Scholar] [CrossRef] [PubMed]
- Nuszkiewicz, J.; Wozniak, A.; Szewczyk-Golec, K. Ionizing radiation as a source of oxidative stress-the protective role of melatonin and vitamin D. Int. J. Mol. Sci. 2020, 21, 5804. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Zhu, Q.; Liu, Y.; Cheng, H.; Zhang, Y.; Li, T. Emodin protects mice against radiation-induced mortality and intestinal injury via inhibition of apoptosis and modulation of p53. Environ. Toxicol. Pharmacol. 2016, 46, 311–318. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Zhu, Q.; Liu, Y.; Cheng, H.; Zhang, Y.; Li, T. Data on the radioprotective effect of emodin in vivo and vitro via inhibition of apoptosis and modulation of p53. Data Brief 2017, 11, 290–295. [Google Scholar] [CrossRef]
- Checker, R.; Bhilwade, H.N.; Nandha, S.R.; Patwardhan, R.S.; Sharma, D.; Sandur, S.K. Withaferin A, a steroidal lactone, selectively protects normal lymphocytes against ionizing radiation induced apoptosis and genotoxicity via activation of ERK/Nrf-2/HO-1 axis. Toxicol. Appl. Pharmacol. 2023, 461, 116389. [Google Scholar] [CrossRef]
- Chen, C.; Mu, X.Y.; Zhou, Y.; Shun, K.; Geng, S.; Liu, J.; Wang, J.W.; Chen, J.; Li, T.Y.; Wang, Y.P. Ginsenoside Rg1 enhances the resistance of hematopoietic stem/progenitor cells to radiation-induced aging in mice. Acta Pharmacol. Sin. 2014, 35, 143–150. [Google Scholar] [CrossRef]
- Jelonek, K.; Widlak, P.; Pietrowska, M. The influence of ionizing radiation on exosome composition, secretion and intercellular communication. Protein Pept. Lett. 2016, 23, 656–663. [Google Scholar] [CrossRef]
- Yang, Z.; Zhong, W.; Yang, L.; Wen, P.; Luo, Y.; Wu, C. The emerging role of exosomes in radiotherapy. Cell Commun. Signal. 2022, 20, 171. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Q.; Yang, Y.; Li, Q.; Wang, Z. Exosome: Function and role in cancer metastasis and drug resistance. Technol. Cancer Res. Treat. 2018, 17, 1533033818763450. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zheng, Z.; Yuan, Y.; Pathak, J.L.; Yang, X.; Wang, L.; Ye, Z.; Cho, W.C.; Zeng, M.; Wu, L. The emerging role of exosomes in oral squamous cell carcinoma. Front. Cell Dev. Biol. 2021, 9, 628103. [Google Scholar] [CrossRef] [PubMed]
- Dhar, R.; Mallik, S.; Devi, A. Exosomal microRNAs (exoMIRs): Micromolecules with macro impact in oral cancer. 3 Biotech 2022, 12, 155. [Google Scholar] [CrossRef]
- Li, Y.; Gao, S.; Hu, Q.; Wu, F. Functional properties of cancer epithelium and stroma-derived exosomes in head and neck squamous cell carcinoma. Life 2022, 12, 757. [Google Scholar] [CrossRef]
- St-Denis-Bissonnette, F.; Khoury, R.; Mediratta, K.; El-Sahli, S.; Wang, L.; Lavoie, J.R. Applications of extracellular vesicles in triple-negative breast cancer. Cancers 2022, 14, 451. [Google Scholar] [CrossRef]
- Lorenc, T.; Klimczyk, K.; Michalczewska, I.; Slomka, M.; Kubiak-Tomaszewska, G.; Olejarz, W. Exosomes in prostate cancer diagnosis, prognosis and therapy. Int. J. Mol. Sci. 2020, 21, 2118. [Google Scholar] [CrossRef]
- Li, X.; Jiang, W.; Gan, Y.; Zhou, W. The application of exosomal microRNAs in the treatment of pancreatic cancer and its research progress. Pancreas 2021, 50, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Babaker, M.A.; Aljoud, F.A.; Alkhilaiwi, F.; Algarni, A.; Ahmed, A.; Khan, M.I.; Saadeldin, I.M.; Alzahrani, F.A. The Therapeutic Potential of Milk Extracellular Vesicles on Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 6812. [Google Scholar] [CrossRef]
- Hashemipour, M.; Boroumand, H.; Mollazadeh, S.; Tajiknia, V.; Nourollahzadeh, Z.; Rohani Borj, M.; Pourghadamyari, H.; Rahimian, N.; Hamblin, M.R.; Mirzaei, H. Exosomal microRNAs and exosomal long non-coding RNAs in gynecologic cancers. Gynecol. Oncol. 2021, 161, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Fu, S.; Wu, S.; Tu, R. Growing evidence of exosomal microRNA-related metastasis of hepatocellular carcinoma. BioMed Res. Int. 2020, 2020, 4501454. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, G.; Liu, J.; Zhang, C.; Yao, Y.; Liao, W. Exosomal cargoes in OSCC: Current findings and potential functions. PeerJ 2020, 8, e10062. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Cao, K.; Liao, Z.; Chen, Y.; Lei, X.; Wei, Q.; Liu, C.; Sun, X.; Yang, Y.; Cai, J.; et al. Monophosphoryl lipid A alleviated radiation-induced testicular injury through TLR4-dependent exosomes. J. Cell. Mol. Med. 2020, 24, 3917–3930. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, Y.; Zhao, B.; Yang, Y.; Wang, J.; Shen, K.; Yang, X.; Hu, D.; Zheng, G.; Han, J. Exosomes derived from adipose-derived mesenchymal stem cells ameliorate radiation-induced brain injury by activating the SIRT1 pathway. Front. Cell Dev. Biol. 2021, 9, 693782. [Google Scholar] [CrossRef] [PubMed]
- Payton, C.; Pang, L.Y.; Gray, M.; Argyle, D.J. Exosomes derived from radioresistant breast cancer cells promote therapeutic resistance in naive recipient cells. J. Pers. Med. 2021, 11, 1310. [Google Scholar] [CrossRef]
- Ni, J.; Bucci, J.; Malouf, D.; Knox, M.; Graham, P.; Li, Y. Exosomes in cancer radioresistance. Front. Oncol. 2019, 9, 869. [Google Scholar] [CrossRef]
- Bach, D.H.; Hong, J.Y.; Park, H.J.; Lee, S.K. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 2017, 141, 220–230. [Google Scholar] [CrossRef]
- Bian, E.B.; Chen, E.F.; Xu, Y.D.; Yang, Z.H.; Tang, F.; Ma, C.C.; Wang, H.L.; Zhao, B. Exosomal lncRNA-ATB activates astrocytes that promote glioma cell invasion. Int. J. Oncol. 2019, 54, 713–721. [Google Scholar] [CrossRef]
- Lafitte, M.; Lecointre, C.; Roche, S. Roles of exosomes in metastatic colorectal cancer. Am. J. Physiol. Cell Physiol. 2019, 317, C869–C880. [Google Scholar] [CrossRef]
- Feng, S.; Lou, K.; Zou, X.; Zou, J.; Zhang, G. The potential role of exosomal proteins in prostate cancer. Front. Oncol. 2022, 12, 873296. [Google Scholar] [CrossRef]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Jan, A.T.; Rahman, S.; Badierah, R.; Lee, E.J.; Mattar, E.H.; Redwan, E.M.; Choi, I. Expedition into exosome biology: A perspective of progress from discovery to therapeutic development. Cancers 2021, 13, 1157. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Desai, N.N.; Qureshi, M.Z.; Librelotto, D.R.N.; Gasparri, M.L.; Bishayee, A.; Nabavi, S.M.; Curti, V.; Daglia, M. Exosome biogenesis, bioactivities and functions as new delivery systems of natural compounds. Biotechnol. Adv. 2018, 36, 328–334. [Google Scholar] [CrossRef]
- Teng, F.; Fussenegger, M. Shedding light on extracellular vesicle biogenesis and bioengineering. Adv. Sci. 2020, 8, 2003505. [Google Scholar] [CrossRef]
- Hu, S.; Liu, Y.; Guan, S.; Qiu, Z.; Liu, D. Natural products exert anti-tumor effects by regulating exosomal ncRNA. Front. Oncol. 2022, 12, 1006114. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef]
- Alnuqaydan, A.M. Targeting micro-RNAs by natural products: A novel future therapeutic strategy to combat cancer. Am. J. Transl. Res. 2020, 12, 3531–3556. [Google Scholar]
- Phuah, N.H.; Nagoor, N.H. Regulation of microRNAs by natural agents: New strategies in cancer therapies. BioMed Res. Int. 2014, 2014, 804510. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Wang, Z. MicroRNAs as biomarkers for ionizing radiation injury. Front. Cell. Dev. Biol. 2022, 10, 861451. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, A.; Anastasov, N.; Angermeier, M.; Winkler, K.; Atkinson, M.J.; Moertl, S. MicroRNA-mediated processes are essential for the cellular radiation response. Radiat. Res. 2011, 176, 575–586. [Google Scholar] [CrossRef]
- Zhou, B.R.; Guo, X.F.; Zhang, J.A.; Xu, Y.; Li, W.; Wu, D.; Yin, Z.Q.; Permatasari, F.; Luo, D. Elevated miR-34c-5p mediates dermal fibroblast senescence by ultraviolet irradiation. Int. J. Biol. Sci. 2013, 9, 743–752. [Google Scholar] [CrossRef]
- Chiba, M.; Monzen, S.; Iwaya, C.; Kashiwagi, Y.; Yamada, S.; Hosokawa, Y.; Mariya, Y.; Nakamura, T.; Wojcik, A. Serum miR-375-3p increase in mice exposed to a high dose of ionizing radiation. Sci. Rep. 2018, 8, 1302. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, H.; Wang, X.; Ren, Z.; Tian, Y.; Zhao, J.; Qi, W.; Wang, H.; Yu, Y.; Gong, R.; et al. Light emitting diodes irradiation regulates miRNA-877-3p to promote cardiomyocyte proliferation. Int. J. Med. Sci. 2022, 19, 1254–1264. [Google Scholar] [CrossRef]
- Wang, L.J.; Li, N.N.; Xu, S.J.; Zhang, F.; Hao, M.H.; Yang, X.J.; Cai, X.H.; Qiu, P.Y.; Ji, H.L.; Xu, P. A new and important relationship between miRNA-147a and PDPK1 in radiotherapy. J. Cell. Biochem. 2018, 119, 3519–3527. [Google Scholar] [CrossRef]
- Zhang, T.; Ma, S.; Liu, C.; Hu, K.; Xu, M.; Wang, R. Rosmarinic acid prevents radiation-induced pulmonary fibrosis through attenuation of ROS/MYPT1/TGFbeta1 signaling via miR-19b-3p. Dose Response 2020, 18, 1559325820968413. [Google Scholar] [CrossRef]
- Leavitt, R.J.; Acharya, M.M.; Baulch, J.E.; Limoli, C.L. Extracellular vesicle-derived miR-124 resolves radiation-induced brain injury. Cancer Res. 2020, 80, 4266–4277. [Google Scholar] [CrossRef]
- Long, D.; Xu, L.; Deng, Z.; Guo, D.; Zhang, Y.; Liu, Z.; Zhang, C. HPV16 E6 enhances the radiosensitivity in HPV-positive human head and neck squamous cell carcinoma by regulating the miR-27a-3p/SMG1 axis. Infect. Agent Cancer 2021, 16, 56. [Google Scholar] [CrossRef]
- Liu, J.; Xue, N.; Guo, Y.; Niu, K.; Gao, L.; Zhang, S.; Gu, H.; Wang, X.; Zhao, D.; Fan, R. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging 2019, 11, 12412–12427. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Liang, B.; Jia, J.; Liang, N.; Xu, H.; Ju, G.; Ma, S.; Liu, X. Differential roles of miR-199a-5p in radiation-induced autophagy in breast cancer cells. FEBS Lett. 2013, 587, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, L.; Du, Y.; Mi, Y.; Wang, L. The HNF1A-AS1/miR-92a-3p axis affects the radiosensitivity of non-small cell lung cancer by competitively regulating the JNK pathway. Cell Biol. Toxicol. 2021, 37, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, L.; Li, D.; Xu, Y.; Zhang, L.; Niu, K.; Kong, R.; Gu, J.; Xu, Z.; Chen, Z.; et al. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1alpha. Cancer Med. 2018, 7, 3834–3847. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Ding, C.; Zhang, H.; Gao, J. Let-7 miRNAs sensitize breast cancer stem cells to radiation-induced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol. Med. Rep. 2016, 14, 3285–3292. [Google Scholar] [CrossRef] [PubMed]
- Tsogbadrakh, B.; Jung, J.A.; Lee, M.; Lee, J.A.; Seo, J.H. Identifying serum miRNA biomarkers for radiation exposure in hematopoietic humanized NSG-SGM3 mice. Biochem. Biophys. Res. Commun. 2022, 599, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Ni, B. hsa-miR-9-5p down-regulates HK2 and confers radiosensitivity to nasopharyngeal carcinoma. Technol. Cancer Res. Treat. 2021, 20, 1533033821997822. [Google Scholar] [CrossRef]
- Xu, S.; Li, X.; Li, L.; Wang, Y.; Geng, C.; Guo, F.; Zhang, T.; Du, A.; Lu, Z.; Hui, H.; et al. CTCF-silenced miR-137 contributes to EMT and radioresistance in esophageal squamous cell carcinoma. Cancer Cell. Int. 2021, 21, 155. [Google Scholar] [CrossRef]
- Du, R.; Jiang, F.; Yin, Y.; Xu, J.; Li, X.; Hu, L.; Wang, X. Knockdown of lncRNA X inactive specific transcript (XIST) radiosensitizes non-small cell lung cancer (NSCLC) cells through regulation of miR-16-5p/WEE1 G2 checkpoint kinase (WEE1) axis. Int. J. Immunopathol. Pharmacol. 2021, 35, 2058738420966087. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, W.; Wu, X.; Liu, W.; Ding, F. miR-16-5p modulates the radiosensitivity of cervical cancer cells via regulating coactivator-associated arginine methyltransferase 1. Pathol. Int. 2020, 70, 12–20. [Google Scholar] [CrossRef]
- Jin, Y.Y.; Chen, Q.J.; Wei, Y.; Wang, Y.L.; Wang, Z.W.; Xu, K.; He, Y.; Ma, H.B. Upregulation of microRNA-98 increases radiosensitivity in esophageal squamous cell carcinoma. J. Radiat. Res. 2016, 57, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Zhou, F.; Gui, S. Excessive miR-30a-5p increases the radiosensitivity of hepatoma cells by inhibiting GRP78. Trop. J. Pharm. Res. 2021, 21, 501–506. [Google Scholar] [CrossRef]
- Ge, Y.; Tu, W.; Li, J.; Chen, X.; Chen, Y.; Xu, Y.; Xu, Y.; Wang, Y.; Liu, Y. MiR-122-5p increases radiosensitivity and aggravates radiation-induced rectal injury through CCAR1. Toxicol. Appl. Pharmacol. 2020, 399, 115054. [Google Scholar] [CrossRef]
- Zhu, L.; Xue, F.; Cui, Y.; Liu, S.; Li, G.; Li, J.; Guan, B.; Zeng, H.; Bian, W.; Yang, C.; et al. miR-155-5p and miR-760 mediate radiation therapy suppressed malignancy of non-small cell lung cancer cells. Biofactors 2019, 45, 393–400. [Google Scholar] [CrossRef]
- Liu, H.M.; Tan, H.Y.; Lin, Y.; Xu, B.N.; Zhao, W.H.; Xie, Y.A. MicroRNA-1271-5p inhibits cell proliferation and enhances radiosensitivity by targeting CDK1 in hepatocellular carcinoma. J. Biochem. 2020, 167, 513–524. [Google Scholar] [CrossRef]
- Ma, Q.; Niu, R.; Huang, W.; Da, L.; Tang, Y.; Jiang, D.; Xi, Y.; Zhang, C. Long noncoding RNA PTPRG antisense RNA 1 reduces radiosensitivity of nonsmall cell lung cancer cells via regulating miR-200c-3p/TCF4. Technol. Cancer Res. Treat. 2020, 19, 1533033820942615. [Google Scholar] [CrossRef]
- Liu, H.; Chen, Q.; Zheng, W.; Zhou, Y.; Bai, Y.; Pan, Y.; Zhang, J.; Shao, C. LncRNA CASC19 enhances the radioresistance of nasopharyngeal carcinoma by regulating the miR-340-3p/FKBP5 axis. Int. J. Mol. Sci. 2023, 24, 3047. [Google Scholar] [CrossRef]
- Pan, D.; Du, Y.; Li, R.; Shen, A.; Liu, X.; Li, C.; Hu, B. miR-29b-3p increases radiosensitivity in stemness cancer cells via modulating oncogenes axis. Front. Cell. Dev. Biol. 2021, 9, 741074. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, H.; Wei, R.; Liu, Z.; Chen, H.; Guan, X.; Zhao, Z.; Wang, X.; Jiang, Z. LncRNA EGOT/miR-211-5p affected radiosensitivity of rectal cancer by competitively regulating ErbB4. OncoTargets Ther. 2021, 14, 2867–2878. [Google Scholar] [CrossRef]
- Ji, D.; Zhan, T.; Li, M.; Yao, Y.; Jia, J.; Yi, H.; Qiao, M.; Xia, J.; Zhang, Z.; Ding, H.; et al. Enhancement of sensitivity to chemo/radiation therapy by using miR-15b against DCLK1 in colorectal cancer. Stem Cell Rep. 2018, 11, 1506–1522. [Google Scholar] [CrossRef]
- Jiang, Y.; Jin, S.; Tan, S.; Xue, Y.; Cao, X. Long noncoding RNA NEAT1 regulates radio-sensitivity via microRNA-27b-3p in gastric cancer. Cancer Cell Int. 2020, 20, 581. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Xie, W.; Sun, W.; Sun, W.; Liao, Y. A concise review of microRNA-383: Exploring the insights of its function in tumorigenesis. J. Cancer 2022, 13, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Si, Z.Z.; Li, T.; Li, J.Q.; Zhang, Z.Q.; Chen, G.S.; Qi, H.Z.; Yao, H.L. MicroRNA-146a-5p enhances radiosensitivity in hepatocellular carcinoma through replication protein A3-induced activation of the DNA repair pathway. Am. J. Physiol. Cell. Physiol. 2019, 316, C299–C311. [Google Scholar] [CrossRef]
- Gong, T.; Jia, B.; Gu, L.; Yu, T. KLF5-trancripted miR-125b-5p is involved in enhancing the radio-sensitivity of breast cancer cells by targeting BRCA1. Mol. Cell. Toxicol. 2022, 18, 101–110. [Google Scholar] [CrossRef]
- Jiang, C.; Liu, F.; Xiao, S.; He, L.; Wu, W.; Zhao, Q. miR-29a-3p enhances the radiosensitivity of oral squamous cell carcinoma cells by inhibiting ADAM12. Eur. J. Histochem. 2021, 65, 3295. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Dong, M.; Liu, Z.; Yang, J.; Shi, Y. MiR-499a-5p inhibits proliferation, invasion, migration, and epithelial-mesenchymal transition, and enhances radiosensitivity of cervical cancer cells via targeting eIF4E. OncoTargets Ther. 2020, 13, 2913–2924. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, D.; Wu, H.; Li, P.; Zhao, W.; Yang, X.; Xing, X.; Li, S.; Li, J. Circular RNA PRKCI silencing represses esophageal cancer progression and elevates cell radiosensitivity through regulating the miR-186-5p/PARP9 axis. Life Sci. 2020, 259, 118168. [Google Scholar] [CrossRef]
- Li, H.; Zhao, S.; Chen, X.; Feng, G.; Chen, Z.; Fan, S. MiR-145 modulates the radiosensitivity of non-small cell lung cancer cells by suppression of TMOD3. Carcinogenesis 2022, 43, 288–296. [Google Scholar] [CrossRef]
- Rana, S.; Espinosa-Diez, C.; Ruhl, R.; Chatterjee, N.; Hudson, C.; Fraile-Bethencourt, E.; Agarwal, A.; Khou, S.; Thomas, C.R., Jr.; Anand, S. Differential regulation of microRNA-15a by radiation affects angiogenesis and tumor growth via modulation of acid sphingomyelinase. Sci. Rep. 2020, 10, 5581. [Google Scholar] [CrossRef]
- Song, Y.; Zuo, Y.; Qian, X.L.; Chen, Z.P.; Wang, S.K.; Song, L.; Peng, L.P. Inhibition of microRNA-21-5p promotes the radiation sensitivity of non-small cell lung cancer through HMSH2. Cell Physiol. Biochem. 2017, 43, 1258–1272. [Google Scholar] [CrossRef]
- Ye, T.; Zhong, L.; Ye, X.; Liu, J.; Li, L.; Yi, H. miR-221-3p and miR-222-3p regulate the SOCS3/STAT3 signaling pathway to downregulate the expression of NIS and reduce radiosensitivity in thyroid cancer. Exp. Ther. Med. 2021, 21, 652. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Li, Y.; He, Y.; Zeng, B.; Yi, C.; Wang, C.; Zhang, X.; Zhao, W.; Yu, D. Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol. Ther. Nucleic Acids 2020, 19, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.J.; Chen, Y.Y.; Dai, J.J.; Gu, D.N.; Mei, Z.; Liu, F.R.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 68. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shen, C.; Zhang, Y.; Hu, C. LncRNA ANRIL negatively regulated chitooligosaccharide-induced radiosensitivity in colon cancer cells by sponging miR-181a-5p. Adv. Clin. Exp. Med. 2021, 30, 55–65. [Google Scholar] [CrossRef]
- Aryankalayil, M.J.; Bylicky, M.A.; Martello, S.; Chopra, S.; Sproull, M.; May, J.M.; Shankardass, A.; MacMillan, L.; Vanpouille-Box, C.; Dalo, J.; et al. Microarray analysis identifies coding and non-coding RNA markers of liver injury in whole body irradiated mice. Sci. Rep. 2023, 13, 200. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Han, X.; Wang, J.; Wang, L.; Xu, Z.; Wei, Q.; Zhang, W.; Wang, H. miR-22 enhances the radiosensitivity of small-cell lung cancer by targeting the WRNIP1. J. Cell. Biochem. 2019, 120, 17650–17661. [Google Scholar] [CrossRef]
- Wang, B.; Wang, K.; Jin, T.; Xu, Q.; He, Y.; Cui, B.; Wang, Y. NCK1-AS1 enhances glioma cell proliferation, radioresistance and chemoresistance via miR-22-3p/IGF1R ceRNA pathway. Biomed. Pharmacother. 2020, 129, 110395. [Google Scholar] [CrossRef]
- Cao, K.; Li, J.; Chen, J.; Qian, L.; Wang, A.; Chen, X.; Xiong, W.; Tang, J.; Tang, S.; Chen, Y.; et al. microRNA-33a-5p increases radiosensitivity by inhibiting glycolysis in melanoma. Oncotarget 2017, 8, 83660–83672. [Google Scholar] [CrossRef]
- Xia, W.; Zhu, J.; Tang, Y.; Wang, X.; Wei, X.; Zheng, X.; Hou, M.; Li, S. PD-L1 inhibitor regulates the miR-33a-5p/PTEN signaling pathway and can be targeted to sensitize glioblastomas to radiation. Front. Oncol. 2020, 10, 821. [Google Scholar] [CrossRef]
- Li, Z.; Ye, L.; Wang, L.; Quan, R.; Zhou, Y.; Li, X. Identification of miRNA signatures in serum exosomes as a potential biomarker after radiotherapy treatment in glioma patients. Ann. Diagn. Pathol. 2020, 44, 151436. [Google Scholar] [CrossRef]
- Chen, F.; Yin, S.; Feng, Z.; Liu, C.; Lv, J.; Chen, Y.; Shen, R.; Wang, J.; Deng, Z. Knockdown of circ_NEK6 Decreased (131)I resistance of differentiated thyroid carcinoma via regulating miR-370-3p/MYH9 axis. Technol. Cancer Res. Treat. 2021, 20, 15330338211004950. [Google Scholar] [CrossRef]
- Qin, H.; Li, X.; Zhang, W.; Ding, Z. LncRNA OGFRP1 promotes cell proliferation and suppresses cell radiosensitivity in gastric cancer by targeting the miR-149-5p/MAP3K3 axis. J. Mol. Histol. 2022, 53, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.C.; Hsu, J.H.; Lai, L.C.; Tsai, M.H.; Chuang, E.Y. MicroRNA-107 enhances radiosensitivity by suppressing granulin in PC-3 prostate cancer cells. Sci. Rep. 2020, 10, 14584. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, H.; Xiong, Y.; Liu, J. Exosomal miR-199a-5p derived from endothelial cells attenuates apoptosis and inflammation in neural cells by inhibiting endoplasmic reticulum stress. Brain Res. 2020, 1726, 146515. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Luan, Y.; Li, J.; Song, H.; Li, Y.; Qi, H.; Sun, B.; Zhang, P.; Wu, X.; Liu, X.; et al. Exosomal miR-199a-5p promotes hepatic lipid accumulation by modulating MST1 expression and fatty acid metabolism. Hepatol. Int. 2020, 14, 1057–1074. [Google Scholar] [CrossRef] [PubMed]
- Mensa, E.; Guescini, M.; Giuliani, A.; Bacalini, M.G.; Ramini, D.; Corleone, G.; Ferracin, M.; Fulgenzi, G.; Graciotti, L.; Prattichizzo, F.; et al. Small extracellular vesicles deliver miR-21 and miR-217 as pro-senescence effectors to endothelial cells. J. Extracell. Vesicles 2020, 9, 1725285. [Google Scholar] [CrossRef]
- Dai, J.; Dong, R.; Han, X.; Li, J.; Gong, X.; Bai, Y.; Kang, F.; Liang, M.; Zeng, F.; Hou, Z.; et al. Osteoclast-derived exosomal let-7a-5p targets Smad2 to promote the hypertrophic differentiation of chondrocytes. Am. J. Physiol. Cell Physiol. 2020, 319, C21–C33. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Ren, J.; Qi, S. Exosomal miR-9-5p secreted by bone marrow-derived mesenchymal stem cells alleviates osteoarthritis by inhibiting syndecan-1. Cell Tissue Res. 2020, 381, 99–114. [Google Scholar] [CrossRef]
- Tao, K.; Liu, J.; Liang, J.; Xu, X.; Xu, L.; Mao, W. Vascular endothelial cell-derived exosomal miR-30a-5p inhibits lung adenocarcinoma malignant progression by targeting CCNE2. Carcinogenesis 2021, 42, 1056–1067. [Google Scholar] [CrossRef]
- Long, R.; Gao, L.; Li, Y.; Li, G.; Qin, P.; Wei, Z.; Li, D.; Qian, C.; Li, J.; Yang, G. M2 macrophage-derived exosomes carry miR-1271-5p to alleviate cardiac injury in acute myocardial infarction through down-regulating SOX6. Mol. Immunol. 2021, 136, 26–35. [Google Scholar] [CrossRef]
- Su, T.; Xiao, Y.; Xiao, Y.; Guo, Q.; Li, C.; Huang, Y.; Deng, Q.; Wen, J.; Zhou, F.; Luo, X.H. Bone marrow mesenchymal stem cells-derived exosomal miR-29b-3p regulates aging-associated insulin resistance. ACS Nano 2019, 13, 2450–2462. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhou, K.; Sun, M.; Shu, R.; Qian, J.; Xie, Y. The miR-223-3p regulates pyroptosis through NLRP3-Caspase 1-GSDMD signal axis in periodontitis. Inflammation 2021, 44, 2531–2542. [Google Scholar] [CrossRef]
- Wei, M.; Li, C.; Yan, Z.; Hu, Z.; Dong, L.; Zhang, J.; Wang, X.; Li, Y.; Zhang, H. Activated microglia exosomes mediated miR-383-3p promotes neuronal necroptosis through inhibiting ATF4 expression in intracerebral hemorrhage. Neurochem. Res. 2021, 46, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Quan, Y.; Wang, Z.; Gong, L.; Peng, X.; Richard, M.A.; Zhang, J.; Fornage, M.; Alcorn, J.L.; Wang, D. Exosome miR-371b-5p promotes proliferation of lung alveolar progenitor type II cells by using PTEN to orchestrate the PI3K/Akt signaling. Stem Cell Res. Ther. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Liang, H.X.; Wu, S.H.; Jiang, H.Q.; Wang, Q.; Yu, Z.J. Overexpressed tumor suppressor exosomal miR-15a-5p in cancer cells inhibits PD1 expression in CD8+T cells and suppresses the hepatocellular carcinoma progression. Front. Oncol. 2021, 11, 622263. [Google Scholar] [CrossRef]
- Li, J.; Yuan, H.; Xu, H.; Zhao, H.; Xiong, N. Hypoxic cancer-secreted exosomal miR-182-5p promotes glioblastoma angiogenesis by targeting kruppel-like factor 2 and 4. Mol. Cancer Res. 2020, 18, 1218–1231. [Google Scholar] [CrossRef]
- Bongolo, C.C.; Thokerunga, E.; Yan, Q.; Yacouba, M.B.M.; Wang, C. Exosomes derived from microRNA-27a-3p overexpressing mesenchymal stem cells inhibit the progression of liver cancer through suppression of golgi membrane protein 1. Stem Cells Int. 2022, 2022, 9748714. [Google Scholar] [CrossRef]
- Lu, X.; Lu, J.; Wang, S.; Zhang, Y.; Ding, Y.; Shen, X.; Jing, R.; Ju, S.; Chen, H.; Cong, H. Circulating serum exosomal miR-92a-3p as a novel biomarker for early diagnosis of gastric cancer. Future Oncol. 2021, 17, 907–919. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, D.; Cui, M.; Qu, F.; Yu, J.; Tang, Z.; Wang, L. Exosome marker proteins of tumor-associated fibroblasts and exosome-derived miR-92a-3p act as potential biomarkers for liver cancer. Crit. Rev. Eukaryot. Gene Expr. 2022, 32, 49–57. [Google Scholar] [CrossRef]
- Zhou, C.F.; Ma, J.; Huang, L.; Yi, H.Y.; Zhang, Y.M.; Wu, X.G.; Yan, R.M.; Liang, L.; Zhong, M.; Yu, Y.H.; et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene 2019, 38, 1256–1268. [Google Scholar] [CrossRef]
- Li, Y.Y.; Tao, Y.W.; Gao, S.; Li, P.; Zheng, J.M.; Zhang, S.E.; Liang, J.; Zhang, Y. Cancer-associated fibroblasts contribute to oral cancer cells proliferation and metastasis via exosome-mediated paracrine miR-34a-5p. EBioMedicine 2018, 36, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.L.; Luo, Y.P.; Lin, M.W.; Peng, X.X.; Liu, M.L.; Wang, Y.C.; Li, S.J.; Yang, D.H.; Yang, Z.X. Serum exosomal miR-16-5p functions as a tumor inhibitor and a new biomarker for PD-L1 inhibitor-dependent immunotherapy in lung adenocarcinoma by regulating PD-L1 expression. Cancer Med. 2022, 11, 2627–2643. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Wise, P.M.; Murtadha, M.; Liu, C.W.; Wu, C.H.; Jong, A.Y.; Seeger, R.C.; Fabbri, M. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res. 2019, 79, 1151–1164. [Google Scholar] [CrossRef]
- Yang, Q.; Wei, B.; Peng, C.; Wang, L.; Li, C. Identification of serum exosomal miR-98-5p, miR-183-5p, miR-323-3p and miR-19b-3p as potential biomarkers for glioblastoma patients and investigation of their mechanisms. Curr. Res. Transl. Med. 2022, 70, 103315. [Google Scholar] [CrossRef]
- Duan, B.; Shi, S.; Yue, H.; You, B.; Shan, Y.; Zhu, Z.; Bao, L.; You, Y. Exosomal miR-17-5p promotes angiogenesis in nasopharyngeal carcinoma via targeting BAMBI. J. Cancer 2019, 10, 6681–6692. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, G.; Xia, Q.; Shao, S.; Fang, H. Exosomal miR-200 family as serum biomarkers for early detection and prognostic prediction of cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2019, 12, 3870–3876. [Google Scholar]
- Han, Q.; Tan, S.; Gong, L.; Li, G.; Wu, Q.; Chen, L.; Du, S.; Li, W.; Liu, X.; Cai, J.; et al. Omental cancer-associated fibroblast-derived exosomes with low microRNA-29c-3p promote ovarian cancer peritoneal metastasis. Cancer Sci. 2023, 114, 1929–1942. [Google Scholar] [CrossRef]
- Yu, F.; Lin, Y.; Tan, G.; Ai, M.; Gong, H.; Liu, W.; Huang, J.; Zou, Z. Tumor-derived exosomal microRNA-15b-5p augments laryngeal cancer by targeting TXNIP. Cell Cycle 2022, 21, 730–740. [Google Scholar] [CrossRef]
- Zhang, B.; Sun, C.; Liu, Y.; Bai, F.; Tu, T.; Liu, Q. Exosomal miR-27b-3p derived from hypoxic cardiac microvascular endothelial cells alleviates rat myocardial ischemia/reperfusion injury through inhibiting oxidative stress-induced pyroptosis via Foxo1/GSDMD signaling. Oxid. Med. Cell. Longev. 2022, 2022, 8215842. [Google Scholar] [CrossRef]
- Ren, W.; Zhang, X.; Li, W.; Feng, Q.; Feng, H.; Tong, Y.; Rong, H.; Wang, W.; Zhang, D.; Zhang, Z.; et al. Exosomal miRNA-107 induces myeloid-derived suppressor cell expansion in gastric cancer. Cancer Manag. Res. 2019, 11, 4023–4040. [Google Scholar] [CrossRef]
- Wu, M.; Tan, X.; Liu, P.; Yang, Y.; Huang, Y.; Liu, X.; Meng, X.; Yu, B.; Wu, Y.; Jin, H. Role of exosomal microRNA-125b-5p in conferring the metastatic phenotype among pancreatic cancer cells with different potential of metastasis. Life Sci. 2020, 255, 117857. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.D.; Xu, Z.Y.; Hu, C.; Lv, H.; Xie, H.X.; Huang, T.; Zhang, Y.Q.; Chen, G.P.; Fu, Y.F.; Cheng, X.D. Exosomal miR-590-5p in serum as a biomarker for the diagnosis and prognosis of gastric cancer. Front. Mol. Biosci. 2021, 8, 636566. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Wang, L.; Wu, J.; Wang, Y.; Wen, H.; Zhu, X.; Wang, B.; Yang, H. miR-370-3p as a novel biomarker promotes breast cancer progression by targeting FBLN5. Stem Cells Int. 2021, 2021, 4649890. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yang, Z.; He, L. Effect of miR-29a-3p in exosomes on glioma cells by regulating the PI3K/AKT/HIF-1alpha pathway. Mol. Med. Rep. 2023, 27, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Cheng, J.; Li, B.; Nie, D.; Li, C.; Gui, S.; Wang, H.; Zhang, Y. Up-regulation of the expressions of MiR-149-5p and MiR-99a-3p in exosome inhibits the progress of pituitary adenomas. Cell Biol. Toxicol. 2021, 37, 633–651. [Google Scholar] [CrossRef]
- He, S.; Li, Z.; Yu, Y.; Zeng, Q.; Cheng, Y.; Ji, W.; Xia, W.; Lu, S. Exosomal miR-499a-5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Exp. Cell Res. 2019, 379, 203–213. [Google Scholar] [CrossRef]
- Huyan, T.; Gao, L.; Gao, N.; Wang, C.; Guo, W.; Zhou, X.; Li, Q. miR-221-5p and miR-186-5p are the critical bladder cancer derived exosomal miRNAs in natural killer cell dysfunction. Int. J. Mol. Sci. 2022, 23, 15177. [Google Scholar] [CrossRef]
- Wan, F.Z.; Chen, K.H.; Sun, Y.C.; Chen, X.C.; Liang, R.B.; Chen, L.; Zhu, X.D. Exosomes overexpressing miR-34c inhibit malignant behavior and reverse the radioresistance of nasopharyngeal carcinoma. J. Transl. Med. 2020, 18, 12. [Google Scholar] [CrossRef]
- Wang, Z.G.; Deng, M.S.; Su, J.Q.; Liu, D.B.; Zhou, Y. Exosomal miR-181a-5p derived from SAOS-2 cells promotes macrophages M2 polarization by targeting RORA. Kaohsiung J. Med. Sci. 2023, 39, 124–133. [Google Scholar] [CrossRef]
- Hang, W.; Feng, Y.; Sang, Z.; Yang, Y.; Zhu, Y.; Huang, Q.; Xi, X. Downregulation of miR-145-5p in cancer cells and their derived exosomes may contribute to the development of ovarian cancer by targeting CT. Int. J. Mol. Med. 2019, 43, 256–266. [Google Scholar] [CrossRef]
- Kim, H.; Rhee, W.J. Exosome-mediated Let7c-5p delivery for breast cancer therapeutic development. Biotechnol. Bioprocess. Eng. 2020, 25, 513–520. [Google Scholar] [CrossRef]
- Cho, H.J.; Eun, J.W.; Baek, G.O.; Seo, C.W.; Ahn, H.R.; Kim, S.S.; Cho, S.W.; Cheong, J.Y. Serum exosomal microRNA, miR-10b-5p, as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. J. Clin. Med. 2020, 9, 281. [Google Scholar] [CrossRef]
- Wu, J. Pancreatic cancer-derived exosomes promote the proliferation, invasion, and metastasis of pancreatic cancer by the miR-3960/TFAP2A axis. J. Oncol. 2022, 2022, 3590326. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.M.; Guan, Z.; Yang, Z.J.; Ma, L.Y.; Dai, Y.J.; Liang, C.; Hu, J.T. Comprehensive analysis of M2 macrophage-derived exosomes facilitating osteogenic differentiation of human periodontal ligament stem cells. BMC Oral Health 2022, 22, 647. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Zhao, C.; Wang, R.; Ren, L.; Qiu, H.; Zou, Z.; Ding, H.; Sun, Z.; Li, J.; Dong, S. Antagonizing exosomal miR-18a-5p derived from prostate cancer cells ameliorates metastasis-induced osteoblastic lesions by targeting Hist1h2bc and activating Wnt/β-catenin pathway. Genes Dis. 2022, 10, 1626–1640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, C. Exosomes miR-22-3p derived from mesenchymal stem cells suppress colorectal cancer cell proliferation and invasion by regulating RAP2B and PI3K/AKT pathway. J. Oncol. 2021, 2021, 3874478. [Google Scholar] [CrossRef]
- Chen, J.; Ding, C.; Yang, X.; Zhao, J. BMSCs-derived exosomal miR-126-3p inhibits the viability of NSCLC cells by targeting PTPN9. J. BUON 2021, 26, 1832–1841. [Google Scholar]
- Jiang, Y.; Liu, J.; Chen, L.; Jin, Y.; Zhang, G.; Lin, Z.; Du, S.; Fu, Z.; Chen, T.; Qin, Y.; et al. Serum secreted miR-137-containing exosomes affects oxidative stress of neurons by regulating OXR1 in Parkinson’s disease. Brain Res. 2019, 1722, 146331. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhang, L.; Li, J.; He, Y.; Zhang, X.; Li, J. Exosomal miR-122-5p inhibits tumorigenicity of gastric cancer by downregulating GIT1. Int. J. Biol. Markers 2021, 36, 36–46. [Google Scholar]
- Ni, Q.; Zhang, H.; Shi, X.; Li, X. Exosomal microRNA-23a-3p contributes to the progression of cholangiocarcinoma by interaction with Dynamin3. Bioengineered 2022, 13, 6208–6221. [Google Scholar]
- Shi, S.S.; Zhang, H.P.; Yang, C.Q.; Li, L.N.; Shen, Y.; Zhang, Y.Q. Exosomal miR-155-5p promotes proliferation and migration of gastric cancer cells by inhibiting TP53INP1 expression. Pathol. Res. Pract. 2020, 216, 152986. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Hosokawa, M.; Miyamoto, T.; Nakagawa, A.; Haruna, M.; Ueda, K.; Iwakawa, S.; Ogawara, K.I. miR-33a-5p in small extracellular vesicles as non-invasive biomarker for oxaliplatin sensitivity in human colorectal cancer cells. Biochem. Biophys. Rep. 2021, 26, 100996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, Q.; Zhu, S.; Tang, Y.; Chen, Y.; Chen, D.; Liang, Z. Chemerin-induced down-regulation of placenta-derived exosomal miR-140-3p and miR-574-3p promotes umbilical vein endothelial cells proliferation, migration, and tube formation in gestational diabetes mellitus. Cells 2022, 11, 3457. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Li, C.; Cao, L.; Zhang, C.H.; Zhang, Z.H. Exosomal miR-132-3p from mesenchymal stem cells alleviated LPS-induced acute lung injury by repressing TRAF6. Autoimmunity 2021, 54, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Chen, Y.; Chen, H.; Zhao, Q.; Sun, Z.; Liu, D.; Li, X.; Zhang, Y.; Wang, J.; Xing, H.R. Exosomal miR-211-5p regulates glucose metabolism, pyroptosis, and immune microenvironment of melanoma through GNA15. Pharmacol. Res. 2023, 188, 106660. [Google Scholar] [CrossRef]
- Mao, S.; Zheng, S.; Lu, Z.; Wang, X.; Wang, Y.; Zhang, G.; Xu, H.; Huang, J.; Lei, Y.; Liu, C.; et al. Exosomal miR-375-3p breaks vascular barrier and promotes small cell lung cancer metastasis by targeting claudin-1. Transl. Lung Cancer Res. 2021, 10, 3155–3172. [Google Scholar] [CrossRef]
- Song, H.; Zhang, X.; Chen, R.; Miao, J.; Wang, L.; Cui, L.; Ji, H.; Liu, Y. Cortical neuron-derived exosomal microRNA-181c-3p inhibits neuroinflammation by downregulating CXCL1 in astrocytes of a rat model with ischemic brain injury. Neuroimmunomodulation 2019, 26, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Yuwen, D.L.; Sheng, B.B.; Liu, J.; Wenyu, W.; Shu, Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2650–2658. [Google Scholar] [PubMed]
- Xiong, Q.H.; Zhao, L.; Wan, G.Q.; Hu, Y.G.; Li, X.L. Engineered BMSCs-derived exosomal miR-542-3p promotes cutaneous wound healing. Endocr. Metab. Immune Disord. Drug Targets 2023, 23, 336–346. [Google Scholar] [PubMed]
- Xie, Y.; Jia, Y.; Cuihua, X.; Hu, F.; Xue, M.; Xue, Y. Urinary exosomal microRNA profiling in incipient type 2 diabetic kidney disease. J. Diabetes Res. 2017, 2017, 6978984. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, K.; Li, L.; Zheng, B.; Zhang, Q.; Zhang, F.; Chen, J.; Wang, S. Plasma exosomal miR-1260a, miR-7977 and miR-192-5p as diagnostic biomarkers in epithelial ovarian cancer. Future Oncol. 2022, 18, 2919–2931. [Google Scholar] [CrossRef]
- Zhao, S.; Li, J.; Zhang, G.; Wang, Q.; Wu, C.; Zhang, Q.; Wang, H.; Sun, P.; Xiang, R.; Yang, S. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell Physiol. Biochem. 2019, 53, 19–35. [Google Scholar]
- Jin, J.; Wang, Y.; Zhao, L.; Zou, W.; Tan, M.; He, Q. Exosomal miRNA-215-5p derived from adipose-derived stem cells attenuates epithelial-mesenchymal transition of podocytes by inhibiting ZEB2. BioMed Res. Int. 2020, 2020, 2685305. [Google Scholar] [CrossRef]
- Gao, X.; Gao, L.F.; Kong, X.Q.; Zhang, Y.N.; Jia, S.; Meng, C.Y. Mesenchymal stem cell-derived extracellular vesicles carrying miR-99b-3p restrain microglial activation and neuropathic pain by stimulating autophagy. Int. Immunopharmacol. 2023, 115, 109695. [Google Scholar] [CrossRef]
- Xuan, Z.; Chen, C.; Tang, W.; Ye, S.; Zheng, J.; Zhao, Y.; Shi, Z.; Zhang, L.; Sun, H.; Shao, C. TKI-resistant renal cancer secretes low-level exosomal miR-549a to induce vascular permeability and angiogenesis to promote tumor metastasis. Front. Cell Dev. Biol. 2021, 9, 689947. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Dong, Z.; Chen, T. MiR-1247-5p functions as a tumor suppressor in human astroglioma cells by targeting CDC14B. Ann. Clin. Lab. Sci. 2020, 50, 182–189. [Google Scholar] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Bult, C.J.; Blake, J.A.; Smith, C.L.; Kadin, J.A.; Richardson, J.E.; Mouse Genome Database Group. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019, 47, D801–D806. [Google Scholar] [CrossRef]
- Sharma, N.K. Modulation of radiation-induced and mitomycin C-induced chromosome damage by apigenin in human lymphocytes in vitro. J. Radiat. Res. 2013, 54, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Eldemerdash, O.M.; Elshaffei, I.M.; Yousef, E.M.; Soliman, A.S.; Senousy, M.A. Apigenin attenuates hippocampal microglial activation and restores cognitive function in methotrexate-treated rats: Targeting the miR-15a/ROCK-1/ERK1/2 pathway. Mol. Neurobiol. 2023, 60, 3770–3787. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, K.S.; Shuaib, M.; Gupta, S.; Kumar, S. Withaferin A mediated changes of miRNA expression in breast cancer-derived mammospheres. Mol. Carcinog. 2022, 61, 876–889. [Google Scholar] [CrossRef]
- Qi, J.; Li, J.; Bie, B.; Shi, M.; Zhu, M.; Tian, J.; Zhu, K.; Sun, J.; Mu, Y.; Li, Z.; et al. miR-3,178 contributes to the therapeutic action of baicalein against hepatocellular carcinoma cells via modulating HDAC10. Phytother. Res. 2023, 37, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Yang, T.Y.; Lu, H.J.; Wan, C.K.; Hsu, S.L.; Wu, C.C. Attenuating role of withaferin A in the proliferation and migration of lung cancer cells via a p53-miR-27a/miR-10b pathway. Oncol. Lett. 2021, 21, 232. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Gu, J.; Yu, Y. Celastrol assuages oxygen-glucose deprivation and reoxygenation-induced damage in human brain microvascular endothelial cells through the circDLGAP4/miR-6085/GDF11 pathway. Metab. Brain Dis. 2023, 38, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.Z.; Moawed, F.S.M.; Elmarkaby, S.M. Protective effect of 5, 7-dihydroxyflavone on brain of rats exposed to acrylamide or gamma-radiation. J. Photochem. Photobiol. B 2017, 175, 149–155. [Google Scholar] [CrossRef]
- Lin, C.M.; Wang, B.W.; Pan, C.M.; Fang, W.J.; Chua, S.K.; Cheng, W.P.; Shyu, K.G. Chrysin boosts KLF2 expression through suppression of endothelial cell-derived exosomal microRNA-92a in the model of atheroprotection. Eur. J. Nutr. 2021, 60, 4345–4355. [Google Scholar] [CrossRef] [PubMed]
- Mohammadian, F.; Pilehvar-Soltanahmadi, Y.; Alipour, S.; Dadashpour, M.; Zarghami, N. Chrysin alters microRNAs expression levels in gastric cancer cells: Possible molecular mechanism. Drug Res. 2017, 67, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Kabacik, S.; Manning, G.; Raffy, C.; Bouffler, S.; Badie, C. Time, dose and ataxia telangiectasia mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiat. Res. 2015, 183, 325–337. [Google Scholar] [CrossRef]
- Cinkilic, N.; Cetintas, S.K.; Zorlu, T.; Vatan, O.; Yilmaz, D.; Cavas, T.; Tunc, S.; Ozkan, L.; Bilaloglu, R. Radioprotection by two phenolic compounds: Chlorogenic and quinic acid, on X-ray induced DNA damage in human blood lymphocytes in vitro. Food Chem. Toxicol. 2013, 53, 359–363. [Google Scholar] [CrossRef]
- Li, J.; Ge, H.; Xu, Y.; Xie, J.; Karim, N.; Yan, F.; Mo, J.; Chen, W. Chlorogenic acid alleviates oxidative damage in hepatocytes by regulating miR-199a-5p/GRP78 axis. Food Biosci. 2023, 53, 102595. [Google Scholar] [CrossRef]
- Nadeem, R.I.; Aboutaleb, A.S.; Younis, N.S.; Ahmed, H.I. Diosmin mitigates gentamicin-induced nephrotoxicity in rats: Insights on miR-21 and -155 expression, Nrf2/HO-1 and p38-MAPK/NF-kappaB pathways. Toxics 2023, 11, 48. [Google Scholar] [CrossRef]
- Jin, Y.; Zou, X.; Feng, X. 3,3′-Diindolylmethane negatively regulates Cdc25A and induces a G2/M arrest by modulation of microRNA 21 in human breast cancer cells. Anticancer Drugs 2010, 21, 814–822. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Huang, M.; Tang, C.; Yue, Y.; Liu, X.; Zheng, Z.; Dong, H.; Liu, D. Dietary daidzein inhibits hepatitis C virus replication by decreasing microRNA-122 levels. Virus Res. 2021, 298, 198404. [Google Scholar] [CrossRef] [PubMed]
- Mahgoub, S.; Sallam, A.O.; Sarhan, H.K.A.; Ammar, A.A.A.; Soror, S.H. Role of Diosmin in protection against the oxidative stress induced damage by gamma-radiation in Wistar albino rats. Regul. Toxicol. Pharmacol. 2020, 113, 104622. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.M.; Kim, S.H.; Kang, B.S. Radioprotective effects of delphinidin on normal human lung cells against proton beam exposure. Nutr. Res. Pract. 2018, 12, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Nonaka, H.; Komatsu, S.; Goto, M.; Morozumi, M.; Yamada, S.; Lin, I.C.; Yamashita, S.; Tachibana, H. Delphinidin prevents muscle atrophy and upregulates miR-23a expression. J. Agric. Food Chem. 2017, 65, 45–50. [Google Scholar] [CrossRef]
- Huang, C.C.; Hung, C.H.; Hung, T.W.; Lin, Y.C.; Wang, C.J.; Kao, S.H. Dietary delphinidin inhibits human colorectal cancer metastasis associating with upregulation of miR-204-3p and suppression of the integrin/FAK axis. Sci. Rep. 2019, 9, 18954. [Google Scholar] [CrossRef]
- Hasan, H.F.; Abdel-Rafei, M.K.; Galal, S.M. Diosmin attenuates radiation-induced hepatic fibrosis by boosting PPAR-gamma expression and hampering miR-17-5p-activated canonical Wnt-beta-catenin signaling. Biochem. Cell Biol. 2017, 95, 400–414. [Google Scholar] [CrossRef]
- Yi, J.; Chen, C.; Liu, X.; Kang, Q.; Hao, L.; Huang, J.; Lu, J. Radioprotection of EGCG based on immunoregulatory effect and antioxidant activity against 60Coγ radiation-induced injury in mice. Food Chem. Toxicol. 2020, 135, 111051. [Google Scholar] [CrossRef]
- Zhang, C.; Gan, X.; Liang, R.; Jian, J. Exosomes derived from epigallocatechin gallate-treated cardiomyocytes attenuated acute myocardial infarction by modulating MicroRNA-30a. Front. Pharmacol. 2020, 11, 126. [Google Scholar] [CrossRef]
- Abdullaev, S.A.; Glukhov, S.I.; Gaziev, A.I. Radioprotective and radiomitigative effects of melatonin in tissues with different proliferative activity. Antioxidants 2021, 10, 1885. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Lu, Z.; Ji, C.; Chen, Y.; Liu, Y.; Lei, Z.; Wang, L.; Zhang, H.T.; Li, X. Melatonin inhibits proliferation and invasion via repression of miRNA-155 in glioma cells. Biomed. Pharmacother. 2017, 93, 969–975. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kang, H.S.; Lee, J.H.; Park, J.H.; Jung, C.H.; Bae, J.H.; Oh, B.C.; Song, D.K.; Baek, W.K.; Im, S.S. Melatonin ameliorates ER stress-mediated hepatic steatosis through miR-23a in the liver. Biochem. Biophys. Res. Commun. 2015, 458, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Teksoy, O.; Sahinturk, V.; Cengiz, M.; Inal, B.; Ayhanci, A. The protective effects of silymarin on thioacetamide-induced liver damage: Measurement of miR-122, miR-192, and miR-194 levels. Appl. Biochem. Biotechnol. 2020, 191, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Shanthakumar, J.; Karthikeyan, A.; Bandugula, V.R.; Rajendra Prasad, N. Ferulic acid, a dietary phenolic acid, modulates radiation effects in Swiss albino mice. Eur. J. Pharmacol. 2012, 691, 268–274. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Guo, Y.; Zhao, J.; Xu, X.; Wang, N.; Liu, Q. Ferulic acid ameliorates lipopolysaccharide-induced barrier dysfunction via microRNA-200c-3p-mediated activation of PI3K/AKT pathway in Caco-2 cells. Front. Pharmacol. 2020, 11, 376. [Google Scholar] [CrossRef]
- Du, K.; Li, Z.; Fang, X.; Cao, T.; Xu, Y. Ferulic acid promotes osteogenesis of bone marrow-derived mesenchymal stem cells by inhibiting microRNA-340 to induce beta-catenin expression through hypoxia. Eur. J. Cell Biol. 2017, 96, 496–503. [Google Scholar] [CrossRef]
- Mahran, Y.F.; Badr, A.M.; Aldosari, A.; Bin-Zaid, R.; Alotaibi, H.N. Carvacrol and thymol modulate the cross-talk between tNF-alpha and IGF-1 signaling in radiotherapy-induced ovarian failure. Oxid. Med. Cell. Longev. 2019, 2019, 3173745. [Google Scholar] [CrossRef]
- Hussein, R.M.; Arafa, E.A.; Raheem, S.A.; Mohamed, W.R. Thymol protects against bleomycin-induced pulmonary fibrosis via abrogation of oxidative stress, inflammation, and modulation of miR-29a/TGF-beta and PI3K/Akt signaling in mice. Life Sci. 2023, 314, 121256. [Google Scholar] [CrossRef]
- Chen, P.; Li, X.; Yu, X.; Yang, M. Ginsenoside Rg1 suppresses non-small-cell lung cancer via microRNA-126-PI3K-AKT-mTOR pathway. Evid. Based Complement. Altern. Med. 2022, 2022, 1244836. [Google Scholar] [CrossRef]
- Maurya, D.K.; Salvi, V.P.; Krishnan Nair, C.K. Radioprotection of normal tissues in tumor-bearing mice by troxerutin. J. Radiat. Res. 2004, 45, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Shu, L.L.; Zhang, W.Z.; Huang, G.C.; Huang, C.; Zhu, X.H.; Su, G.; Xu, J. Troxerutin attenuates myocardial cell apoptosis following myocardial ischemia-reperfusion injury through inhibition of miR-146a-5p expression. J. Cell. Physiol. 2019, 234, 9274–9282. [Google Scholar] [CrossRef]
- Hu, Y.J.; Song, G.Y.; Zhang, F.; Zhang, N.; Wang, F.; Wang, J.L.; Wang, X.; Wang, T.Y.; Li, Y.F.; Yan, Y.D.; et al. Activation of long-non-coding RNA NEAT1 sponging microRNA-147 inhibits radiation damage by targeting PDPK1 in troxerutin radioprotection. iScience 2023, 26, 105932. [Google Scholar] [CrossRef]
- Sato, T.; Kinoshita, M.; Yamamoto, T.; Ito, M.; Nishida, T.; Takeuchi, M.; Saitoh, D.; Seki, S.; Mukai, Y. Treatment of irradiated mice with high-dose ascorbic acid reduced lethality. PLoS ONE 2015, 10, e0117020. [Google Scholar] [CrossRef]
- Kolhe, R.; Mondal, A.K.; Pundkar, C.; Periyasamy-Thandavan, S.; Mendhe, B.; Hunter, M.; Isales, C.M.; Hill, W.D.; Hamrick, M.W.; Fulzele, S. Modulation of miRNAs by vitamin C in human bone marrow stromal cells. Nutrients 2018, 10, 186. [Google Scholar] [CrossRef]
- Zheng, H.; Wang, S.; Zhou, P.; Liu, W.; Ni, F. Effects of Ligustrazine on DNA damage and apoptosis induced by irradiation. Environ. Toxicol. Pharmacol. 2013, 36, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, S.; Xia, L. Ligustrazine inhibits the proliferation and migration of ovarian cancer cells via regulating miR-211. Biosci. Rep. 2021, 41, BSR20200199. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, L.; Huang, X.; Sun, Q. Ligustrazine promotes hypoxia/reoxygenation-treated trophoblast cell proliferation and migration by regulating the microRNA-27a-3p/ATF3 axis. Arch. Biochem. Biophys. 2023, 737, 109522. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhao, L.; Wang, X.Y.; Lian, F.; Cai, Y. Ligustrazine-induced microRNA-16-5p inhibition alleviates preeclampsia through IGF-2. Reproduction 2020, 160, 905–917. [Google Scholar] [CrossRef]
- Ruknarong, L.; Boonthongkaew, C.; Chuangchot, N.; Jumnainsong, A.; Leelayuwat, N.; Jusakul, A.; Gaudieri, S.; Leelayuwat, C. Vitamin C supplementation reduces expression of circulating miR-451a in subjects with poorly controlled type 2 diabetes mellitus and high oxidative stress. PeerJ 2021, 9, e10776. [Google Scholar] [CrossRef]
- Srinivasan, M.; Devipriya, N.; Kalpana, K.B.; Menon, V.P. Lycopene: An antioxidant and radioprotector against gamma-radiation-induced cellular damages in cultured human lymphocytes. Toxicology 2009, 262, 43–49. [Google Scholar] [CrossRef]
- Li, D.; Chen, L.; Zhao, W.; Hao, J.; An, R. MicroRNA-let-7f-1 is induced by lycopene and inhibits cell proliferation and triggers apoptosis in prostate cancer. Mol. Med. Rep. 2016, 13, 2708–2714. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.Y.; Li, X.N.; Zhao, Y.; Dai, X.Y.; Guo, J.Y.; Li, J.L. Lycopene ameliorate atrazine-induced oxidative damage in the B cell zone via targeting the miR-27a-3p/Foxo1 axis. J. Agric. Food Chem. 2022, 70, 12502–12512. [Google Scholar] [CrossRef] [PubMed]
- Rao, B.N.; Archana, P.R.; Aithal, B.K.; Rao, B.S. Protective effect of zingerone, a dietary compound against radiation induced genetic damage and apoptosis in human lymphocytes. Eur. J. Pharmacol. 2011, 657, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Mou, R.; Li, Y.; Yang, T. Zingerone promotes osteoblast differentiation via miR-200c-3p/smad7 regulatory axis in human bone mesenchymal stem cells. Med. Sci. Monit. 2020, 26, e919309. [Google Scholar] [CrossRef]
- Ghelishli, N.; Ghasemi, A.; Hosseinimehr, S.J. The influence of piperine on the radioprotective effect of curcumin in irradiated human lymphocytes. Turk. J. Pharm. Sci. 2019, 16, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Rajarajan, D.; Natesh, J.; Penta, D.; Meeran, S.M. Dietary piperine suppresses obesity-associated breast cancer growth and metastasis by regulating the miR-181c-3p/PPARα Axis. J. Agric. Food Chem. 2021, 69, 15562–15574. [Google Scholar] [CrossRef]
- Ding, W.; Gu, Q.; Liu, M.; Zou, J.; Sun, J.; Zhu, J. Astrocytes-derived exosomes pre-treated by berberine inhibit neuroinflammation after stroke via miR-182-5p/Rac1 pathway. Int. Immunopharmacol. 2023, 118, 110047. [Google Scholar] [CrossRef]
- Abdelhamid, A.M.; Selim, A.; Zaafan, M.A. The hepatoprotective effect of piperine against thioacetamide-induced liver fibrosis in mice: The involvement of miR-17 and TGF-beta/Smads pathways. Front. Mol. Biosci. 2021, 8, 754098. [Google Scholar] [CrossRef]
- Guo, X.; Hu, S.; Liu, J.J.; Huang, L.; Zhong, P.; Fan, Z.X.; Ye, P.; Chen, M.H. Piperine protects against pyroptosis in myocardial ischaemia/reperfusion injury by regulating the miR-383/RP105/AKT signalling pathway. J. Cell. Mol. Med. 2021, 25, 244–258. [Google Scholar] [CrossRef]
- Eder-Czembirek, C.; Erovic, B.M.; Czembirek, C.; Brunner, M.; Selzer, E.; Potter, R.; Thurnher, D. Betulinic acid a radiosensitizer in head and neck squamous cell carcinoma cell lines. Strahlenther. Onkol. 2010, 186, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Chintharlapalli, S.; Papineni, S.; Lei, P.; Pathi, S.; Safe, S. Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 2011, 11, 371. [Google Scholar] [CrossRef]
- Carsten, R.E.; Bachand, A.M.; Bailey, S.M.; Ullrich, R.L. Resveratrol reduces radiation-induced chromosome aberration frequencies in mouse bone marrow cells. Radiat. Res. 2008, 169, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Alharris, E.; Alghetaa, H.; Seth, R.; Chatterjee, S.; Singh, N.P.; Nagarkatti, M.; Nagarkatti, P. Resveratrol attenuates allergic asthma and associated inflammation in the lungs through regulation of miRNA-34a that targets FoxP3 in mice. Front. Immunol. 2018, 9, 2992. [Google Scholar] [CrossRef] [PubMed]
- Venkatadri, R.; Muni, T.; Iyer, A.K.; Yakisich, J.S.; Azad, N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Dis. 2016, 7, e2104. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, Z.; Li, B.; Nie, J.; Tong, J.; Zhang, Z. Radioprotection of vitamin D on mice injured by irradiation. J. Radiat. Res. Radiat. Process. 2008, 26, 122–124. [Google Scholar]
- Mohamed, D.I.; Abou-Bakr, D.A.; Ezzat, S.F.; El-Kareem, H.F.A.; Nahas, H.H.A.; Saad, H.A.; Mehana, A.E.; Saied, E.M. Vitamin D3 prevents the deleterious effects of testicular torsion on testis by targeting miRNA-145 and ADAM17: In silico and in vivo study. Pharmaceuticals 2021, 14, 1222. [Google Scholar] [CrossRef]
- Chang, S.; Gao, Z.; Yang, Y.; He, K.; Wang, X.; Wang, L.; Gao, N.; Li, H.; He, X.; Huang, C. miR-99b-3p is induced by vitamin D3 and contributes to its antiproliferative effects in gastric cancer cells by targeting HoxD3. Biol. Chem. 2019, 400, 1079–1086. [Google Scholar] [CrossRef]
- Fu, R.; Liu, S.; Zhu, M.; Zhu, J.; Chen, M. Apigenin reduces the suppressive effect of exosomes derived from irritable bowel syndrome patients on the autophagy of human colon epithelial cells by promoting ATG14. World J. Surg. Oncol. 2023, 21, 95. [Google Scholar] [CrossRef]
- Yildiz, O.G.; Soyuer, S.; Saraymen, R.; Eroglu, C. Protective effects of caffeic acid phenethyl ester on radiation induced lung injury in rats. Clin. Investig. Med. 2008, 31, E242–E247. [Google Scholar] [CrossRef]
- Mo, F.; Luo, Y.; Fan, D.; Zeng, H.; Zhao, Y.; Luo, M.; Liu, X.; Ma, X. Integrated analysis of mRNA-seq and miRNA-seq to identify c-MYC, YAP1 and miR-3960 as major players in the anticancer effects of caffeic acid phenethyl ester in human small cell lung cancer cell line. Curr. Gene Ther. 2020, 20, 15–24. [Google Scholar] [CrossRef]
- Liang, Y.; Yang, A.Y.; Liu, M.; Cheng, Y.J.; Zang, S.B.; Huang, J.; Tang, Y.Y.; Huang, Z.P. Effect of carvacrol on the biological behavior of leukemia cells and its mechanism. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2022, 30, 393–399. [Google Scholar]
- Xiao, Z.; Su, Y.; Yang, S.; Yin, L.; Wang, W.; Yi, Y.; Fenton, B.M.; Zhang, L.; Okunieff, P. Protective effect of esculentoside A on radiation-induced dermatitis and fibrosis. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Okunieff, P.; Xu, J.; Hu, D.; Liu, W.; Zhang, L.; Morrow, G.; Pentland, A.; Ryan, J.L.; Ding, I. Curcumin protects against radiation-induced acute and chronic cutaneous toxicity in mice and decreases mRNA expression of inflammatory and fibrogenic cytokines. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 890–898. [Google Scholar] [CrossRef] [PubMed]
- Brockmueller, A.; Samuel, S.M.; Mazurakova, A.; Büsselberg, D.; Kubatka, P.; Shakibaei, M. Curcumin, calebin A and chemosensitization: How are they linked to colorectal cancer? Life Sci. 2023, 38, 121504. [Google Scholar] [CrossRef]
- Yang, J.; Cao, Y.; Sun, J.; Zhang, Y. Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med. Oncol. 2010, 27, 1114–1118. [Google Scholar] [CrossRef]
- Liu, W.L.; Chang, J.M.; Chong, I.W.; Hung, Y.L.; Chen, Y.H.; Huang, W.T.; Kuo, H.F.; Hsieh, C.C.; Liu, P.L. Curcumin inhibits LIN-28A through the activation of miRNA-98 in the lung cancer cell line A549. Molecules 2017, 22, 929. [Google Scholar] [CrossRef]
- Pan, L.; Sha, J.; Lin, W.; Wang, Y.; Bian, T.; Guo, J. Curcumin inhibits prostate cancer progression by regulating the miR-30a-5p/PCLAF axis. Exp. Ther. Med. 2021, 22, 969. [Google Scholar] [CrossRef] [PubMed]
- Son, E.W.; Mo, S.J.; Rhee, D.K.; Pyo, S. Inhibition of ICAM-1 expression by garlic component, allicin, in gamma-irradiated human vascular endothelial cells via downregulation of the JNK signaling pathway. Int. Immunopharmacol. 2006, 6, 1788–1795. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, T.; Ti, X.; Shi, J.; Wu, C.; Ren, X.; Yin, H. Curcumin promotes apoptosis in A549/DDP multidrug-resistant human lung adenocarcinoma cells through an miRNA signaling pathway. Biochem. Biophys. Res. Commun. 2010, 399, 1–6. [Google Scholar] [CrossRef]
- Bebb, D.G.; Steele, P.P.; Warrington, P.J.; Moffat, J.A.; Glickman, B.W. Caffeine does not potentiate gamma-radiation induced DNA damage in ataxia telangiectasia lymphoblastoid cells. Mutat. Res. 1998, 401, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Meng, Q.; Xu, J.; Jiao, Y.; Zhao, L.; Zhang, X.; Sarkar, F.H.; Brown, M.L.; Dritschilo, A.; Rosen, E.M. DIM (3,3′-diindolylmethane) confers protection against ionizing radiation by a unique mechanism. Proc. Natl. Acad. Sci. USA 2013, 110, 18650–18655. [Google Scholar] [CrossRef] [PubMed]
- Geric, M.; Gajski, G.; Mihaljevic, B.; Miljanic, S.; Domijan, A.M.; Garaj-Vrhovac, V. Radioprotective properties of food colorant sodium copper chlorophyllin on human peripheral blood cells in vitro. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019, 845, 403027. [Google Scholar] [CrossRef]
- Li, N.; Wang, C.; Zhang, P.; You, S. Emodin inhibits pancreatic cancer EMT and invasion by up-regulating microRNA-1271. Mol. Med. Rep. 2018, 18, 3366–3374. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, J.; Moon, C.; Kim, S.H.; Hyun, J.W.; Park, J.W.; Shin, T. Radioprotective effects of fucoidan in mice treated with total body irradiation. Phytother. Res. 2008, 22, 1677–1681. [Google Scholar] [CrossRef]
- Yan, M.D.; Yao, C.J.; Chow, J.M.; Chang, C.L.; Hwang, P.A.; Chuang, S.E.; Whang-Peng, J.; Lai, G.M. Fucoidan elevates microRNA-29b to regulate DNMT3B-MTSS1 axis and inhibit EMT in human hepatocellular carcinoma cells. Mar. Drugs 2015, 13, 6099–6116. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Wu, A.T.; Yuan, K.S.; Liu, S.H. Brown seaweed fucoidan inhibits cancer progression by dual regulation of miR-29c/ADAM12 and miR-17-5p/PTEN axes in human breast cancer cells. J. Cancer 2016, 7, 2408–2419. [Google Scholar] [CrossRef]
- Malhotra, P.; Gupta, A.K.; Singh, D.; Mishra, S.; Singh, S.K.; Kumar, R. N-Acetyl-tryptophan glucoside (NATG) protects J774A.1 murine macrophages against gamma radiation-induced cell death by modulating oxidative stress. Mol. Cell. Biochem. 2018, 447, 9–19. [Google Scholar] [CrossRef]
- El-Far, Y.M.; Khodir, A.E.; Emarah, Z.A.; Ebrahim, M.A.; Al-Gayyar, M.M.H. Fucoidan ameliorates hepatocellular carcinoma induced in rats: Effect on miR143 and inflammation. Nutr. Cancer 2021, 73, 1498–1510. [Google Scholar] [CrossRef]
- Nair, G.G.; Nair, C.K. Radioprotective effects of gallic acid in mice. BioMed Res. Int. 2013, 2013, 953079. [Google Scholar] [CrossRef]
- Jabbari, N.; Feghhi, M.; Esnaashari, O.; Soraya, H.; Rezaie, J. Inhibitory effects of gallic acid on the activity of exosomal secretory pathway in breast cancer cell lines: A possible anticancer impact. Bioimpacts 2022, 12, 549–559. [Google Scholar] [CrossRef]
- Liang, W.; Li, X.; Li, Y.; Li, C.; Gao, B.; Gan, H.; Li, S.; Shen, J.; Kang, J.; Ding, S.; et al. Gallic acid induces apoptosis and inhibits cell migration by upregulating miR-518b in SW1353 human chondrosarcoma cells. Int. J. Oncol. 2014, 44, 91–98. [Google Scholar] [CrossRef]
- Sinha, P.; Annamalai, S.K.; Arunachalam, K.D. A wee study on behavioural, organ somatic index and histological alterations of the fresh water fish Pangasius sutchi in response to protection studies, exposed to gamma radiation perceived by genotoxic assays. Int. J. Pharm. Sci. Rev. Res. 2018, 50, 18–30. [Google Scholar]
- Davis, T.A.; Mungunsukh, O.; Zins, S.; Day, R.M.; Landauer, M.R. Genistein induces radioprotection by hematopoietic stem cell quiescence. Int. J. Radiat. Biol. 2008, 84, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Hidaka, H.; Majid, S.; Saini, S.; Arora, S.; Deng, G.; Shahryari, V.; Chang, I.; et al. Genistein up-regulates tumor suppressor microRNA-574-3p in prostate cancer. PLoS ONE 2013, 8, e58929. [Google Scholar] [CrossRef]
- Yang, H.J.; Youn, H.; Seong, K.M.; Yun, Y.J.; Kim, W.; Kim, Y.H.; Lee, J.Y.; Kim, C.S.; Jin, Y.W.; Youn, B. Psoralidin, a dual inhibitor of COX-2 and 5-LOX, regulates ionizing radiation (IR)-induced pulmonary inflammation. Biochem. Pharmacol. 2011, 82, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xiang, J.; Shen, J.; Zou, X.; Zhai, S.; Yin, Y.; Li, P.; Wang, X.; Sun, Q. Oncogenic MicroRNA-27a is a target for genistein in ovarian cancer cells. Anticancer Agents Med. Chem. 2013, 13, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- de la Parra, C.; Castillo-Pichardo, L.; Cruz-Collazo, A.; Cubano, L.; Redis, R.; Calin, G.A.; Dharmawardhane, S. Soy isoflavone genistein-mediated downregulation of miR-155 contributes to the anticancer effects of genistein. Nutr. Cancer 2016, 68, 154–164. [Google Scholar] [CrossRef]
- Ma, J.; Cheng, L.; Liu, H.; Zhang, J.; Shi, Y.; Zeng, F.; Miele, L.; Sarkar, F.H.; Xia, J.; Wang, Z. Genistein down-regulates miR-223 expression in pancreatic cancer cells. Curr. Drug Targets 2013, 14, 1150–1156. [Google Scholar] [CrossRef]
- Hosseinimehr, S.J.; Mahmoudzadeh, A.; Ahmadi, A.; Mohamadifar, S.; Akhlaghpoor, S. Radioprotective effects of hesperidin against genotoxicity induced by gamma-irradiation in human lymphocytes. Mutagenesis 2009, 24, 233–235. [Google Scholar] [CrossRef]
- Magura, J.; Moodley, R.; Mackraj, I. The effect of hesperidin and luteolin isolated from Eriocephalus africanus on apoptosis, cell cycle and miRNA expression in MCF-7. J. Biomol. Struct. Dyn. 2022, 40, 1791–1800. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Dai, L.; Tan, P.; Liu, W.; Mu, Y.; Wang, J.; Huang, X.; Hou, A. Hesperidin administration suppresses the proliferation of lung cancer cells by promoting apoptosis via targeting the miR-132/ZEB2 signalling pathway. Int. J. Mol. Med. 2020, 46, 2069–2077. [Google Scholar] [CrossRef] [PubMed]
- Keshava, C.; Keshava, N.; Ong, T.M.; Nath, J. Protective effect of vanillin on radiation-induced micronuclei and chromosomal aberrations in V79 cells. Mutat. Res. 1998, 397, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, P.; Zhao, H.; Dong, S.; Yang, Y.; Cui, J.; Gao, F.; Cai, J.; Liu, C. The mechanism for the radioprotective effects of zymosan-A in mice. J. Cell. Mol. Med. 2018, 22, 2413–2421. [Google Scholar] [CrossRef]
- Piao, M.J.; Kang, K.A.; Hyun, J.W. Protective effect of dieckol on γ-ray radiation-induced V79-4 lung fibroblast cell damage involved in modulation of reactive oxygen species. J. Med. Life Sci. 2009, 6, 368–372. [Google Scholar] [CrossRef]
- Xiao, J.; Liu, L.; Zhong, Z.; Xiao, C.; Zhang, J. Mangiferin regulates proliferation and apoptosis in glioma cells by induction of microRNA-15b and inhibition of MMP-9 expression. Oncol. Rep. 2015, 33, 2815–2820. [Google Scholar] [CrossRef]
- Chi, X.J.; Meng, J.J.; Lin, C.Y.; Su, Q.S.; Qin, Y.Y.; Wei, R.H.; Lan, D.; Huang, C. Mangiferin inhibits human lung adenocarcinoma by suppressing MiR-27b and MiR-92a. Evid. Based Complement. Altern. Med. 2021, 2021, 2822950. [Google Scholar] [CrossRef]
- Park, E.; Ahn, G.N.; Lee, N.H.; Kim, J.M.; Yun, J.S.; Hyun, J.W.; Jeon, Y.J.; Wie, M.B.; Lee, Y.J.; Park, J.W.; et al. Radioprotective properties of eckol against ionizing radiation in mice. FEBS Lett. 2008, 582, 925–930. [Google Scholar] [CrossRef]
- Li, J.; Xu, J.; Lu, Y.; Qiu, L.; Xu, W.; Lu, B.; Hu, Z.; Chu, Z.; Chai, Y.; Zhang, J. MASM, a matrine derivative, offers radioprotection by modulating lethal total-body irradiation-induced multiple signaling pathways in wistar rats. Molecules 2016, 21, 649. [Google Scholar] [CrossRef]
- Wei, Y.P.; Wang, X.H.; Liu, G.; Zhang, J.F.; Yang, Y.X.; Zhang, J.; Song, X.L.; Li, Z.D.; Zhao, L.D. Matrine exerts inhibitory effects in melanoma through the regulation of miR-19b-3p/PTEN. Int. J. Oncol. 2018, 53, 791–800. [Google Scholar] [CrossRef]
- Moon, C.; Kim, S.H.; Kim, J.C.; Hyun, J.W.; Lee, N.H.; Park, J.W.; Shin, T. Protective effect of phlorotannin components phloroglucinol and eckol on radiation-induced intestinal injury in mice. Phytother. Res. 2008, 22, 238–242. [Google Scholar] [CrossRef]
- Mendonca, M.S.; Chin-Sinex, H.; Gomez-Millan, J.; Datzman, N.; Hardacre, M.; Comerford, K.; Nakshatri, H.; Nye, M.; Benjamin, L.; Mehta, S.; et al. Parthenolide sensitizes cells to X-ray-induced cell killing through inhibition of NF-kappaB and split-dose repair. Radiat. Res. 2007, 168, 689–697. [Google Scholar] [CrossRef]
- Moeng, S.; Seo, H.A.; Hwang, C.Y.; Cipolla, G.A.; Lee, D.J.; Kuh, H.J.; Park, J.K. MicroRNA-107 targets IKBKG and sensitizes A549 cells to parthenolide. Anticancer Res. 2018, 38, 6309–6316. [Google Scholar] [CrossRef]
- Kang, K.A.; Zhang, R.; Lee, K.H.; Chae, S.; Kim, B.J.; Kwak, Y.S.; Park, J.W.; Lee, N.H.; Hyun, J.W. Protective effect of triphlorethol-A from Ecklonia cava against ionizing radiation in vitro. J. Radiat. Res. 2006, 47, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Moorthy, R.K.; Jayamohan, S.; Kannan, M.K.; Arockiam, A.J.V. Parthenolide induces apoptosis and cell cycle arrest by the suppression of miR-375 through nucleolin in prostate cancer. J. Pharm. Res. Int. 2021, 33, 215–227. [Google Scholar] [CrossRef]
- Horton, J.A.; Li, F.; Chung, E.J.; Hudak, K.; White, A.; Krausz, K.; Gonzalez, F.; Citrin, D. Quercetin inhibits radiation-induced skin fibrosis. Radiat. Res. 2013, 180, 205–215. [Google Scholar] [CrossRef]
- Kim, D.H.; Khan, H.; Ullah, H.; Hassan, S.T.S.; Smejkal, K.; Efferth, T.; Mahomoodally, M.F.; Xu, S.; Habtemariam, S.; Filosa, R.; et al. MicroRNA targeting by quercetin in cancer treatment and chemoprotection. Pharmacol. Res. 2019, 147, 104346. [Google Scholar] [CrossRef]
- Ahn, M.; Moon, C.; Yang, W.; Ko, E.J.; Hyun, J.W.; Joo, H.G.; Jee, Y.; Lee, N.H.; Park, J.W.; Ko, R.K.; et al. Diphlorethohydroxycarmalol, isolated from the brown algae Ishige okamurae, protects against radiation-induced cell damage in mice. Food Chem. Toxicol. 2011, 49, 864–870. [Google Scholar] [CrossRef]
- Thabet, N.M.; Moustafa, E.M. Protective effect of rutin against brain injury induced by acrylamide or gamma radiation: Role of PI3K/AKT/GSK-3beta/NRF-2 signalling pathway. Arch. Physiol. Biochem. 2018, 124, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, G.; Hu, J.; Zhu, Y.; Lan, H.; Shen, X.; Lv, Y.; Huang, L. Rutin attenuates sorafenib-induced chemoresistance and autophagy in hepatocellular carcinoma by regulating BANCR/miRNA-590-5P/OLR1 axis. Int. J. Biol. Sci. 2021, 17, 3595–3607. [Google Scholar] [CrossRef]
- Huo, M.; Xia, A.; Cheng, W.; Zhou, M.; Wang, J.; Shi, T.; Cai, C.; Jin, W.; Zhou, M.; Liao, Y.; et al. Rutin promotes pancreatic cancer cell apoptosis by upregulating miRNA-877-3p expression. Molecules 2022, 27, 2293. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xu, D.; Gu, Z.; Li, T.; Huang, P.; Ren, L. Rutin restrains the growth and metastasis of mouse breast cancer cells by regulating the microRNA-129-1-3p-mediated calcium signaling pathway. J. Biochem. Mol. Toxicol. 2021, 35, e22794. [Google Scholar] [CrossRef] [PubMed]
- Malyarenko, O.S.; Usoltseva, R.V.; Zvyagintseva, T.N.; Ermakova, S.P. Laminaran from brown alga Dictyota dichotoma and its sulfated derivative as radioprotectors and radiosensitizers in melanoma therapy. Carbohydr. Polym. 2019, 206, 539–547. [Google Scholar] [CrossRef]
- Prasad, N.R.; Menon, V.P.; Vasudev, V.; Pugalendi, K.V. Radioprotective effect of sesamol on gamma-radiation induced DNA damage, lipid peroxidation and antioxidants levels in cultured human lymphocytes. Toxicology 2005, 209, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wang, J.; Hu, G.; Chen, Y.; Hu, X.; Zhu, Y.; Ding, L.; Ning, S. Sesamol epigenetically induces estrogen receptor alpha re-expression by upregulating miR-370-3p in estrogen receptor alpha-negative breast cancer. J. Agric. Food Chem. 2021, 69, 8737–8746. [Google Scholar] [CrossRef]
- Ben-Amotz, A.; Yatziv, S.; Sela, M.; Greenberg, S.; Rachmilevich, B.; Shwarzman, M.; Weshler, Z. Effect of natural beta-carotene supplementation in children exposed to radiation from the Chernobyl accident. Radiat. Environ. Biophys. 1998, 37, 187–193. [Google Scholar] [CrossRef]
- Wang, H.; Sim, M.K.; Loke, W.K.; Chinnathambi, A.; Alharbi, S.A.; Tang, F.R.; Sethi, G. Potential protective effects of ursolic acid against gamma irradiation-induced damage are mediated through the modulation of diverse inflammatory mediators. Front. Pharmacol. 2017, 8, 352. [Google Scholar] [CrossRef]
- Xiang, F.; Fan, Y.; Ni, Z.; Liu, Q.; Zhu, Z.; Chen, Z.; Hao, W.; Yue, H.; Wu, R.; Kang, X. Ursolic acid reverses the chemoresistance of breast cancer cells to paclitaxel by targeting miRNA-149-5p/MyD88. Front. Oncol. 2019, 9, 501. [Google Scholar] [CrossRef]
- Lu, Q.; Chen, W.; Ji, Y.; Liu, Y.; Xue, X. Ursolic acid enhances cytotoxicity of doxorubicin-resistant triple-negative breast cancer cells via ZEB1-AS1/miR-186-5p/ABCC1 axis. Cancer Biother. Radiopharm. 2022, 37, 673–683. [Google Scholar] [CrossRef]
- Wei, X.; Lan, Y.; Nong, Z.; Li, C.; Feng, Z.; Mei, X.; Zhai, Y.; Zou, M. Ursolic acid represses influenza A virus-triggered inflammation and oxidative stress in A549 cells by modulating the miR-34c-5p/TLR5 axis. Cytokine 2022, 157, 155947. [Google Scholar] [CrossRef]
- Xue, X.L.; Han, X.D.; Li, Y.; Chu, X.F.; Miao, W.M.; Zhang, J.L.; Fan, S.J. Astaxanthin attenuates total body irradiation-induced hematopoietic system injury in mice via inhibition of oxidative stress and apoptosis. Stem Cell Res. Ther. 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Gamit, N.; Varier, L.; Dharmarajan, A.; Warrier, S. Inhibition of breast cancer stem-like cells by a triterpenoid, ursolic acid, via activation of Wnt antagonist, sFRP4 and suppression of miRNA-499a-5p. Life Sci. 2021, 265, 118854. [Google Scholar] [CrossRef]
- Aminin, D.L.; Zaporozhets, T.S.; Adryjashchenko, P.V.; Avilov, S.A.; Kalinin, V.I.; Stonik, V.A. Radioprotective properties of Cumaside, a complex of triterpene glycosides from the sea cucumber Cucumaria japonica and cholesterol. Nat. Prod. Commun. 2011, 6, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Guo, Y.; Cao, J. Matrine triggers colon cancer cell apoptosis and G0/G1 cell cycle arrest via mediation of microRNA-22. Phytother. Res. 2020, 34, 1619–1628. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Cui, S. Matrine inhibits TPC-1 human thyroid cancer cells via the miR-21/PTEN/Akt pathway. Oncol. Lett. 2018, 16, 2965–2970. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Duan, H.; Wang, W.; Zhao, S.; Khan, G.J.; Wang, M.; Zhang, Y.; Thakur, K.; Fang, X.; Wu, C.; et al. Ginsenoside Rg1 ameliorates blood-brain barrier disruption and traumatic brain injury via attenuating macrophages derived exosomes miR-21 release. Acta Pharm. Sin. B 2021, 11, 3493–3507. [Google Scholar] [CrossRef]
- Li, Y.; Lin, Q.; Chang, S.; Zhang, R.; Wang, J. Vitamin D3 mediates miR-15a-5p inhibition of liver cancer cell proliferation via targeting E2F3. Oncol. Lett. 2020, 20, 292–298. [Google Scholar] [CrossRef]
- Oh, J.Y.; Fernando, I.P.S.; Jeon, Y.J. Potential applications of radioprotective phytochemicals from marine algae. Algae 2016, 31, 403–414. [Google Scholar] [CrossRef]
- Dutta, S.; Wadekar, R.R.; Roy, T. Radioprotective natural products as alternative complements in oncological radiotherapy. Bol. Latinoam Caribe Plant Med. Aromat. 2021, 20, 101–122. [Google Scholar] [CrossRef]
- Abraham, R.E.; Alghazwi, M.; Liang, Q.; Zhang, W. Advances on marine-derived natural radioprotection compounds: Historic development and future perspective. Mar. Life Sci. Technol. 2021, 3, 474–487. [Google Scholar] [CrossRef]
- Heo, S.J.; Ko, S.C.; Cha, S.H.; Kang, D.H.; Park, H.S.; Choi, Y.U.; Kim, D.; Jung, W.K.; Jeon, Y.J. Effect of phlorotannins isolated from Ecklonia cava on melanogenesis and their protective effect against photo-oxidative stress induced by UV-B radiation. Toxicol. In Vitro 2009, 23, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Lyons, N.M.; O’Brien, N.M. Modulatory effects of an algal extract containing astaxanthin on UVA-irradiated cells in culture. J. Dermatol. Sci. 2002, 30, 73–84. [Google Scholar] [CrossRef]
- Lin, M.Y.; Chang, Y.C.; Wang, S.Y.; Yang, M.H.; Chang, C.H.; Hsiao, M.; Kitsis, R.N.; Lee, Y.J. OncomiR miR-182-5p enhances radiosensitivity by inhibiting the radiation-induced antioxidant effect through SESN2 in head and neck cancer. Antioxidants 2021, 10, 1808. [Google Scholar] [CrossRef] [PubMed]
- Munson, P.B.; Hall, E.M.; Farina, N.H.; Pass, H.I.; Shukla, A. Exosomal miR-16-5p as a target for malignant mesothelioma. Sci. Rep. 2019, 9, 11688. [Google Scholar] [CrossRef] [PubMed]
- Vicic, I.; Belev, B. The pathogenesis of bone metastasis in solid tumors: A review. Croat. Med. J. 2021, 62, 270–282. [Google Scholar] [CrossRef]
- Giudice, V.; Banaszak, L.G.; Gutierrez-Rodrigues, F.; Kajigaya, S.; Panjwani, R.; Ibanez, M.; Rios, O.; Bleck, C.K.; Stempinski, E.S.; Raffo, D.Q.; et al. Circulating exosomal microRNAs in acquired aplastic anemia and myelodysplastic syndromes. Haematologica 2018, 103, 1150–1159. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Akter, R.; Najda, A.; Rahman, M.H.; Shah, M.; Wesolowska, S.; Hassan, S.S.U.; Mubin, S.; Bibi, P.; Saeeda, S. Potential role of natural products to combat radiotherapy and their future perspectives. Molecules 2021, 26, 5997. [Google Scholar] [CrossRef]
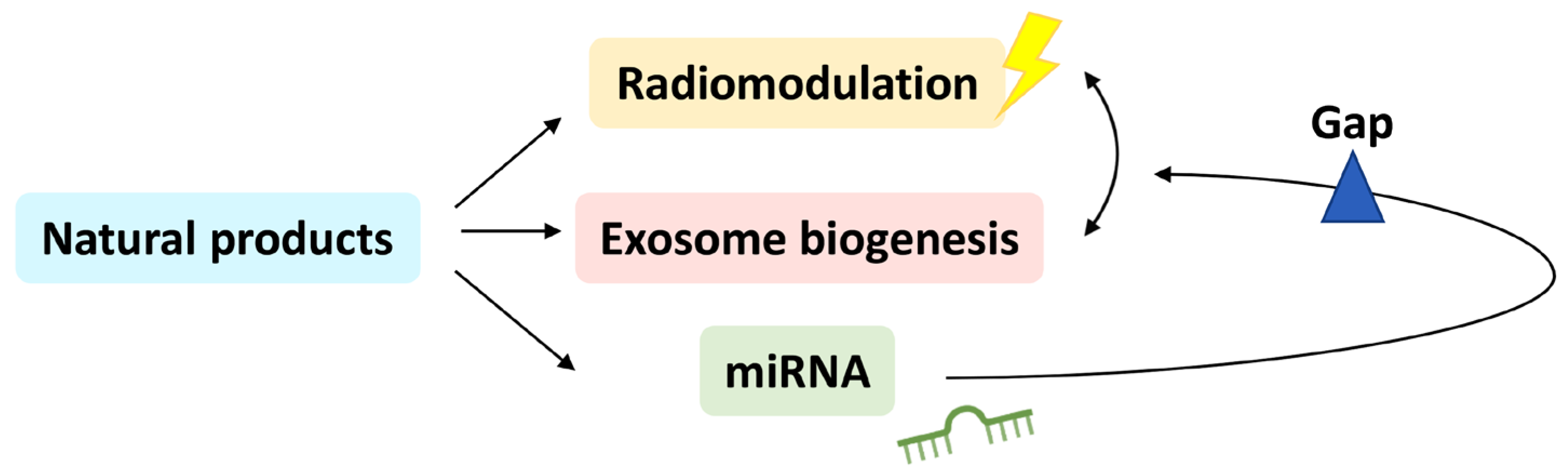
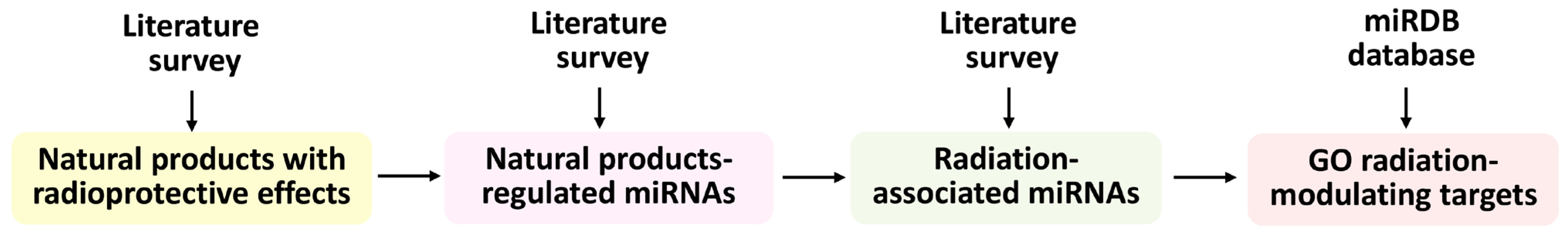

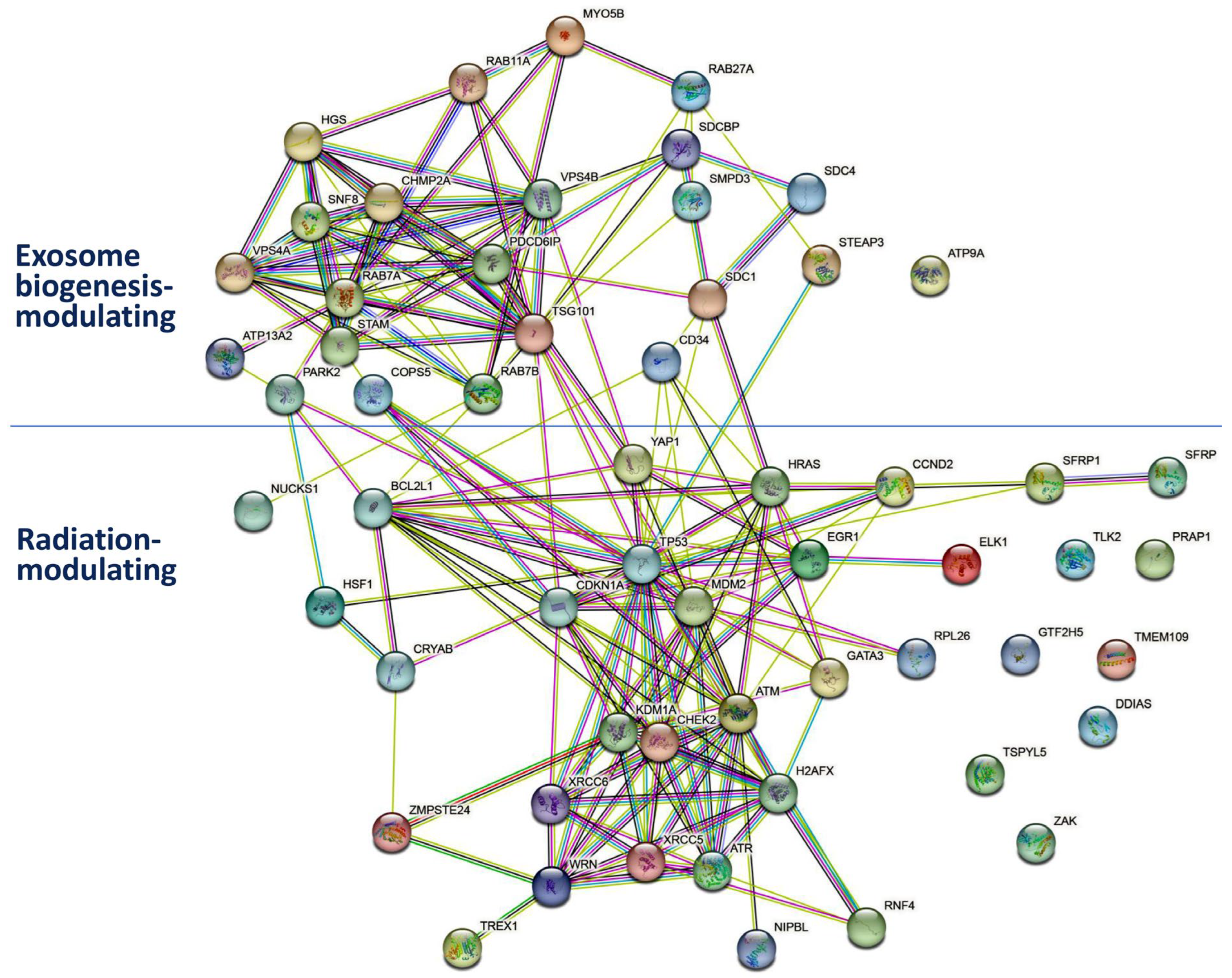
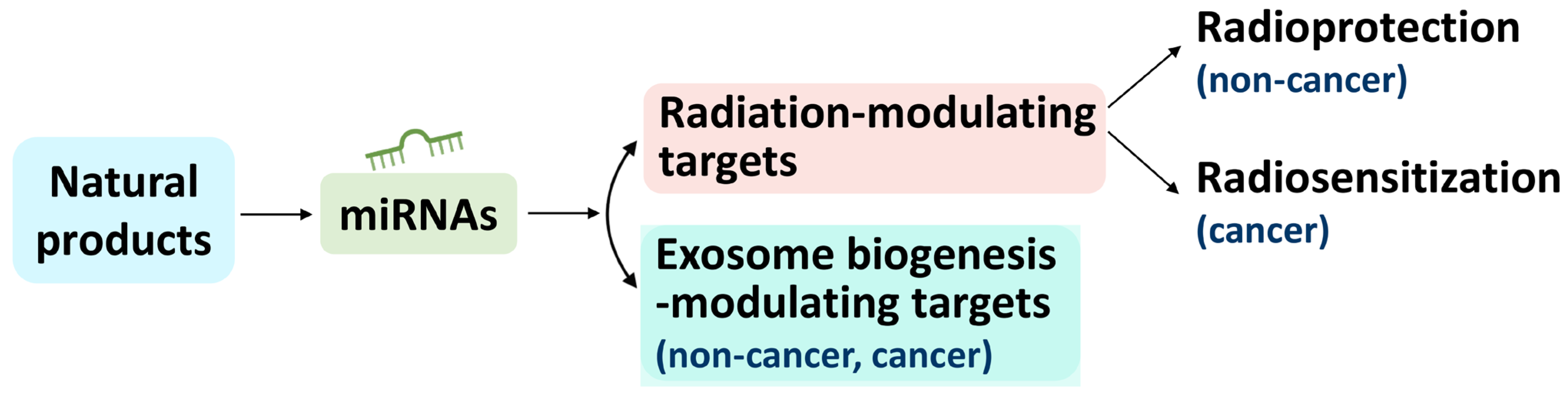
| Radioprotective Natural Product | Natural-Product-Regulated miRNAs | Radioprotective Natural Product | Natural-Product-Regulated miRNAs |
|---|---|---|---|
| Apigenin [169] | miR-15a-5p [170] | Withaferin A [16] | let-7c-5p, let-7a-5p [171] |
| Baicalein [10] | miR-3178 [172] | miR-549a-5p, miR-1247-5p, miR-124-5p, miR-137-5p [171], miR-10b-5p, miR-27a-3p [173] | |
| Celastrol [12] | miR-6085 [174] | Chrysin [175] | miR-92a-3p [176], let-7a-5p, miR-9-5p, miR-22-3p [177], miR-34a-5p [177,178], miR-126-3p [177] |
| Chlorogenic acid [179] | miR-199a-5p [180] | miR-18a-5p [177], miR-21-5p [181,182], miR-221-3p [177] | |
| Daidzein [5] | miR-122-5p [183] | ||
| Diosmin [184] | miR-21-5p [181,182] | Delphinidin [185] | miR-23a-3p [186], miR-204-3p [187] |
| miR-155-5p [181], miR-17-5p [188] | Epigallocatechin gallate (EGCG) [189] | miR-30a-5p [190] | |
| Melatonin [191] | miR-155-5p [192], miR-23a-3p [193] | ||
| Silymarin [5] | miR-122-5p, miR-192-5p, miR-194-5p [194] | Ferulic acid [195] | miR-200c-3p [196] |
| miR-340-3p [197] | |||
| Thymol [198] | miR-29a-3p [199] | Ginsenoside Rg1 [17] | miR-126-3p [200] |
| Troxerutin [201] | miR-146a-5p [202], miR-147a [203] | miR-21-5p [181,182] | |
| Vitamin C [204] | miR-215-3p, miR-215-5p, miR-371b-5p, miR-181a-5p [205] | Ligustrazine [206] | miR-211-5p [207], miR-27a-3p [208] |
| miR-16-5p [209] | |||
| miR-29b-1, miR-589-5p, miR-451a [210] | Lycopene [211] | let-7f-1-3p [212] | |
| miR-27a-3p [213] | |||
| Zingerone [214] | miR-200c-3p [215] | Piperine [216] | miR-181c-3p [217] |
| Berberine [9] | miR-182-5p [218] | miR-17-5p [219], miR-383-3p, miR-383-5p [220] | |
| Betulinic acid [221] | miR-27a-3p [222] | Resveratrol [223] | miR-34a-5p [224], miR-542-3p, miR-125b-5p [225] |
| Vitamin D [226] | miR-145-5p [227], miR-99b-3p [228], miR-15a-5p [170,228,229] | ||
| CAPE [230] | miR-3960 [231] | ||
| Carvacrol [198] | miR-217-3p, miR-217-5p [232] | Esculentoside A [233] | |
| Curcumin [234] | miR-137-3p, miR-137-5p [235], miR-16-5p [236], miR-98-5p [237], miR-30a-5p [238] | ||
| Allicin [239] | |||
| miR-186-3p [240] | Caffeine [241] | ||
| 3,3′-Diindolylmethane [242] | miR-21-5p [181,182] | Chlorophyllin [243] | |
| Emodin [15] | miR-1271-5p [244] | Dehydrozingerone [8] | |
| Fucoidan [245] | miR-29b-3p [246], miR-29c-3p [247] | N-Acetyl tryptophan glucopyranoside [248] | |
| miR-17-5p [249] | |||
| Gallic acid [250] | miR-182-5p [251], miR-518b [252] | Gymnemagenin [253] | |
| miR-21-5p [181,182] | |||
| Genistein [254] | miR-574-3p [255] | Psoralidin [256] | |
| miR-27a-3p [257], miR-155-5p [258], miR-223-3p [259] | Quinic acid [179] | ||
| Hesperidin [260] | miR-16-5p, miR-34a-5p [261], miR-132-3p [262] | Vanillin [263] | |
| Zymosan A [264] | |||
| miR-21-5p [181,182] | Dieckol [265] | ||
| Mangiferin [11] | miR-15b-5p [266], miR-27b-3p, miR-92a-3p [267] | Eckol [268] | |
| Matrine [269] | miR-22-3p [177], miR-19b-3p [270] | Phloroglucinol [271] | |
| miR-21-5p [181,182] | |||
| Parthenolide [272] | miR-107 [273] | Triphlorethol-A [274] | |
| miR-375-3p, miR-375-5p [275] | |||
| Quercetin [276] | let-7a-5p [277], miR-146a-5p [277] | Diphlorethohydroxycarmalol [278] | |
| miR-21-5p [181,182] | |||
| Rutin [279] | miR-590-5p [280], miR-877-3p [281], miR-129-1-3p [282] | ||
| Laminaran [283] | |||
| Sesamol [284] | miR-370-3p [285] | β-carotene [286] | |
| Ursolic acid [287] | miR-149-5p [288], miR-186-5p [289], miR-34c-5p [290] | Astaxanthin [291] | |
| miR-499a-5p [292] | Cumaside [293] |
| Natural-Product-Regulated miRNA | Radiation-Associated miRNA | Radiation-Modulating Targets | Natural-Product-Regulated miRNA | Radiation-Associated miRNA | Radiation-Modulating Targets |
|---|---|---|---|---|---|
| miR-17-5p | [52] | DDIAS, MAP3K20, CCND2 | miR-221-3p | [91] | RNF4, NIPBL, MDM2 |
| miR-518b | [53] | EGR1 | miR-23a-3p | [92] | MAP3K20, EGR1, RNF4 |
| miR-223-3p | [52] | MDM2 | miR-194-5p | [93] | SFRP2, YAP1, RNF4 |
| miR-34c-5p | [54] | TMEM109, GATA3 | miR-181a-5p | [94] | ATM, NIPBL, NUCKS1 |
| miR-375-3p | [55] | MAP3K20 | miR-34a-5p | [7,95] | TMEM109, GATA3 |
| miR-877-3p | [56] | GTF2H5, KDM1A, WRN | miR-107 | [103] | TSPYL5 |
| miR-147a | [57] | MDM2 | miR-3960 | [95] | |
| miR-19b-3p | [58] | CCND2, MAP3K20 | miR-217-3p | [61] | |
| miR-124-5p | [59] | DDIAS, YAP1, TLK2 | miR-22-3p | [96,97] | |
| miR-182-5p | [178,303] | MAP3K20, CCND2 | miR-126-3p | [52] | |
| miR-27a-3p | [60] | SFRP1, TLK2, GATA3, YAP1 | miR-137-5p | [68] | |
| miR-217-5p | [61] | NIPBL, ZMPSTE24 | miR-33a-5p | [98,99] | |
| miR-199a-5p | [62] | NUCKS1 | miR-574-3p | [100] | |
| miR-92a-3p | [63] | RNF4, MAP3K20, NIPBL | miR-370-3p | [101] | |
| miR-18a-5p | [64] | ATM, RNF4, CCND2 | miR-149-5p | [102] | |
| let-7a-5p | [65,66] | CCND2, TSPYL5, BCL2L1 | miR-451a | [52] | |
| miR-9-5p | [67] | TMEM109, MAP3K20 | miR-186-3p | GTF2H5, TSPYL5 | |
| miR-137-3p | [68] | MAP3K20, RNF4, KDM1A, NIPBL, NUCKS1 | miR-204-3p | MAP3K20 | |
| miR-16-5p | [69,70] | CCND2, YAP1, NUCKS1 | miR-29c-3p | CCND2, MDM2 | |
| miR-98-5p | [71] | CCND2, TSPYL5, BCL2L1 | miR-132-3p | MAP3K20, EGR1, TSPYL5, NUCKS1 | |
| miR-30a-5p | [72] | NUCKS1 | let-7f-1-3p | GATA3, SFRP2, MAP3K20, MDM2 | |
| miR-122-5p | [73] | MAP3K20 | miR-542-3p | SFRP1 | |
| miR-155-5p | [74] | MAP3K20, GTF2H5, GATA3, MDM2 | miR-590-5p | YAP1 | |
| miR-1271-5p | [75] | NIPBL, MAP3K20, CCND2 | miR-192-5p | GTF2H5, NIPBL | |
| miR-200c-3p | [76] | YAP1 | miR-215-3p | TLK2, MAP3K20 | |
| miR-340-3p | [77] | MDM2 | miR-215-5p | GTF2H5, NIPBL | |
| miR-29b-3p | [78] | CCND2, MDM2 | miR-371b-5p | MDM2, KDM1A | |
| miR-211-5p | [79] | CCND2, NIPBL, GTF2H5, EGR1 | miR-589-5p | RNF4 | |
| miR-15b-5p | [80] | CCND2, YAP1, NUCKS1 | let-7c-5p | CCND2, TSPYL5, BCL2L1 | |
| miR-27b-3p | [81] | SFRP1, TLK2, GATA3, YAP1 | miR-10b-5p | GATA3 | |
| miR-383-3p | [82] | CRYAB, CCND2, GATA3, NUCKS1, MAP3K20, MDM2 | miR-3178 | ||
| miR-383-5p | [82] | ATR | miR-6085 | ||
| miR-146a-5p | [83] | NUCKS1, YAP1, RNF4 | miR-375-5p | ||
| miR-125b-5p | [84] | TLK2 | miR-181c-3p | ||
| miR-29a-3p | [85] | CCND2, MDM2 | miR-129-1-3p | ||
| miR-499a-5p | [86] | GATA3 | miR-29b-1 | ||
| miR-186-5p | [87] | YAP1, NIPBL, NUCKS1 | miR-99b-3p | ||
| miR-145-5p | [88] | MDM2 | miR-549a-5p | ||
| miR-15a-5p | [89] | CCND2, YAP1, NUCKS1 | miR-1247-5p | ||
| miR-21-5p | [90] | YAP1 | miR-137-5p |
| Natural-Product-Regulated miRNA | Exosome Biogenesis-Associated miRNA | Exosome Biogenesis-Modulating Targets | Natural-Product-Regulated miRNA | Exosome Biogenesis-Associated miRNA | Exosome Biogenesis-Modulating Targets |
|---|---|---|---|---|---|
| miR-199a-5p | [104,105] | ATP13A2, RAB7A, ATP9A | miR-3960 | [143] | |
| miR-21-5p | [106] | RAB11A, MYO5B | miR-6085 | [144] | |
| let-7a-5p | [107] | STEAP3, MYO5B | miR-18a-5p | [145] | |
| miR-9-5p | [108] | STEAP3, PDCD6IP, STAM, SDC1, SMPD3, CD34 | miR-22-3p | [146] | |
| miR-30a-5p | [109] | RAB11A | miR-126-3p | [147] | |
| miR-1271-5p | [110] | RAB27A, RAB7A, MYO5B | miR-137-3p | [148] | |
| miR-29b-3p | [111] | SMPD3 | miR-122-5p | [149] | |
| miR-223-3p | [112] | MYO5B, STAM | miR-23a-3p | [150] | |
| miR-383-3p | [113] | SMPD3, TSG101, SDCBP | miR-155-5p | [151] | |
| miR-371b-5p | [114] | RAB11A, STAM, SDCBP | miR-33a-5p | [152] | |
| miR-15a-5p | [115] | MYO5B, VPS4A | miR-574-3p | [153] | |
| miR-182-5p | [116] | RAB7A, ATP9A, SDC1 | miR-132-3p | [154] | |
| miR-27a-3p | [117] | SMPD3 | miR-211-5p | [155] | |
| miR-217-3p | [106] | VPS4B | miR-375-3p | [156] | |
| miR-92a-3p | [118,119] | VPS4B | miR-181c-3p | [157] | |
| miR-221-3p | [120] | PDCD6IP | miR-146a-5p | [158] | |
| miR-34a-5p | [121] | VPS4A | miR-542-3p | [159] | |
| miR-16-5p | [122] | MYO5B, VPS4A | miR-877-3p | [160] | |
| miR-186-3p | [123] | VPS4B | miR-192-5p | [161] | |
| miR-98-5p | [124] | STEAP3, MYO5B | miR-451a | [162] | |
| miR-17-5p | [125] | TSG101, MYO5B | miR-215-5p | [163] | |
| miR-200c-3p | [126] | PRKN, STAM | miR-99b-3p | [164] | |
| miR-29c-3p | [127] | SMPD3 | miR-549a-5p | [165] | |
| miR-15b-5p | [128] | MYO5B, VPS4A | miR-1247-5p | [166] | |
| miR-27b-3p | [129] | SMPD3 | miR-217-5p | ATP9A, STEAP3, PDCD6IP | |
| miR-19b-3p | [124] | SDC1, VPS4B, MYO5B | miR-204-3p | RAB11A | |
| miR-107 | [130] | VPS4A, SDCBP | miR-340-3p | RAB11A | |
| miR-125b-5p | [131] | VPS4B | let-7f-1-3p | SDCBP, RAB7A | |
| miR-590-5p | [132] | RAB11A, MYO5B | miR-147a | ATP9A | |
| miR-370-3p | [133] | ATP9A, RAB11A, RAB7A | miR-29b-1 | COPS5 | |
| miR-194-5p | [93] | SDC4 | miR-124-5p | PDCD6IP | |
| miR-29a-3p | [134] | SMPD3 | miR-3178 | ||
| miR-149-5p | [135] | VPS4A, CD34 | miR-137-5p | ||
| miR-499a-5p | [136] | ATP9A | miR-518b | ||
| miR-186-5p | [137] | VPS4B, ATP9A, RAB27A, STAM | miR-375-5p | ||
| miR-34c-5p | [138] | VPS4A | miR-383-5p | ||
| miR-181a-5p | [139] | PDCD6IP, PRKN | miR-129-1-3p | ||
| miR-145-5p | [140] | STAM | miR-215-3p | ||
| let-7c-5p | [141] | STEAP3, MYO5B | miR-589-5p | ||
| miR-10b-5p | [142] | SDC1, SMPD3 | miR-137-5p |
| Natural Product | miRNA | Radiation-Modulated Gene | Natural Product | miRNA | Radiation-Modulated Gene | Natural Product | miRNA | Radiation-Modulated Gene |
|---|---|---|---|---|---|---|---|---|
| Chrysin | miR-18a-5p | ATM | Lycopene | let-7f-1-3p | MAP3K20 | Curcumin | miR-137-3p | RNF4 |
| Vitamin C | miR-181a-5p | Daidzein | miR-122-5p | Quercetin | miR-146a-5p | |||
| Piperine | miR-383-5p | ATR | Silymarin | miR-122-5p | Troxerutin | miR-146a-5p | ||
| Chrysin | let-7a-5p | BCL2L1 | Emodin | miR-1271-5p | Chrysin | miR-18a-5p | ||
| Quercetin | let-7a-5p | Hesperidin | miR-132-3p | Silymarin | miR-194-5p | |||
| Withaferin A | let-7a-5p | Curcumin | miR-137-3p | Chrysin | miR-221-3p | |||
| Withaferin A | let-7c-5p | Diosmin | miR-155-5p | Delphinidin | miR-23a-3p | |||
| Curcumin | miR-98-5p | Genistein | miR-155-5p | Melatonin | miR-23a-3p | |||
| Chrysin | let-7a-5p | CCND2 | Melatonin | miR-155-5p | Vitamin C | miR-589-5p | ||
| Quercetin | let-7a-5p | Diosmin | miR-17-5p | Chrysin | miR-92a-3p | |||
| Withaferin A | let-7a-5p | Fucoidan | miR-17-5p | Mangiferin | miR-92a-3p | |||
| Withaferin A | let-7c-5p | Piperine | miR-17-5p | Lycopene | let-7f-1-3p | SFRP1 | ||
| Emodin | miR-1271-5p | Berberine | miR-182-5p | Silymarin | miR-194-5p | |||
| Apigenin | miR-15a-5p | Gallic acid | miR-182-5p | Betulinic acid | miR-27a-3p | |||
| Vitamin D | miR-15a-5p | Matrine | miR-19b-3p | Genistein | miR-27a-3p | |||
| Mangiferin | miR-15b-5p | Delphinidin | miR-204-3p | Lycopene | miR-27a-3p | |||
| Curcumin | miR-16-5p | Vitamin C | miR-215-3p | Ligustrazine | miR-27a-3p | |||
| Hesperidin | miR-16-5p | Delphinidin | miR-23a-3p | Withaferin A | miR-27a-3p | |||
| Ligustrazine | miR-16-5p | Melatonin | miR-23a-3p | Mangiferin | miR-27b-3p | |||
| Diosmin | miR-17-5p | Parthenolide | miR-375-3p | Resveratrol | miR-542-3p | |||
| Fucoidan | miR-17-5p | Piperine | miR-383-3p | Withaferin A | miR-124-5p | TLK2 | ||
| Piperine | miR-17-5p | Chrysin | miR-92a-3p | Resveratrol | miR-125b-5p | |||
| Berberine | miR-182-5p | Mangiferin | miR-92a-3p | Vitamin C | miR-215-3p | |||
| Chrysin | miR-18a-5p | Chrysin | miR-9-5p | Betulinic acid | miR-27a-3p | |||
| Matrine | miR-19b-3p | Lycopene | let-7f-1-3p | MDM2 | Genistein | miR-27a-3p | ||
| Ligustrazine | miR-211-5p | Vitamin D | miR-145-5p | Ligustrazine | miR-27a-3p | |||
| Thymol | miR-29a-3p | Troxerutin | miR-147a | Lycopene | miR-27a-3p | |||
| Fucoidan | miR-29b-3p | Diosmin | miR-155-5p | Withaferin A | miR-27a-3p | |||
| Fucoidan | miR-29c-3p | Genistein | miR-155-5p | Mangiferin | miR-27b-3p | |||
| Piperine | miR-383-3p | Melatonin | miR-155-5p | Chrysin | let-7a-5p | TSPYL5 | ||
| Curcumin | miR-98-5p | Chrysin | miR-221-3p | Quercetin | let-7a-5p | |||
| Diosmin | miR-17-5p | DDIAS | Genistein | miR-223-3p | Withaferin A | let-7a-5p | ||
| Fucoidan | miR-17-5p | Thymol | miR-29a-3p | Withaferin A | let-7c-5p | |||
| Piperine | miR-17-5p | Fucoidan | miR-29b-3p | Parthenolide | miR-107 | |||
| Withaferin A | miR-124-5p | Fucoidan | miR-29c-3p | Hesperidin | miR-132-3p | |||
| Hesperidin | miR-132-3p | EGR1 | Ferulic acid | miR-340-3p | Curcumin | miR-186-3p | ||
| Ligustrazine | miR-211-5p | Vitamin C | miR-371b-5p | Curcumin | miR-98-5p | |||
| Delphinidin | miR-23a-3p | Piperine | miR-383-3p | Withaferin A | miR-124-5p | YAP1 | ||
| Melatonin | miR-23a-3p | Emodin | miR-1271-5p | NIPBL | Quercetin | miR-146a-5p | ||
| Gallic acid | miR-518b | Curcumin | miR-137-3p | Troxerutin | miR-146a-5p | |||
| Lycopene | let-7f-1-3p | GATA3 | Vitamin C | miR-181a-5p | Apigenin | miR-15a-5p | ||
| Withaferin A | miR-10b-5p | Ursolic Acid | miR-186-5p | Vitamin D | miR-15a-5p | |||
| Diosmin | miR-155-5p | Silymarin | miR-192-5p | Mangiferin | miR-15b-5p | |||
| Genistein | miR-155-5p | Ligustrazine | miR-211-5p | Curcumin | miR-16-5p | |||
| Melatonin | miR-155-5p | Vitamin C | miR-215-5p | Hesperidin | miR-16-5p | |||
| Betulinic acid | miR-27a-3p | Carvacrol | miR-217-5p | Ligustrazine | miR-16-5p | |||
| Genistein | miR-27a-3p | Chrysin | miR-221-3p | Ursolic Acid | miR-186-5p | |||
| Lycopene | miR-27a-3p | Chrysin | miR-92a-3p | Silymarin | miR-194-5p | |||
| Ligustrazine | miR-27a-3p | Mangiferin | miR-92a-3p | Ferulic acid | miR-200c-3p | |||
| Withaferin A | miR-27a-3p | Hesperidin | miR-132-3p | NUCKS1 | Zingerone | miR-200c-3p | ||
| Mangiferin | miR-27b-3p | Curcumin | miR-137-3p | 3,3′-Diindolylmethane | miR-21-5p | |||
| Chrysin | miR-34a-5p | Quercetin | miR-146a-5p | Diosmin | miR-21-5p | |||
| Gallic acid | miR-34a-5p | Troxerutin | miR-146a-5p | Gallic acid | miR-21-5p | |||
| Hesperidin | miR-34a-5p | Apigenin | miR-15a-5p | Ginsenoside Rg1 | miR-21-5p | |||
| Resveratrol | miR-34a-5p | Vitamin D | miR-15a-5p | Hesperidin | miR-21-5p | |||
| Ursolic acid | miR-34c-5p | Mangiferin | miR-15b-5p | Matrine | miR-21-5p | |||
| Piperine | miR-383-3p | Curcumin | miR-16-5p | Quercetin | miR-21-5p | |||
| Ursolic acid | miR-499a-5p | Hesperidin | miR-16-5p | Chrysin | miR-21-5p | |||
| Diosmin | miR-155-5p | GTF2H5 | Ligustrazine | miR-16-5p | Betulinic acid | miR-27a-3p | ||
| Genistein | miR-155-5p | Vitamin C | miR-181a-5p | Ligustrazine | miR-27a-3p | |||
| Melatonin | miR-155-5p | Ursolic acid | miR-186-5p | Genistein | miR-27a-3p | |||
| Curcumin | miR-186-3p | Chlorogenic acid | miR-199a-5p | Lycopene | miR-27a-3p | |||
| Silymarin | miR-192-5p | Curcumin | miR-30a-5p | Withaferin A | miR-27a-3p | |||
| Ligustrazine | miR-211-5p | EGCG | miR-30a-5p | Mangiferin | miR-27b-3p | |||
| Vitamin C | miR-215-5p | Piperine | miR-383-3p | Rutin | miR-590-5p | |||
| Rutin | miR-877-3p | Chrysin | miR-34a-5p | TMEM109 | Carvacrol | miR-217-5p | ZMPSTE24 | |
| Curcumin | miR-137-3p | KDM1A | Hesperidin | miR-34a-5p | Rutin | miR-877-3p | WRN | |
| Rutin | miR-877-3p | Resveratrol | miR-34a-5p | Piperine | miR-383-3p | CRYAB | ||
| Vitamin C | miR-371b-5p | Ursolic acid | miR-34c-5p | |||||
| Chrysin | miR-9-5p |
| Natural Product | miRNA | Exosome Biogenesis-Modulated Gene | Natural Product | miRNA | Exosome Biogenesis-Modulated Gene | Natural Product | miRNA | Exosome Biogenesis-Modulated Gene |
|---|---|---|---|---|---|---|---|---|
| Troxerutin | miR-147a | ATP9A | Delphinidin | miR-204-3p | RAB11A | Vitamin D | miR-145-5p | STAM |
| Berberine | miR-182-5p | 3,3′-Diindolylmethane | miR-21-5p | Ursolic acid | miR-186-5p | |||
| Gallic acid | miR-182-5p | Diosmin | miR-21-5p | Ferulic acid | miR-200c-3p | |||
| Ursolic acid | miR-186-5p | Gallic acid | miR-21-5p | Zingerone | miR-200c-3p | |||
| Chlorogenic acid | miR-199a-5p | Ginsenoside Rg1 | miR-21-5p | Genistein | miR-223-3p | |||
| Carvacrol | miR-217-5p | Hesperidin | miR-21-5p | Vitamin C | miR-371b-5p | |||
| Sesamol | miR-370-3p | Matrine | miR-21-5p | Chrysin | miR-9-5p | |||
| Ursolic acid | miR-499a-5p | Quercetin | miR-21-5p | Chrysin | let-7a-5p | STEAP3 | ||
| Chrysin | let-7a-5p | MYO5B | Chrysin | miR-21-5p | Quercetin | let-7a-5p | ||
| Quercetin | let-7a-5p | Curcumin | miR-30a-5p | Withaferin A | let-7a-5p | |||
| Withaferin A | let-7a-5p | EGCG | miR-30a-5p | Withaferin A | let-7c-5p | |||
| Withaferin A | let-7c-5p | Ferulic acid | miR-340-3p | Carvacrol | miR-217-5p | |||
| Emodin | miR-1271-5p | Sesamol | miR-370-3p | Chrysin | miR-9-5p | |||
| Apigenin | miR-15a-5p | Vitamin C | miR-371b-5p | Curcumin | miR-98-5p | |||
| Vitamin D | miR-15a-5p | Rutin | miR-590-5p | Diosmin | miR-17-5p | TSG101 | ||
| Mangiferin | miR-15b-5p | Emodin | miR-1271-5p | RAB27A | Fucoidan | miR-17-5p | ||
| Curcumin | miR-16-5p | Ursolic acid | miR-186-5p | Piperine | miR-17-5p | |||
| Hesperidin | miR-16-5p | Lycopene | let-7f-1-3p | RAB7A | Piperine | miR-383-3p | ||
| Ligustrazine | miR-16-5p | Emodin | miR-1271-5p | Parthenolide | miR-107 | VPS4A | ||
| Diosmin | miR-17-5p | Berberine | miR-182-5p | Ursolic acid | miR-149-5p | |||
| Fucoidan | miR-17-5p | Gallic acid | miR-182-5p | Apigenin | miR-15a-5p | |||
| Piperine | miR-17-5p | Chlorogenic acid | miR-199a-5p | Vitamin D | miR-15a-5p | |||
| Matrine | miR-19b-3p | Sesamol | miR-370-3p | Mangiferin | miR-15b-5p | |||
| Diosmin | miR-21-5p | Withaferin A | miR-10b-5p | SDC1 | Curcumin | miR-16-5p | ||
| Gallic acid | miR-21-5p | Berberine | miR-182-5p | Hesperidin | miR-16-5p | |||
| Ginsenoside Rg1 | miR-21-5p | Gallic acid | miR-182-5p | Ligustrazine | miR-16-5p | |||
| Hesperidin | miR-21-5p | Matrine | miR-19b-3p | Chrysin | miR-34a-5p | |||
| Matrine | miR-21-5p | Chrysin | miR-9-5p | Hesperidin | miR-34a-5p | |||
| Quercetin | miR-21-5p | Silymarin | miR-194-5p | SDC4 | Resveratrol | miR-34a-5p | ||
| Chrysin | miR-21-5p | Withaferin A | miR-10b-5p | SMPD3 | Ursolic acid | miR-34c-5p | ||
| Genistein | miR-223-3p | Betulinic acid | miR-27a-3p | Resveratrol | miR-125b-5p | VPS4B | ||
| Rutin | miR-590-5p | Mangiferin | miR-27a-3p | Curcumin | miR-186-3p | |||
| Curcumin | miR-98-5p | Ligustrazine | miR-27a-3p | Ursolic acid | miR-186-5p | |||
| Carvacrol | miR-217-5p | PDCD6IP | Genistein | miR-27a-3p | Matrine | miR-19b-3p | ||
| Chrysin | miR-221-3p | Withaferin A | miR-27a-3p | Carvacrol | miR-217-3p | |||
| Chrysin | miR-9-5p | Mangiferin | miR-27b-3p | Chrysin | miR-92a-3p | |||
| Vitamin C | miR-181a-5p | Silymarin | miR-29a-3p | Mangiferin | miR-92a-3p | |||
| Withaferin A | miR-124-5p | Fucoidan | miR-29b-3p | Chrysin | miR-9-5p | CD34 | ||
| Lycopene | let-7f-1-3p | SDCBP | Fucoidan | miR-29c-3p | Ursolic acid | miR-149-5p | ||
| Parthenolide | miR-107 | Piperine | miR-383-3p | Vitamin C | miR-181a-5p | PRKN | ||
| Piperine | miR-383-3p | Chrysin | miR-9-5p | Ferulic acid | miR-200c-3p | |||
| Vitamin C | miR-371b-5p | Vitamin C | miR-29b-1 | COPS5 | Zingerone | miR-200c-3p | ||
| Chlorogenic acid | miR-199a-5p | ATP13A2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.-Y.; Chuang, Y.-T.; Shiau, J.-P.; Yen, C.-Y.; Chang, F.-R.; Tsai, Y.-H.; Farooqi, A.A.; Chang, H.-W. Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis. Int. J. Mol. Sci. 2023, 24, 12449. https://doi.org/10.3390/ijms241512449
Tang J-Y, Chuang Y-T, Shiau J-P, Yen C-Y, Chang F-R, Tsai Y-H, Farooqi AA, Chang H-W. Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis. International Journal of Molecular Sciences. 2023; 24(15):12449. https://doi.org/10.3390/ijms241512449
Chicago/Turabian StyleTang, Jen-Yang, Ya-Ting Chuang, Jun-Ping Shiau, Ching-Yu Yen, Fang-Rong Chang, Yi-Hong Tsai, Ammad Ahmad Farooqi, and Hsueh-Wei Chang. 2023. "Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis" International Journal of Molecular Sciences 24, no. 15: 12449. https://doi.org/10.3390/ijms241512449
APA StyleTang, J.-Y., Chuang, Y.-T., Shiau, J.-P., Yen, C.-Y., Chang, F.-R., Tsai, Y.-H., Farooqi, A. A., & Chang, H.-W. (2023). Connection between Radiation-Regulating Functions of Natural Products and miRNAs Targeting Radiomodulation and Exosome Biogenesis. International Journal of Molecular Sciences, 24(15), 12449. https://doi.org/10.3390/ijms241512449







