SFPQ and Its Isoform as Potential Biomarker for Non-Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Result
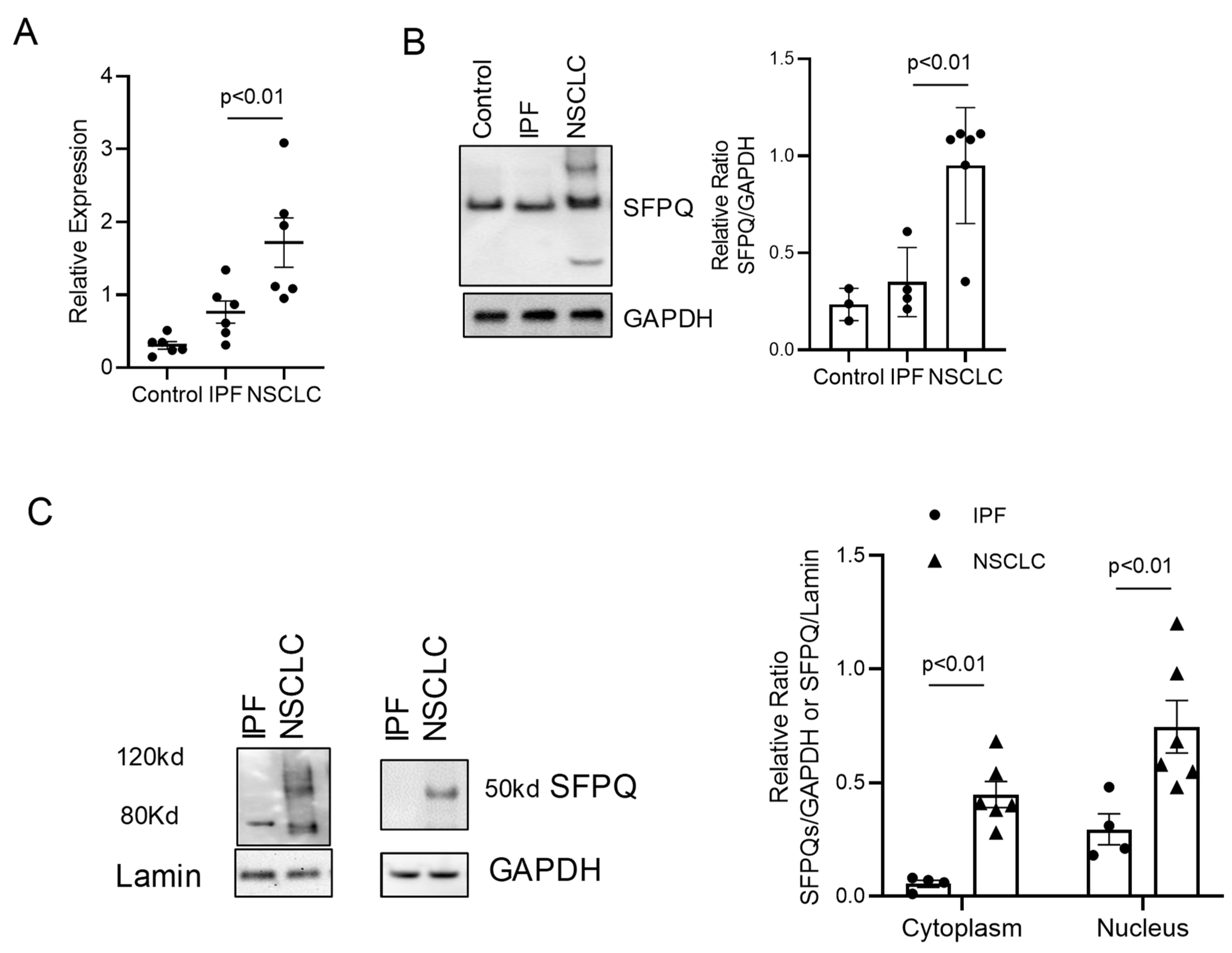
2.1. SFPQ Has Highest Expression in a Panel of Overexpressed Proteins in Lung Cancer Compared to IPF and Other Cellular Controls
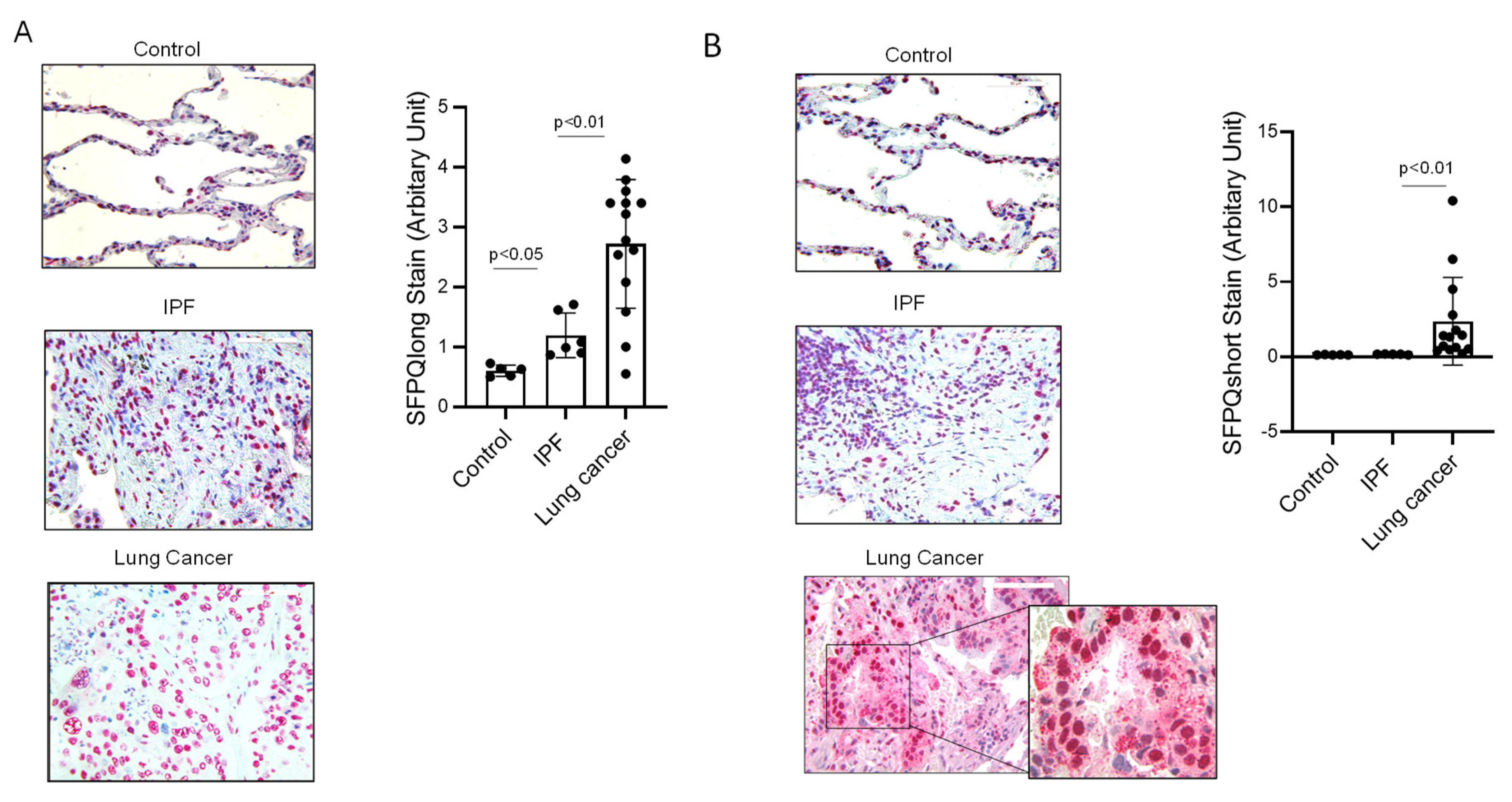
2.2. SFPQ Isoform Levels Are Different between Control and Lung Cancer Cells
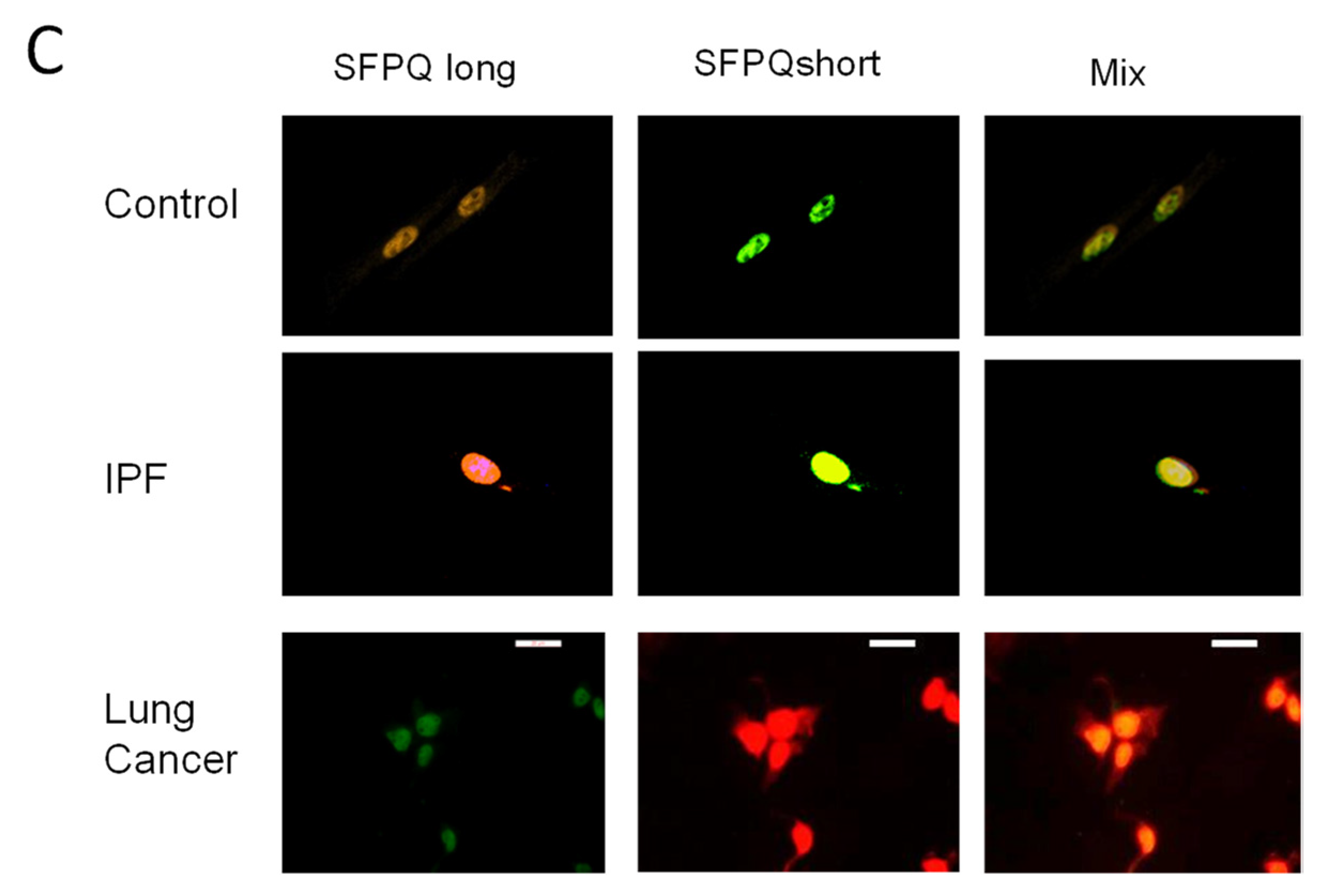
2.3. Short SFPQ Isoform Is Only Located in the Cytoplasm of Lung Cancer Cells
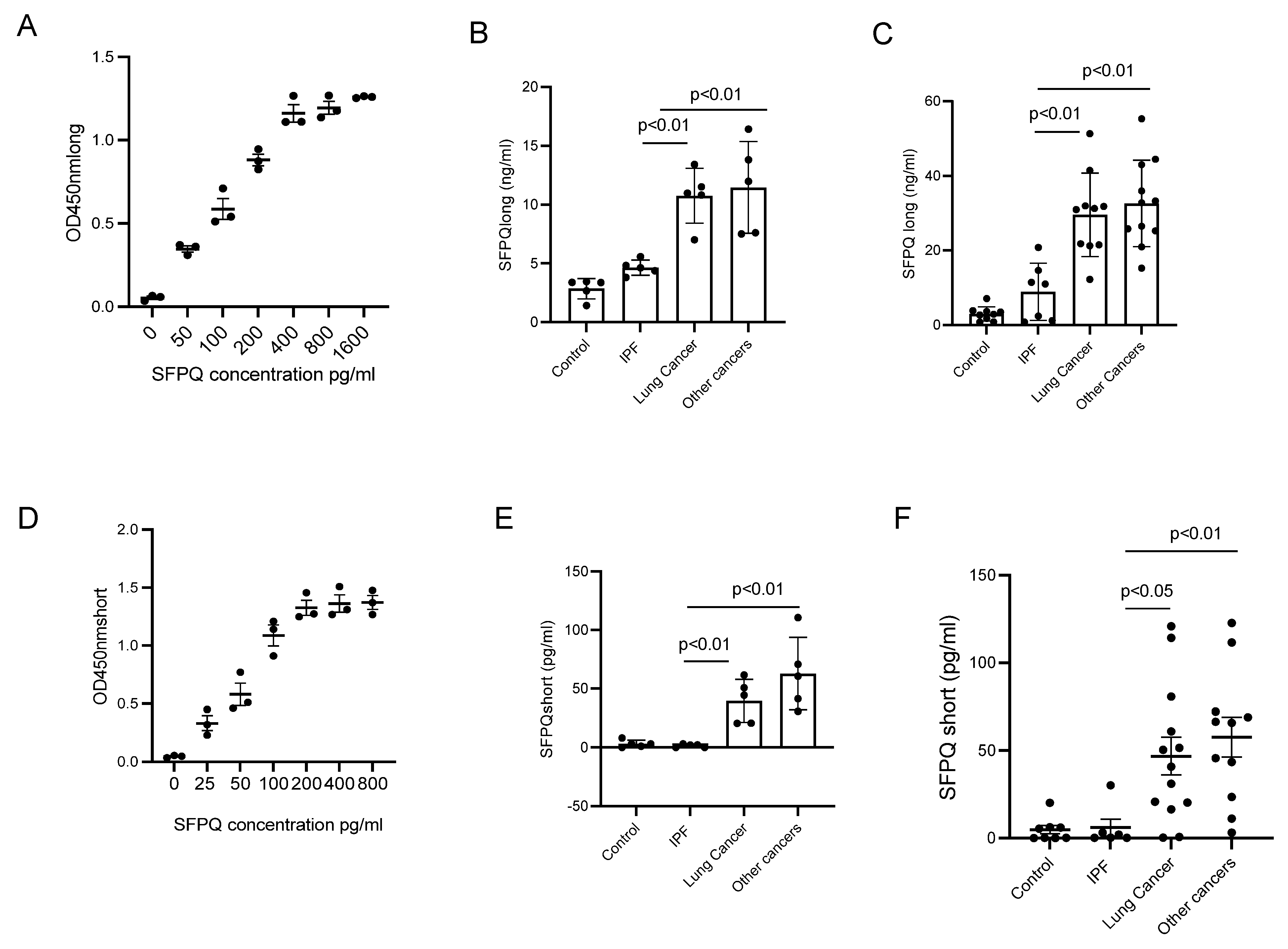
2.4. ELISA Assay for SFPQ Isoforms
2.5. SFPQ Is Elevated in Cancer Serum
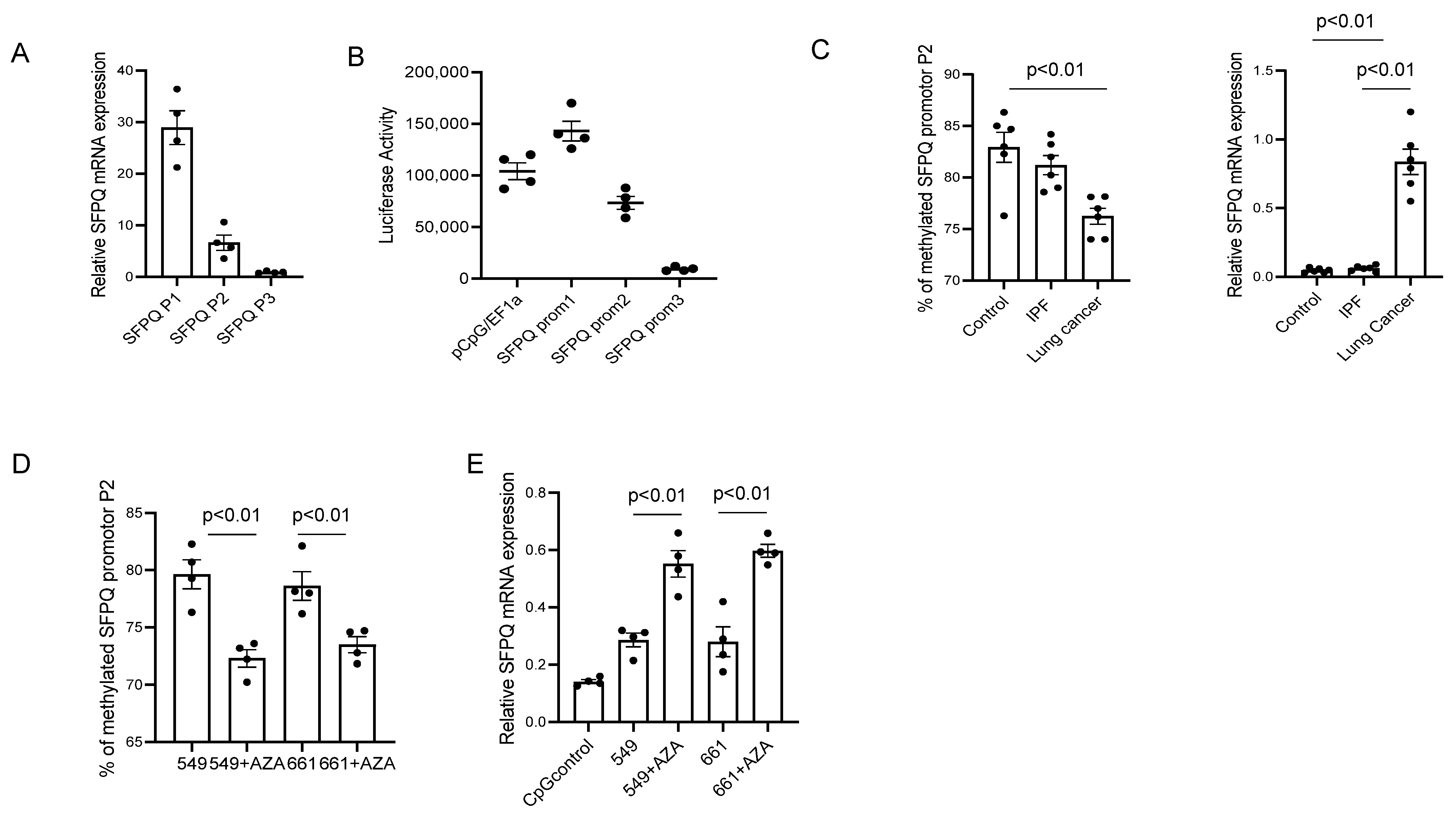
2.6. DNA Methylation Affects SFPQ Expression in Lung Cancer and Other Cancers
3. Discussion
4. Materials and Methods
4.1. Primary Cell Lines, Patient Tissue Sections and Serum
4.2. Cell Cultures and FACS Sorting
4.3. Isolation of Cell Nucleus
4.4. Ingenuity Pathway Analysis (IPA)
4.5. Assay for SFPQ Isoforms
4.6. IHC (Immunohistochemistry) Staining for SFPQ
4.7. DNA Methylation Analysis
4.8. Luciferase Reporter Assay
4.9. Western Blot Analysis
4.10. Real-Time Reverse Transcription PCR
| GAPDH Forward: 5′-TGTTGCCATCAATGACCCCTT-3′ | |
| GAPDH Reverse: 5′-CTCCACGACGTACTCAGCG-3′ | |
| Rictor Forward: 5′-TGTATGCAAGAGCCAAGCAC-3′ | |
| Rictor Reverse: 5′-CTGATTCCTGCTTTCCACAAG-3′ | |
| RTN4 Forward: 5′- GGCTCAGTGGATGAGACCCT-3′ | |
| RTN4 Reverse: 5′- TGTTACCTGGCTGCTCCTTC-3′ | |
| HELLS Forward: 5′-TAGAGAGTCGACAGAAATTCGG-3′ | |
| HELLS Reverse: 5′-CCTCATAACTGGCTTCTCTTCA-3′ | |
| LARP6 Forward: 5′- TTACACGGGACTGGAGAACC-3′ | |
| LARP6 Reverse: 5′- GTCCCAAAAAGCTTGAGCAG-3′ | |
| SFPQ Forward: 5′-GATCTACAGGGAAAGGCATTGTTG-3′ | |
| SFPQ Reverse: 5′-GATACATTGGATTCTTCTGGGCA-3′ | |
| SFPQ isoform PCR primers: | |
| SFPQ P1 Forward: 5′-CTCCACGACTTCCGTTCTCC-3′ | |
| Forward: 5′-CTGAGGAGGTGTGGTAGGGA-3′ | |
| SFPQ P2 Forward: 5′-GTCCCTACCACACCTCCTCA-3′ | |
| Forward: 5′-TCCGAGATCTTCTCCTCGCT-3′ | |
| SFPQ P3 Forward: 5′-GCAGCGAGGAGAAGATCTCG-3′ | |
| Forward: 5′-CTGTCGTCTCATCAGATAGGTCTT-3′ | |
| SFPQ Promotor1 | |
| Forward: 5′-GCCTCAATCAGAATCGCGG-3′ | |
| Reverse: 5′-GGTCGGAGTCGGGCCT-3′ | GGTCGGAGTCGGGCCT |
| SFPQ Promotor 2 | |
| Forward: 5′-GGTCCCAAAGGCGGCAAAAT-3′ | |
| Reverse: 5′-CCATCTTAGGGGAGCCGAC-3′ | |
| SFPQ Promotor 3 | |
| Forward: 5′-GGTCATTTTGTTGCATTTCCCC-3′ | |
| Reverse: 5′-TTTGCCCAACAGAAGTAGCAC-3′ |
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, X.; Zhu, Z.; Shi, Y. Metastasis mechanism and gene/protein expression in gastric cancer with distant organs metastasis. Bull. Cancer 2014. [Google Scholar] [CrossRef] [PubMed]
- Pachmayr, E.; Treese, C.; Stein, U. Underlying Mechanisms for Distant Metastasis—Molecular Biology. Visc. Med. 2017, 33, 11–20. [Google Scholar] [CrossRef]
- Zhu, T.; Bao, X.; Chen, M.; Lin, R.; Zhuyan, J.; Zhen, T.; Xing, K.; Zhou, W.; Zhu, S. Mechanisms and Future of Non-Small Cell Lung Cancer Metastasis. Front. Oncol. 2020, 10, 585284. [Google Scholar] [CrossRef] [PubMed]
- Hudeckova, M.; Koucky, V.; Rottenberg, J.; Gal, B. Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer-A Systematic Review. Biomedicines 2021, 9, 1548. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, Y.; Fu, S.; Yang, L.; Lin, S.; Fan, Q.; Wen, Q. The diagnostic role of DNA methylation in sporadic endometrial cancer: A systematic review and meta-analysis. Oncotarget 2018, 9, 8642–8652. [Google Scholar] [CrossRef] [Green Version]
- Ogden, G.R.; Macluskey, M. An overview of the prevention of oral cancer and diagnostic markers of malignant change: 1. Prevention. Dent. Update 2000, 27, 95–99. [Google Scholar] [CrossRef] [Green Version]
- Raja, V.; Farajzadegan, Z.; Mansourian, M.; Ghasemi, K.; Aboutalebi, M.S.; Nouri, R.; Mokarian, F. Diagnostic Value of Nonacid Nucleic Blood Tumor Marker Panels in Early Diagnosing Breast Cancer: A Systematic Review and Network Meta-Analysis. Dis. Markers 2022, 2022, 4119345. [Google Scholar] [CrossRef]
- Hou, X.; Yang, L.; Wang, K.; Zhou, Y.; Li, Q.; Kong, F.; Liu, X.; He, J. HELLS, a chromatin remodeler is highly expressed in pancreatic cancer and downregulation of it impairs tumor growth and sensitizes to cisplatin by reexpressing the tumor suppressor TGFBR3. Cancer Med. 2021, 10, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Kollarovic, G.; Topping, C.E.; Shaw, E.P.; Chambers, A.L. The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin. Nucleic Acids Res. 2020, 48, 1872–1885. [Google Scholar] [CrossRef]
- Robinson, M.H.; Maximov, V.; Lallani, S.; Farooq, H.; Taylor, M.D.; Read, R.D.; Kenney, A.M. Upregulation of the chromatin remodeler HELLS is mediated by YAP1 in Sonic Hedgehog Medulloblastoma. Sci. Rep. 2019, 9, 13611. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Dong, Z.; Prager, B.C.; Kim, L.J.; Wu, Q.; Gimple, R.C.; Wang, X.; Bao, S.; Hamerlik, P.; Rich, J.N. Chromatin remodeler HELLS maintains glioma stem cells through E2F3 and MYC. JCI Insight 2019, 4, e126140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, C.; Liu, N.; Yue, L.; Qi, W.W.; Xu, L.L.; Qiu, W.S. RTN4/Nogo is an independent prognostic marker for gastric cancer: Preliminary results. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 241–246. [Google Scholar]
- Guo, Z.; Zhang, X.; Zhu, H.; Zhong, N.; Luo, X.; Zhang, Y.; Tu, F.; Zhong, J.; Wang, X.; He, J.; et al. TELO2 induced progression of colorectal cancer by binding with RICTOR through mTORC2. Oncol. Rep. 2021, 45, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Lou, X.; Zou, Y.; Hu, D.; Liu, J.; Ning, J.; Jiao, Y.; Zhang, Z.; Yang, F.; Fan, L.; et al. Overexpression of Rictor protein and Rictor-H. pylori interaction has impact on tumor progression and prognosis in patients with gastric cancer. Folia Histochem. Cytobiol. 2020, 58, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Parte, S.C.; Batra, S.K.; Kakar, S.S. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J. Ovarian Res. 2018, 11, 69. [Google Scholar] [CrossRef] [PubMed]
- Noto, Z.; Yoshida, T.; Okabe, M.; Koike, C.; Fathy, M.; Tsuno, H.; Tomihara, K.; Arai, N.; Noguchi, M.; Nikaido, T. CD44 and SSEA-4 positive cells in an oral cancer cell line HSC-4 possess cancer stem-like cell characteristics. Oral Oncol. 2013, 49, 787–795. [Google Scholar] [CrossRef]
- Yang, L.; Yang, J.; Jacobson, B.; Gilbertsen, A.; Smith, K.; Higgins, L.; Guerrero, C.; Xia, H.; Henke, C.A.; Lin, J. SFPQ Promotes Lung Cancer Malignancy via Regulation of CD44 v6 Expression. Front. Oncol. 2022, 12, 862250. [Google Scholar] [CrossRef]
- Ji, Q.; Zhang, L.; Liu, X.; Zhou, L.; Wang, W.; Han, Z.; Sui, H.; Tang, Y.; Wang, Y.; Liu, N.; et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br. J. Cancer 2014, 111, 736–748. [Google Scholar] [CrossRef] [Green Version]
- Katano-Toki, A.; Yoshino, S.; Nakajima, Y.; Tomaru, T.; Nishikido, A.; Ishida, E.; Horiguchi, K.; Saito, T.; Ozawa, A.; Satoh, T.; et al. SFPQ associated with a co-activator for PPARgamma, HELZ2, regulates key nuclear factors for adipocyte differentiation. Biochem. Biophys. Res. Commun. 2021, 562, 139–145. [Google Scholar] [CrossRef]
- Klotz-Noack, K.; Klinger, B.; Rivera, M.; Bublitz, N.; Uhlitz, F.; Riemer, P.; Luthen, M.; Sell, T.; Kasack, K.; Gastl, B.; et al. SFPQ Depletion Is Synthetically Lethal with BRAF(V600E) in Colorectal Cancer Cells. Cell Rep. 2020, 32, 108184. [Google Scholar] [CrossRef]
- Lu, D.Y.; Mao, X.H.; Zhou, Y.H.; Yan, X.L.; Wang, W.P.; Zheng, Y.B.; Xiao, J.J.; Zhang, P.; Wang, J.G.; Ashwani, N.; et al. RTN4 3′-UTR insertion/deletion polymorphism and susceptibility to non-small cell lung cancer in Chinese Han population. Asian Pac. J. Cancer Prev. 2014, 15, 5249–5252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.; Zou, Y.; Ross, J.S.; Wang, K.; Liu, X.; Halmos, B.; Ali, S.M.; Liu, H.; Verma, A.; Montagna, C.; et al. RICTOR Amplification Defines a Novel Subset of Patients with Lung Cancer Who May Benefit from Treatment with mTORC1/2 Inhibitors. Cancer Discov. 2015, 5, 1262–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakre, N.; Wildey, G.; Behtaj, M.; Kresak, A.; Yang, M.; Fu, P.; Dowlati, A. RICTOR amplification identifies a subgroup in small cell lung cancer and predicts response to drugs targeting mTOR. Oncotarget 2017, 8, 5992–6002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, K.M.; Hellerbrand, C.; Ruemmele, P.; Michalski, C.W.; Kong, B.; Kroemer, A.; Hackl, C.; Schlitt, H.J.; Geissler, E.K.; Lang, S.A. Inhibition of mTORC2 component RICTOR impairs tumor growth in pancreatic cancer models. Oncotarget 2017, 8, 24491–24505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, F.F.; Li, X.Y.; Li, Y.Y.; He, S.; Xu, X.Y.; Liu, Y.H.; Liu, L.; Wu, S.H. Expression of Raptor and Rictor and their relationships with angiogenesis in colorectal cancer. Neoplasma 2020, 67, 501–508. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.K.; Lambert, A.W.; Ozturk, S.; Papageorgis, P.; Lopez, D.; Shen, N.; Sen, Z.; Abdolmaleky, H.M.; Gyorffy, B.; Feng, H.; et al. Targeting RICTOR Sensitizes SMAD4-Negative Colon Cancer to Irinotecan. Mol. Cancer Res. 2020, 18, 414–423. [Google Scholar] [CrossRef]
- Chen, L.; Su, Y.; Yin, B.; Li, S.; Cheng, X.; He, Y.; Jia, C. LARP6 Regulates Keloid Fibroblast Proliferation, Invasion, and Ability to Synthesize Collagen. J. Investig. Dermatol. 2022, 142, 2395–2405.e7. [Google Scholar] [CrossRef]
- Sheel, A.; Shao, R.; Brown, C.; Johnson, J.; Hamilton, A.; Sun, D.; Oppenheimer, J.; Smith, W.; Visconti, P.E.; Markstein, M.; et al. Acheron/Larp6 Is a Survival Protein That Protects Skeletal Muscle From Programmed Cell Death During Development. Front. Cell Dev. Biol. 2020, 8, 622. [Google Scholar] [CrossRef]
- Stefanovic, B.; Manojlovic, Z.; Vied, C.; Badger, C.D.; Stefanovic, L. Discovery and evaluation of inhibitor of LARP6 as specific antifibrotic compound. Sci. Rep. 2019, 9, 326. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Stefanovic, B. mTORC1 phosphorylates LARP6 to stimulate type I collagen expression. Sci. Rep. 2017, 7, 41173. [Google Scholar] [CrossRef]
- Liu, X.; Hou, X.; Zhou, Y.; Li, Q.; Kong, F.; Yan, S.; Lei, S.; Xiong, L.; He, J. Downregulation of the Helicase Lymphoid-Specific (HELLS) Gene Impairs Cell Proliferation and Induces Cell Cycle Arrest in Colorectal Cancer Cells. OncoTargets Ther. 2019, 12, 10153–10163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waseem, A.; Ali, M.; Odell, E.W.; Fortune, F.; Teh, M.T. Downstream targets of FOXM1: CEP55 and HELLS are cancer progression markers of head and neck squamous cell carcinoma. Oral Oncol. 2010, 46, 536–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, Y.W.; James, D.; Huang, J.; Lee, M. The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 7151. [Google Scholar] [CrossRef] [PubMed]
- Grasso, D.; Bintz, J.; Lomberk, G.; Molejon, M.I.; Loncle, C.; Garcia, M.N.; Lopez, M.B.; Urrutia, R.; Iovanna, J.L. Pivotal Role of the Chromatin Protein Nupr1 in Kras-Induced Senescence and Transformation. Sci. Rep. 2015, 5, 17549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanazawa, T.; Misawa, K.; Misawa, Y.; Uehara, T.; Fukushima, H.; Kusaka, G.; Maruta, M.; Carey, T.E. G-Protein-Coupled Receptors: Next Generation Therapeutic Targets in Head and Neck Cancer? Toxins 2015, 7, 2959–2984. [Google Scholar] [CrossRef]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Draht, M.X.G.; Goudkade, D.; Koch, A.; Grabsch, H.I.; Weijenberg, M.P.; van Engeland, M.; Melotte, V.; Smits, K.M. Prognostic DNA methylation markers for sporadic colorectal cancer: A systematic review. Clin. Epigenetics 2018, 10, 35. [Google Scholar] [CrossRef]
- Gurung, P.M.S.; Barnett, A.R.; Wilson, J.S.; Hudson, J.; Ward, D.G.; Messing, E.M.; Bryan, R.T. Prognostic DNA Methylation Biomarkers in High-risk Non-muscle-invasive Bladder Cancer: A Systematic Review to Identify Loci for Prospective Validation. Eur. Urol. Focus 2020, 6, 683–697. [Google Scholar] [CrossRef]
- Nowacka-Zawisza, M.; Wisnik, E. DNA methylation and histone modifications as epigenetic regulation in prostate cancer (Review). Oncol. Rep. 2017, 38, 2587–2596. [Google Scholar] [CrossRef] [Green Version]
- Ahmed Aglan, S.; Mohamad Zaki, A.; Sobhy El Sedfy, A.; Gaber El-Sheredy, H.; Hussein Elgaddar, O. O6-Methylguanine-DNA Methyltransferase and ATP-Binding Cassette Membrane Transporter G2 Promotor Methylation: Can Predict the Response to Chemotherapy in Advanced Breast Cancer? Rep. Biochem. Mol. Biol. 2022, 11, 20–29. [Google Scholar] [CrossRef]
- Michalowska-Sawczyn, M.; Grzywacz, A.; Masiak, J.; Chmielowiec, K.; Chmielowiec, J.; Chycki, J.; Maculewicz, E.; Cieszczyk, P. Associations Between Physical Effort and DNA Methylation in the Promotor Region of the Dopamine Transporter Gene (DAT1). J. Hum. Kinet. 2021, 77, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, O.G.; Aslan, D.; Akalin, H.; Erdem, Y.; Canoz, O.; Aytekin, A.; Ozoner, S.; Dundar, M. The Effects of O(6)-methyl Guanine DNA-methyl Transferase Promotor Methylation and CpG1, CpG2, CpG3 and CpG4 Methylation on Treatment Response and their Prognostic Significance in Patients with Glioblastoma. Balk. J. Med. Genet. 2020, 23, 33–41. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; Pietersma, H.; Cordes, M.; Kuipers, O.P.; Kok, J. PePPER: A webserver for prediction of prokaryote promoter elements and regulons. BMC Genom. 2012, 13, 299. [Google Scholar] [CrossRef] [Green Version]
- Satterstrom, F.K.; Haigis, M.C. Luciferase-based reporter to monitor the transcriptional activity of the SIRT3 promoter. Methods Enzymol. 2014, 543, 141–163. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Li, X.; Kang, J. Advances in the study of serum tumor markers of lung cancer. J. Cancer Res. Ther. 2014, 10 (Suppl. 3), C95–C101. [Google Scholar] [CrossRef]
- Molina, R.; Marrades, R.M.; Auge, J.M.; Escudero, J.M.; Vinolas, N.; Reguart, N.; Ramirez, J.; Filella, X.; Molins, L.; Agusti, A. Assessment of a Combined Panel of Six Serum Tumor Markers for Lung Cancer. Am. J. Respir. Crit. Care Med. 2016, 193, 427–437. [Google Scholar] [CrossRef]
- Zhang, X.; Hu, M.; Lyu, X.; Li, C.; Thannickal, V.J.; Sanders, Y.Y. DNA methylation regulated gene expression in organ fibrosis. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 2389–2397. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Shao, X.; Liu, H.; Liu, S.; Wu, Q.; Zhang, Y. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics 2012, 99, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Bar-Nur, O.; Russ, H.A.; Efrat, S.; Benvenisty, N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet beta cells. Cell Stem Cell 2011, 9, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Calvanese, V.; Horrillo, A.; Hmadcha, A.; Suarez-Alvarez, B.; Fernandez, A.F.; Lara, E.; Casado, S.; Menendez, P.; Bueno, C.; Garcia-Castro, J.; et al. Cancer genes hypermethylated in human embryonic stem cells. PLoS ONE 2008, 3, e3294. [Google Scholar] [CrossRef] [Green Version]
- Rayner, S.L.; Cheng, F.; Hogan, A.L.; Grima, N.; Yang, S.; Ke, Y.D.; Au, C.G.; Morsch, M.; De Luca, A.; Davidson, J.M.; et al. ALS/FTD-causing mutation in cyclin F causes the dysregulation of SFPQ. Hum. Mol. Genet. 2021, 30, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.; Hamid, F.; Fielding, T.; Gordon, P.M.; Maloney, M.; Makeyev, E.V.; Houart, C. Prematurely terminated intron-retaining mRNAs invade axons in SFPQ null-driven neurodegeneration and are a hallmark of ALS. Nat. Commun. 2022, 13, 6994. [Google Scholar] [CrossRef] [PubMed]
- Widagdo, J.; Udagedara, S.; Bhembre, N.; Tan, J.Z.A.; Neureiter, L.; Huang, J.; Anggono, V.; Lee, M. Familial ALS-associated SFPQ variants promote the formation of SFPQ cytoplasmic aggregates in primary neurons. Open Biol. 2022, 12, 220187. [Google Scholar] [CrossRef]
- Zhang, D.G.; Jiang, A.G.; Lu, H.Y.; Zhang, L.X.; Gao, X.Y. Isolation, cultivation and identification of human lung adenocarcinoma stem cells. Oncol. Lett. 2015, 9, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xia, H.; Smith, K.; Gilbertsen, A.; Beisang, D.; Kuo, J.; Bitterman, P.B.; Henke, C.A. A CD44/Brg1 nuclear complex confers mesenchymal progenitor cells with enhanced fibrogenicity in idiopathic pulmonary fibrosis. JCI Insight 2021, 6, e144652. [Google Scholar] [CrossRef]
- Zheng, C.; Sun, Y.H.; Ye, X.L.; Chen, H.Q.; Ji, H.B. Establishment and characterization of primary lung cancer cell lines from Chinese population. Acta Pharmacol. Sin. 2011, 32, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Gu, X.; Yi, S. Ingenuity Pathway Analysis of Gene Expression Profiles in Distal Nerve Stump following Nerve Injury: Insights into Wallerian Degeneration. Front. Cell. Neurosci. 2016, 10, 274. [Google Scholar] [CrossRef] [Green Version]
- Bashtrykov, P.; Jeltsch, A. DNA Methylation Analysis by Bisulfite Conversion Coupled to Double Multiplexed Amplicon-Based Next-Generation Sequencing (NGS). Methods Mol. Biol. 2018, 1767, 367–382. [Google Scholar] [CrossRef]
- Claus, R.; Lucas, D.M.; Stilgenbauer, S.; Ruppert, A.S.; Yu, L.; Zucknick, M.; Mertens, D.; Buhler, A.; Oakes, C.C.; Larson, R.A.; et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J. Clin. Oncol. 2012, 30, 2483–2491. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, L.; Gilbertsen, A.; Jacobson, B.; Pham, J.; Fujioka, N.; Henke, C.A.; Kratzke, R.A. SFPQ and Its Isoform as Potential Biomarker for Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2023, 24, 12500. https://doi.org/10.3390/ijms241512500
Yang L, Gilbertsen A, Jacobson B, Pham J, Fujioka N, Henke CA, Kratzke RA. SFPQ and Its Isoform as Potential Biomarker for Non-Small-Cell Lung Cancer. International Journal of Molecular Sciences. 2023; 24(15):12500. https://doi.org/10.3390/ijms241512500
Chicago/Turabian StyleYang, Libang, Adam Gilbertsen, Blake Jacobson, Jenny Pham, Naomi Fujioka, Craig A. Henke, and Robert A. Kratzke. 2023. "SFPQ and Its Isoform as Potential Biomarker for Non-Small-Cell Lung Cancer" International Journal of Molecular Sciences 24, no. 15: 12500. https://doi.org/10.3390/ijms241512500




