Proteomic and Mutant Analysis of Hydrogenase Maturation Protein Gene hypE in Symbiotic Nitrogen Fixation of Mesorhizobium huakuii
Abstract
:1. Introduction
2. Results
2.1. Bioinformation Analysis of the M. huakuii hypE Gene
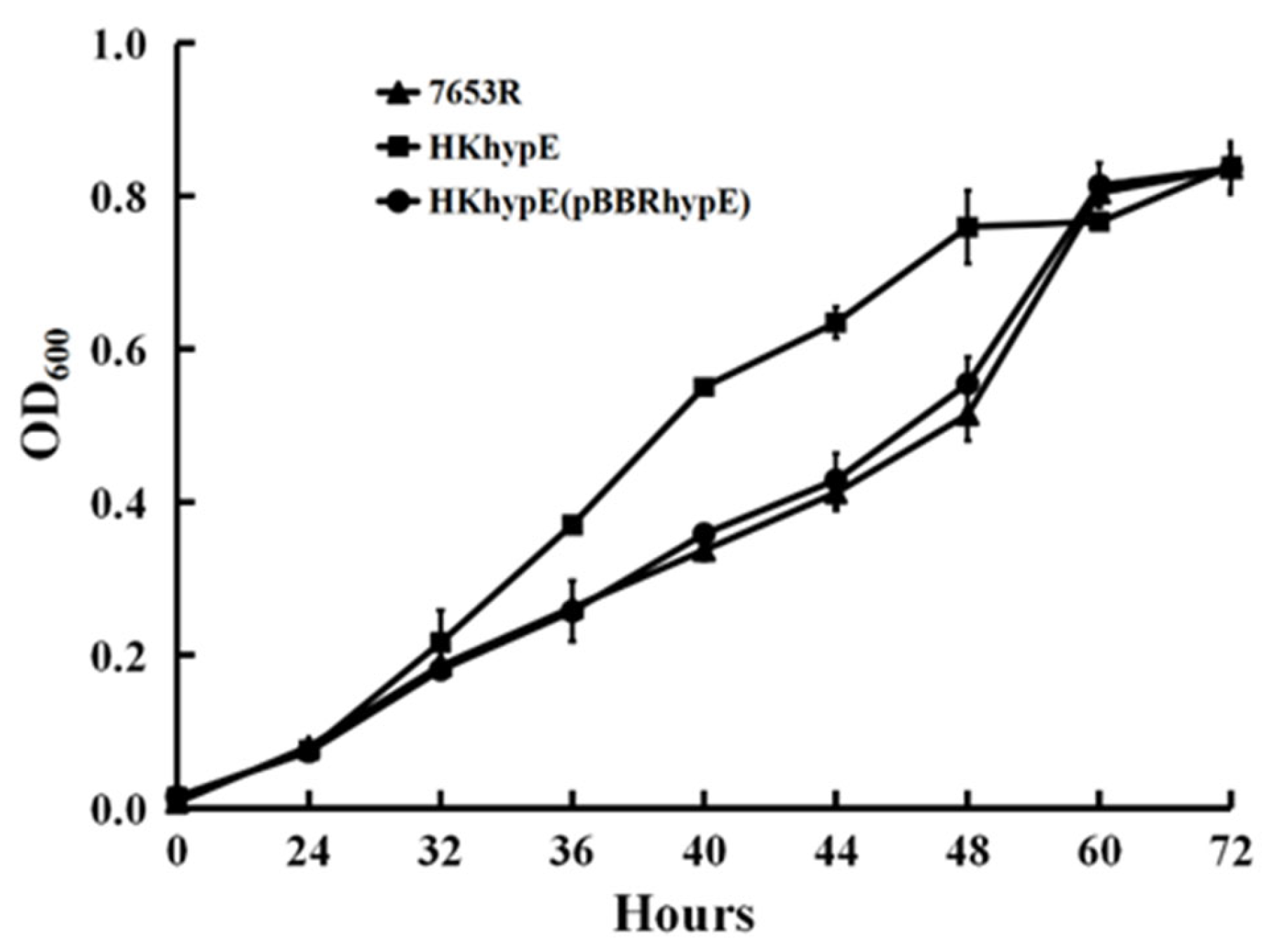
2.2. Growth and Antioxidative Activity in M. huakii hypE Mutant in Free Living Condition
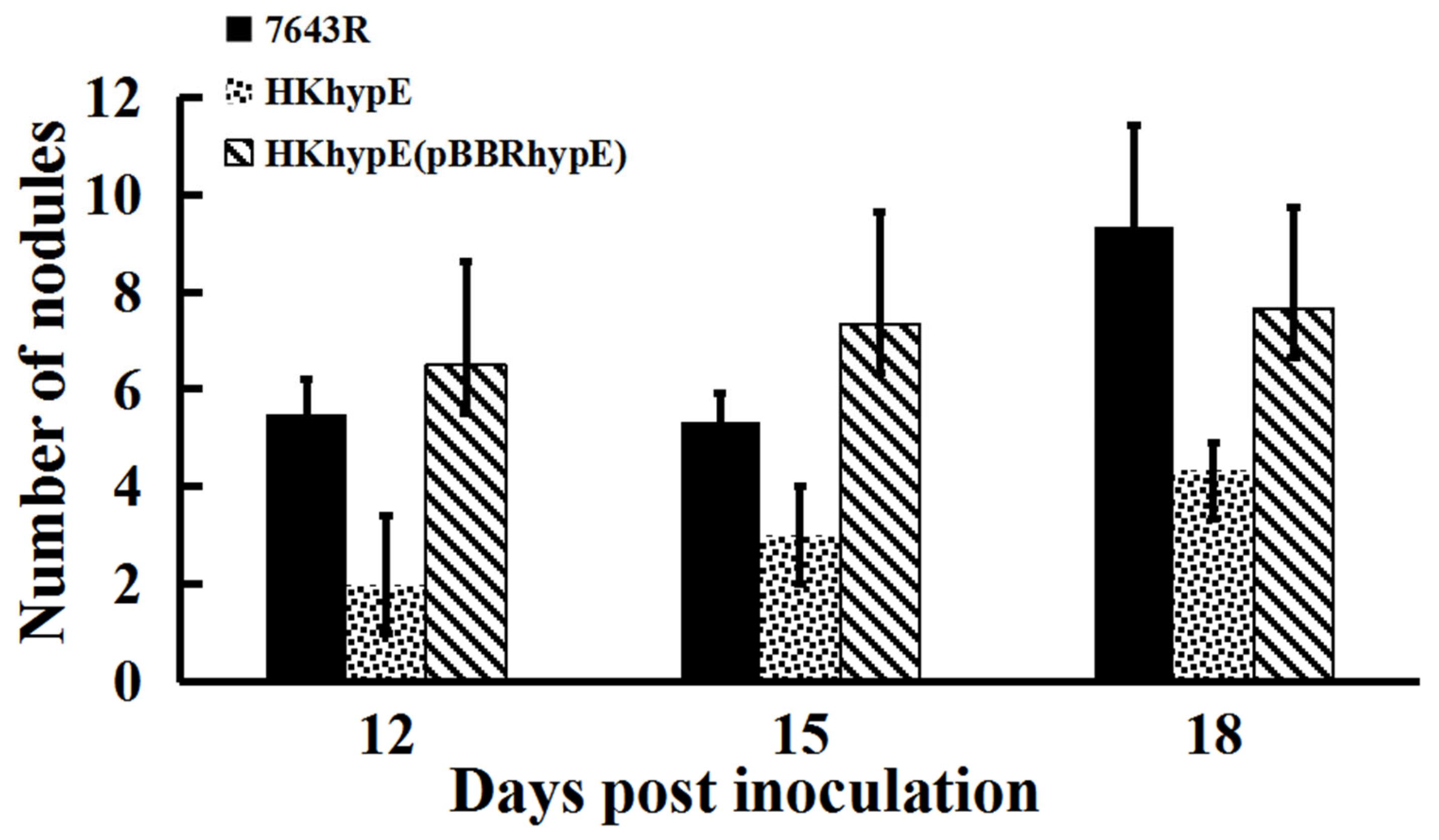
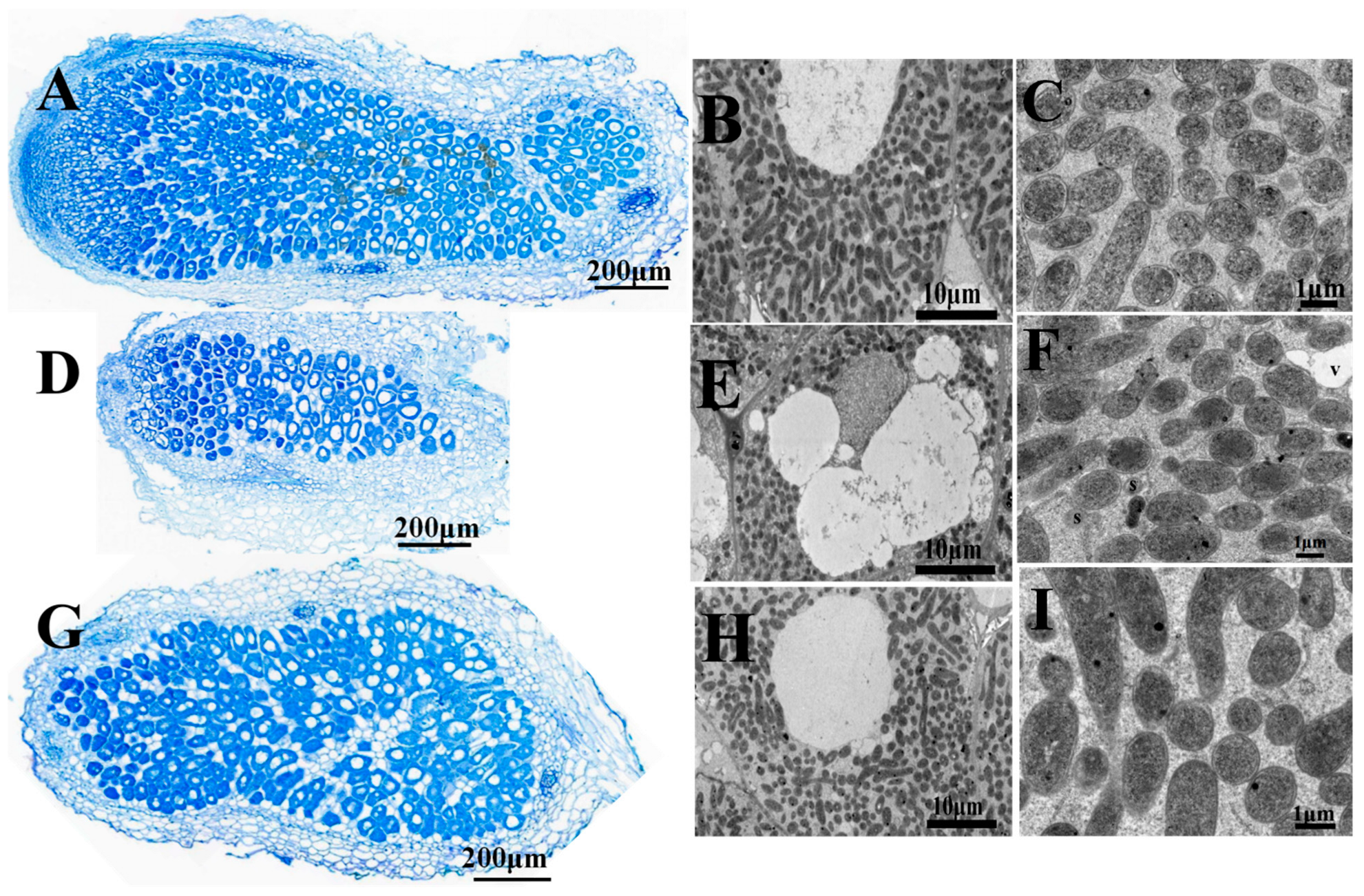
2.3. Symbiotic Properties of hypE Mutant Strain
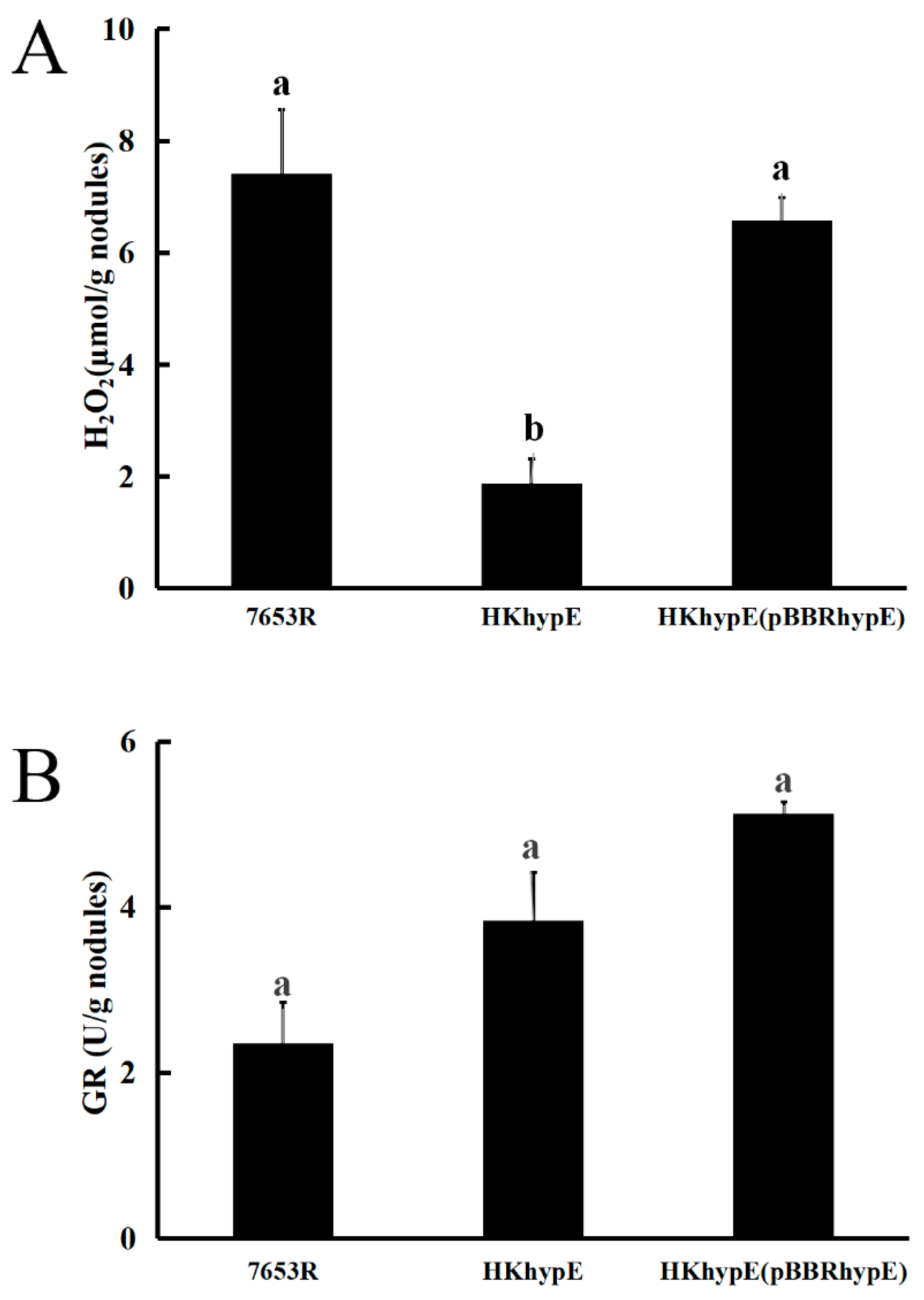
2.4. Effect of hypE Deletion on H2O2 Concentration and Glutathione Reductase Activity in Nodules
2.5. Rhizosphere Colonization and Competition by M. huakuii Strains
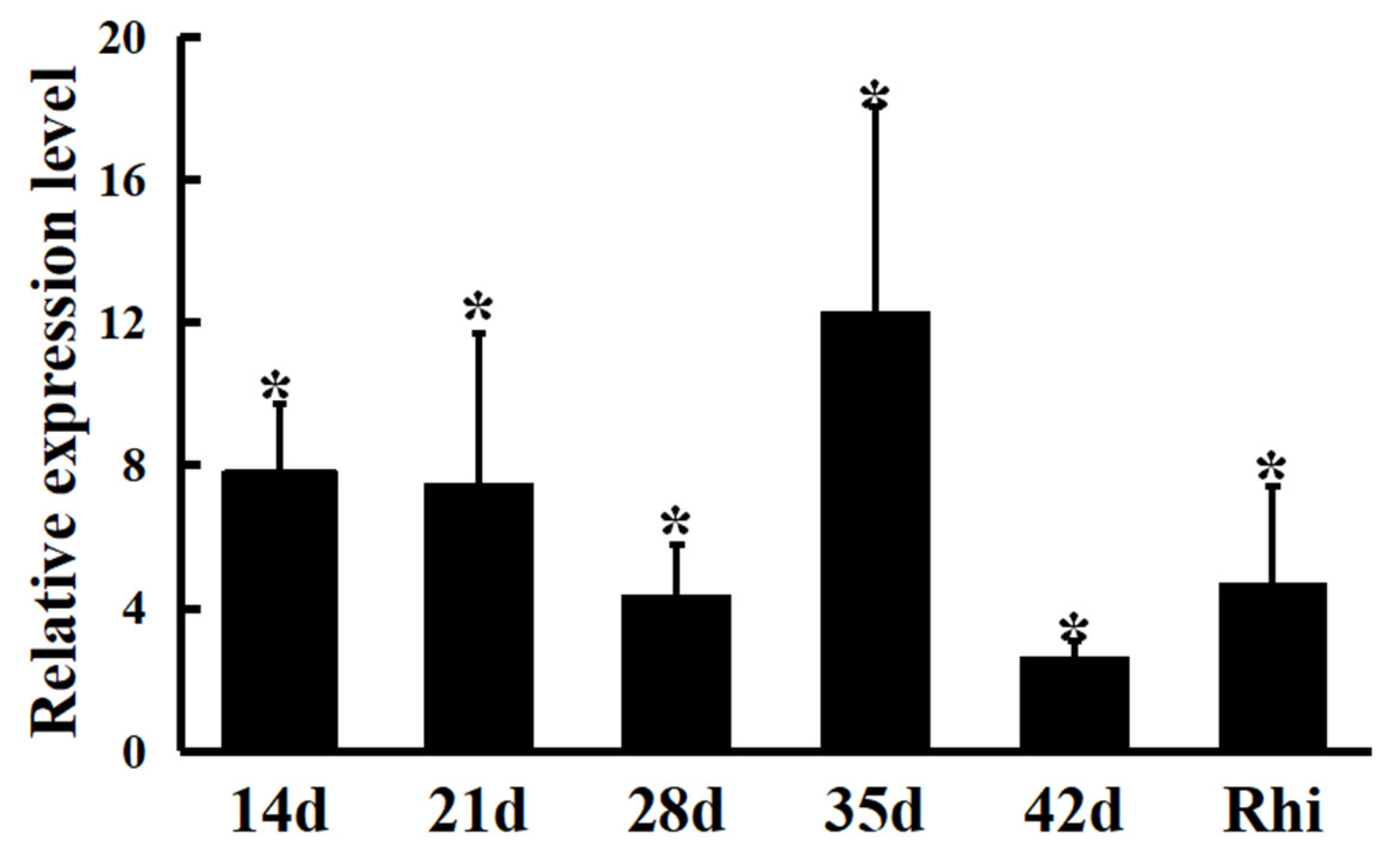
2.6. mRNA Expression Levels of HypE Gene in Nodules Induced by M. huakuii 7653R
2.7. Proteomic Analysis of Differential Protein Expression in Nodule Bacteroids
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Primers, and Culture Conditions
4.2. Construction and Complementation of a hypE Mutant of M. huakuii 7653R
4.3. Cellular Sensitivity to H2O2
4.4. Plant Experiments and Microscope Study of Nodules
4.5. Measurement of H2O2 Concentration and Glutathione Reductase Activity in the Nodules
4.6. Rhizosphere Colonization
4.7. RNA Isolation and Quantitative Reverse Transcription-PCR (qRT-PCR)
4.8. Protein Extraction, Digestion, and Peptide Labeling
4.9. Fractionation of Tryptic Peptides and LC-MS/MS Analysis
4.10. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maeda, T.; Vardar, G.; Self, W.T.; Wood, T.K. Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC. Biotechnol. 2007, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sickerman, N.S.; Hu, Y. Hydrogenases. Methods Mol. Biol. 2019, 1876, 65–88. [Google Scholar]
- Calusinska, M.; Happe, T.; Joris, B.; Wilmotte, A. The surprising diversity of clostridial hydrogenases: A comparative genomic perspective. Microbiology 2010, 156, 1575–1588. [Google Scholar] [CrossRef] [Green Version]
- Forzi, L.; Sawers, R.G. Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 2007, 20, 565–578. [Google Scholar] [CrossRef]
- Lubitz, W.; Ogata, H.; Rüdiger, O.; Reijerse, E. Hydrogenases. Chem. Rev. 2014, 114, 4081–4148. [Google Scholar] [CrossRef] [PubMed]
- Shomura, Y.; Hagiya, K.; Yoont, K.S.; Nishihara, H.; Higuchi, Y. Crystallization and preliminary X-ray diffraction analysis of membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 827–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terpolilli, J.J.; Hood, G.A.; Poole, P.S. What determines the efficiency of N(2)-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 2012, 60, 325–389. [Google Scholar]
- Jensen, B.B.; Cox, R.P. Direct measurements of steady-state kinetics of cyanobacterial n(2) uptake by membrane-leak mass spectrometry and comparisons between nitrogen fixation and acetylene reduction. Appl. Environ. Microbiol. 1983, 45, 1331–1337. [Google Scholar] [CrossRef]
- Brito, B.; Palacios, J.M.; Imperial, J.; Ruiz-Argüeso, T. Engineering the Rhizobium leguminosarum bv. viciae hydrogenase system for expression in free-living microaerobic cells and increased symbiotic hydrogenase activity. Appl. Environ. Microbiol. 2002, 68, 2461–2467. [Google Scholar] [CrossRef] [Green Version]
- Annan, H.; Golding, A.L.; Zhao, Y.; Dong, Z. Choice of hydrogen uptake (Hup) status in legume-rhizobia symbioses. Ecol. Evol. 2012, 2, 2285–2290. [Google Scholar] [CrossRef]
- Brito, B.; Prieto, R.I.; Cabrera, E.; Mandrand-Berthelot, M.A.; Palacios, J.M. Rhizobium leguminosarum hupe encodes a nickel transporter required for hydrogenase activity. J. Bacteriol. 2009, 192, 925–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benoit, S.; Mehta, N.; Wang, G.; Gatlin, M.; Maier, R.J. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb. Pathog. 2004, 36, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Shomura, Y.; Komori, H.; Miyabe, N.; Tomiyama, M.; Shibata, N.; Higuchi, Y. Crystal structures of hydrogenase maturation protein HypE in the Apo and ATP-bound forms. J. Mol. Biol. 2007, 372, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Blokesch, M.; Paschos, A.; Bauer, A.; Reissmann, S.; Drapal, N.; Böck, A. Analysis of the transcarbamoylation-dehydration reaction catalyzed by the hydrogenase maturation proteins HypF and HypE. Eur. J. Biochem. 2004, 271, 3428–3436. [Google Scholar] [CrossRef]
- Miki, K.; Atomi, H.; Watanabe, S. Structural insight into [NiFe] hydrogenase maturation by transient complexes between Hyp proteins. Acc. Chem. Res. 2020, 53, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Blokesch, M.; Albracht, S.P.; Matzanke, B.F.; Drapal, N.M.; Jacobi, A.; Böck, A. The complex between hydrogenase-maturation proteins HypC and HypD is an intermediate in the supply of cyanide to the active site iron of [NiFe]-hydrogenases. J. Mol. Biol. 2004, 344, 155–167. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Zeikus, J.G. Extracellular iron reduction is mediated in part by neutral red and hydrogenase in Escherichia coli. Appl. Environ. Microbiol. 2004, 70, 3467–3474. [Google Scholar] [CrossRef] [Green Version]
- Rey, L.; Imperial, J.; Palacios, J.M.; Ruiz-Argüeso, T. Purification of Rhizobium leguminosarum HypB, a nickel-binding protein required for hydrogenase synthesis. J. Bacteriol. 1994, 176, 6066–6073. [Google Scholar] [CrossRef] [Green Version]
- Brito, B.; Martínez, M.; Fernández, D.; Rey, L.; Cabrera, E.; Palacios, J.M.; Imperial, J.; Ruiz-Argüeso, T. Hydrogenase genes from Rhizobium leguminosarum bv. viciae are controlled by the nitrogen fixation regulatory protein nifA. Proc. Natl. Acad. Sci. USA 1997, 94, 6019–6024. [Google Scholar] [CrossRef]
- Hernando, Y.; Palacios, J.; Imperial, J.; Ruiz-Argüeso, T. Rhizobium leguminosarum bv. viciae hypA gene is specifically expressed in pea (Pisum sativum) bacteroids and required for hydrogenase activity and processing. FEMS Microbiol. Lett. 1998, 169, 295–302. [Google Scholar] [CrossRef] [Green Version]
- Olson, J.W.; Fu, C.; Maier, R.J. The HypB protein from Bradyrhizobium japonicum can store nickel and is required for the nickel-dependent transcriptional regulation of hydrogenase. Mol. Microbiol. 1997, 24, 119–128. [Google Scholar] [CrossRef]
- Albareda, M.; Pacios, L.F.; Manyani, H.; Rey, L.; Brito, B.; Imperial, J.; Ruiz-Argüeso, T.; Palacios, J.M. Maturation of Rhizobium leguminosarum hydrogenase in the presence of oxygen requires the interaction of the chaperone HypC and the scaffolding protein HupK. J. Biol. Chem. 2014, 289, 21217–21229. [Google Scholar] [CrossRef] [Green Version]
- Albareda, M.; Manyani, H.; Imperial, J.; Brito, B.; Ruiz-Argüeso, T.; Böck, A.; Palacios, J.M. Dual role of HupF in the biosynthesis of [NiFe] hydrogenase in Rhizobium leguminosarum. BMC Microbiol. 2012, 12, 256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, G.; Karunakaran, R.; East, A.K.; Munozazcarate, O.; Poole, P.S. Glutathione affects the transport activity of Rhizobium leguminosarum 3841 and is essential for efficient nodulation. FEMS Microbiol. Lett. 2017, 364, fnx045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahiri, S.; Cole, B.; Pulakat, L.; Gavini, N. The NifX Protein is Involved in the Final Stages of FeMo-cofactor Transport to the MoFe Protein. Am. J. Biochem. Biotechnol. 2007, 3, 92–102. [Google Scholar] [CrossRef]
- Brear, E.M.; Day, D.A.; Smith, P.M. Iron: An essential micronutrient for the legume-rhizobium symbiosis. Front. Plant. Sci. 2013, 4, 359. [Google Scholar] [CrossRef] [Green Version]
- Hood, G.; Karunakaran, R.; Downie, J.A.; Poole, P. MgtE from Rhizobium leguminosarum is a Mg2+ channel essential for growth at low pH and N2 fixation on specific plants. Mol. Plant Microbe Interact. 2015, 28, 1281–1287. [Google Scholar] [CrossRef] [Green Version]
- Lindblad, A.; Nordlund, S. Electron transport to nitrogenase in Rhodospirillum rubrum: Role of energization of the chromatophore membrane. Photosynth. Res. 1997, 53, 23–28. [Google Scholar] [CrossRef]
- Eady, R.R.; Postgate, J.R. Nitrogenase. Nature 1974, 249, 805–810. [Google Scholar] [CrossRef]
- Kim, M.S.; Baek, J.S.; Lee, J.K. Comparison of H2 accumulation by Rhodobacter sphaeroides KD131 and its uptake hydrogenase and PHB synthase deficient mutant. Int. J. Hydrogen Energy 2006, 31, 121–127. [Google Scholar] [CrossRef]
- Aguilar, O.M.; Yates, M.G.; Postgate, J.R. The beneficial effect of Hydrogenase in Azotobacter chroococcum under Nitrogen-fixing, carbon-limiting conditions in continuous and batch cultures. Microbiology 1985, 131, 3141–3145. [Google Scholar] [CrossRef] [Green Version]
- Beatrix, S.; Ortwin, M. Characterization of hydrogenase activities associated with the molybdenum CO dehydrogenase from oligotropha carboxidovorans. FEMS Microbiol. Lett. 1996, 136, 157–162. [Google Scholar]
- Wang, V.C.; Esmieu, C.; Redman, H.J.; Berggren, G.; Hammarström, L. The reactivity of molecular oxygen and reactive oxygen species with [FeFe] hydrogenase biomimetics: Reversibility and the role of the second coordination sphere. Dalton Trans. 2020, 49, 858–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seiji, O.; Bunsho, K.; Hidetaka, N.; Yoshihito, W.; Shunichi, F. Why do nitrogenases waste electrons by evolving dihydrogen? Appl. Organomet. Chem. 2004, 18, 589–594. [Google Scholar]
- Appel, J.; Phunpruch, S.; Steinmüller, K.; Schulz, R. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 2000, 173, 333–338. [Google Scholar] [CrossRef]
- Puppo, A.; Pauly, N.; Boscari, A.; Mandon, K.; Brouquisse, R. Hydrogen peroxide and nitric oxide: Key regulators of the Legume-Rhizobium and mycorrhizal symbioses. Antioxid. Redox. Sign. 2013, 18, 2202–2219. [Google Scholar] [CrossRef]
- Long, J.Y.; Song, K.L.; He, X.; Zhang, B.; Cui, X.F.; Song, C.F. Mutagenesis of PhaR, a regulator gene of polyhydroxyalkanoate biosynthesis of Xanthomonas oryzae pv. oryzae-caused pleiotropic phenotype changes. Front. Microbiol. 2018, 9, 3046. [Google Scholar] [CrossRef]
- Gray, J.X.; Rolfe, B.G. Exopolysaccharide production in Rhizobium and its role in invasion. Mol. Microbiol. 1990, 4, 1425–1431. [Google Scholar] [CrossRef]
- Van Slooten, J.C.; Bhuvanasvari, T.V.; Bardin, S.; Stanley, J. Two C4-dicarboxylate transport systems in Rhizobium sp. NGR234: Rhizobial dicarboxylate transport is essential for nitrogen fixation in tropical legume symbioses. Mol. Plant Microbe Interact. 1992, 5, 179–186. [Google Scholar] [CrossRef]
- Maki-Yonekura, S.; Matsuoka, R.; Yamashita, Y.; Shimizu, H.; Tanaka, M.; Iwabuki, F.; Yonekura, K. Hexameric and pentameric complexes of the ExbBD energizer in the Ton system. eLife 2018, 7, e35419. [Google Scholar] [CrossRef]
- Glazebrook, J.; Ichige, A.; Walker, G.C. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 1993, 7, 1485–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, A.; Overlöper, A.; Schlüter, J.P.; Reinkensmeier, J.; Robledo, M.; Giegerich, R.; Narberhaus, F.; Evguenieva-Hackenberg, E. Riboregulation in plant-associated α-proteobacteria. RNA Biol. 2014, 11, 550–562. [Google Scholar] [CrossRef] [Green Version]
- Boubakri, H.; Chihaoui, S.A.; Najjar, E.; Gargouri, M.; Barhoumi, F.; Jebara, M. Genome-wide analysis and expression profiling of H-type Trx family in Phaseolus vulgaris revealed distinctive isoforms associated with symbiotic N2-fixing performance and abiotic stress response. J. Plant Physiol. 2021, 260, 153410. [Google Scholar] [CrossRef]
- Bentchikou, E.; Servant, P.; Coste, G.; Sommer, S. A major role of the RecFOR pathway in DNA double-strand-break repair through ESDSA in Deinococcus radiodurans. PLoS Genet. 2010, 6, e1000774. [Google Scholar] [CrossRef] [Green Version]
- Satoh, K.; Kikuchi, M.; Ishaque, A.M.; Ohba, H.; Yamada, M.; Tejima, K.; Onodera, T.; Narumi, I. The role of Deinococcus radiodurans RecFOR proteins in homologous recombination. DNA Repair 2012, 11, 410–418. [Google Scholar] [CrossRef]
- Beringer, J.E.; Hopwood, D.A. Chromosomal recombination and mapping in Rhizobium leguminosarum. Nature 1976, 264, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Poole, P.S.; Blyth, A.; Reid, C.; Walters, K. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv. viciae. Microbiology 1994, 140, 2787–2795. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.J.; Li, Y.G.; Zhou, J.C. Cloning and identification of opa22, a new gene involved in nodule formation by Mesorhizobium huakuii. FEMS Microbiol. Lett. 2006, 257, 152–157. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, A.; Tauch, A.; Jäger, W.; Kalinowski, J.; Thierbach, G.; Pühler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef]
- Kovach, M.E.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M., 2nd; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, R.; Haag, A.F.; East, A.K.; Ramachandran, V.K.; Prell, J.; James, E.K.; Scocchi, M.; Ferguson, G.P.; Poole, P.S. BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 2010, 192, 2920–2928. [Google Scholar] [CrossRef] [Green Version]
- Zou, Q.; Zhou, Y.; Cheng, G.; Peng, Y.; Luo, S.; Wu, H.; Yan, C.; Li, X.; He, D. Antioxidant ability of glutaredoxins and their role in symbiotic nitrogen fifixation in Rhizobium leguminosarum bv. viciae 3841. Appl. Environ. Microbiol. 2021, 87, e01956-20. [Google Scholar] [CrossRef] [PubMed]
- Allaway, B.; Lodwig, E.; Crompton, L.A.; Wood, M.; Parsons, R.; Wheeler, T.R.; Poole, P.S. Identification of alanine dehydrogenase and its role in mixed secretion of ammonium and alanine by pea bacteroids. Mol. Microbiol. 2000, 36, 508–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunakaran, R.; Ebert, K.; Harvey, S.; Leonard, M.E.; Ramachandran, V.; Poole, P.S. Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 2006, 188, 6661–6668. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Strain | Diameter (cm) | |||
|---|---|---|---|---|
| 25 | 100 | 250 | 1000 | |
| 7653R | 0.96 ± 0.09 a | 2.12 ± 0.17 a | 3.04 ± 0.21 a | 4.22 ± 0.17 a |
| HKhypE | 2.13 ± 0.25 b | 2.75 ± 0.12 b | 3.88 ± 0.18 b | 4.73 ± 0.11 b |
| HKhypE(pBBRhypE) | 1.40 ± 0.26 a | 2.60 ± 0.28 ab | 3.47 ± 0.51 ab | 5.30 ± 0.61 b |
| Strain M. huakuii | The Aboveground Fresh Weight per Plant (mg) β | Number of Total Nodules per Plant β | Acetylene Reduction Activity (nmol of Ethylene/Plant/h) β | Nodule Fresh Weight per Plant (mg of Plant) β | Fresh Weight (mg of Plant) β |
|---|---|---|---|---|---|
| 7653R | 96.41 ± 23.97 a | 17.5 ± 3.5 a | 64.87 ± 10.9 a | 12.0 3± 1.18 a | 129.05 ± 16.55 a |
| HKhypE | 77.56 ± 20.53 a | 11.7 ± 3.8 a | 34.20 ± 0.72 b | 10.10 ± 1.56 a | 83.10 ± 20.68 ab |
| HKhypE(pBBRhypE) | 98.70 ± 18.52 a | 13.7 ± 2.1 a | 52.70 ± 7.50 a | 17.53 ± 9.28 a | 124.03 ± 21.77 a |
| Control γ | 0 | 0 | 0 | 46.08 ± 13.60 b |
| Gene ID | Gene Name | Protein Description | MW [kDa] | Ratio | p Value | qRT-PCR |
|---|---|---|---|---|---|---|
| Stress response and virulence | ||||||
| MCHK_6427 | Formate dehydrogenase | 17.09 | 0.67 | 0.000082 | ||
| MCHK_0466 | Isopenicillin N synthase family oxygenase | 37.22 | 0.67 | 0.000192 | ||
| MCHK_4591 | Oxidoreductase | 27.82 | 0.67 | 0.000901 | ||
| MCHK_4254 | Accessory factor | 10.76 | 0.67 | 0.000011 | ||
| MCHK_10150 | Glutathione S-transferase | 24.74 | 0.66 | 0.001701 | ||
| MCHK_6266 | sfnG | Dimethyl sulfone monooxygenase | 40.68 | 0.66 | 0.000086 | |
| MCHK_1994 | Thioredoxin domain-containing protein | 28.47 | 0.66 | 0.000105 | ||
| MCHK_4344 | Antibiotic biosynthesis monooxygenase | 10.83 | 0.66 | 0.018067 | ||
| MCHK_8201 * | Cold-shock protein | 7.46 | 0.65 | 0.007124 | ||
| MCHK_6140 | trxA | Thioredoxin | 11.45 | 0.66 | 0.000015 | |
| MCHK_4761 | Copper chaperone PCu(A)C | 18.79 | 0.63 | 0.000415 | ||
| MCHK_1937 | Competence/damage-inducible protein | 26.35 | 0.63 | 0.000727 | ||
| MCHK_4145 | Cold-shock protein | 7.36 | 0.62 | 0.000013 | ||
| MCHK_6264 | ssuD | Alkanesulfonate monooxygenase | 42.28 | 0.61 | 0.000139 | |
| MCHK_3694 | Universal stress protein | 15.68 | 0.61 | 0.001310 | ||
| MCHK_1730 | bamE | Outer membrane protein assembly factor BamE | 18.38 | 0.60 | 0.000001 | |
| MCHK_3913 | Response regulator | 22.20 | 0.58 | 0.012234 | ||
| MCHK_6272 | Sulfur acquisition oxidoreductase | 44.72 | 0.58 | 0.000003 | ||
| MCHK_4242 | Cold-shock protein | 7.36 | 0.56 | 0.000046 | ||
| MCHK_5101 | Alkyl hydroperoxide reductase | 24.02 | 0.55 | 0.000007 | ||
| MCHK_3970 | Blue-light-activated histidine kinase | 38.90 | 0.31 | 0.000327 | ||
| Electron transport and nitrogen metabolism β | ||||||
| MCHK_2003 | Peptide chain release factor 2 | 41.93 | 1.59 | 0.000011 | ||
| MCHK_5582 | asd | Aspartate-semialdehyde dehydrogenase | 37.68 | 1.56 | 0.000005 | |
| MCHK_8172 * | nifE | Nitrogenase iron-molybdenum cofactor biosynthesis protein | 54.25 | 0.76 | 0.000009 | |
| MCHK_8175 * | nifD | Nitrogenase protein alpha chain | 55.52 | 0.75 | 0.000001 | |
| MCHK_11255 * | nifX | Nitrogen fixation protein NifX | 18.30 | 0.74 | 0.000258 | 0.19 |
| MCHK_8174 * | nifK | Nitrogenase molybdenum-iron protein beta chain | 57.54 | 0.73 | 0.000005 | |
| MCHK_1461 | Glycine-zipper protein | 10.77 | 0.67 | 0.007390 | ||
| MCHK_4867 | argJ | Arginine biosynthesis bifunctional protein ArgJ | 43.53 | 0.67 | 0.000191 | |
| MCHK_1729 | hppA | K(+)-insensitive pyrophosphate-energized proton pump | 72.53 | 0.67 | 0.000133 | |
| MCHK_2808 | TonB-dependent hemoglobin/transferrin/lactoferrin receptor | 78.12 | 0.63 | 0.000009 | ||
| MCHK_4872 | Parvulin-like PPIase | 32.62 | 0.63 | 0.000006 | ||
| MCHK_8220 * | Ferredoxin-like protein | 11.22 | 0.63 | 0.000046 | ||
| MCHK_4952 | Iron–sulfur metabolism protein | 11.03 | 0.61 | 0.000100 | ||
| MCHK_4282 | Cytochrome b | 48.57 | 0.60 | 0.000122 | ||
| MCHK_5860 | Lipoprotein | 30.60 | 0.60 | 0.000001 | ||
| MCHK_2579 | NAD(P)H nitroreductase | 21.17 | 0.59 | 0.004028 | ||
| MCHK_5859 | ATP-binding cassette protein | 39.11 | 0.57 | 0.001336 | ||
| MCHK_12735 * | Nif11-like leader peptide family natural product | 12.54 | 0.50 | 0.000007 | ||
| Transporter activity | ||||||
| MCHK_2240 | mgtE | Magnesium transporter | 49.34 | 1.59 | 0.004613 | |
| MCHK_6148 | PTS fructose transporter | 14.15 | 0.66 | 0.000040 | ||
| MCHK_5406 | exbB | Biopolymer transport protein | 26.06 | 0.65 | 0.000174 | |
| MCHK_0751 | dctA | C4-dicarboxylate transport protein | 46.13 | 0.65 | 0.004547 | 0.30 |
| MCHK_5366 | MFS transporter | 48.53 | 0.64 | 0.002941 | ||
| MCHK_0068 | Sugar ABC transporter permease | 44.82 | 0.64 | 0.000491 | ||
| MCHK_0065 | Transporter substrate-binding protein | 27.37 | 0.64 | 0.000205 | ||
| MCHK_5842 | Extracellular solute-binding protein | 45.46 | 0.60 | 0.000002 | ||
| MCHK_1547 | ABC transporter substrate-binding protein | 34.13 | 0.59 | 0.000174 | ||
| MCHK_1725 | ABC transporter substrate-binding protein | 31.80 | 0.56 | 0.000000 | ||
| MCHK_4896 | Extracellular solute-binding protein | 35.20 | 0.56 | 0.000000 | ||
| MCHK_0900 | sbmA | Peptide antibiotic transporter | 47.59 | 0.55 | 0.000059 | |
| MCHK_0625 | Transporter substrate-binding domain-containing protein | 34.91 | 0.52 | 0.000281 | ||
| MCHK_5677 | tauA | Taurine ABC transporter substrate-binding protein | 35.37 | 0.51 | 0.000039 | |
| MCHK_3276 | Sulfate ABC transporter substrate-binding protein | 36.02 | 0.51 | 0.000002 | ||
| Carbohydrate metabolism | ||||||
| MCHK_4339 | Anthranilate synthase | 81.05 | 2.32 | 0.000014 | ||
| MCHK_3715 | N-acetyltransferase | 17.31 | 1.58 | 0.018854 | ||
| MCHK_6064 | Phospho-2-dehydro-3-deoxyheptonate aldolase | 38.82 | 1.53 | 0.000117 | ||
| MCHK_5496 | leuC | 2-methyl-cis-aconitate hydratase | 50.89 | 1.50 | 0.000007 | |
| MCHK_1778 | Enoyl-CoA hydratase | 29.06 | 0.67 | 0.000008 | ||
| MCHK_4672 | Tripartite tricarboxylate transporter substrate binding protein | 33.48 | 0.66 | 0.000291 | ||
| MCHK_4592 | Dehydratase | 17.62 | 0.65 | 0.017197 | ||
| MCHK_5108 | Fructose-bisphosphate aldolase | 36.32 | 0.63 | 0.000001 | ||
| MCHK_5188 | phaR | Polyhydroxyalkanoate synthesis repressor PhaR | 23.01 | 0.54 | 0.000042 | 0.45 |
| Nucleotide metabolism | ||||||
| MCHK_5898 | Ribosome biogenesis GTP-binding protein | 23.76 | 0.67 | 0.002586 | ||
| MCHK_3722 | Ester cyclase | 14.75 | 0.67 | 0.014111 | ||
| MCHK_5344 | Pyridoxal phosphate homeostasis protein | 23.62 | 0.66 | 0.001619 | ||
| MCHK_4655 | cdd | Cytidine deaminase | 13.98 | 0.66 | 0.000198 | |
| MCHK_6508 | recR | Recombination protein RecR | 21.46 | 0.65 | 0.000427 | |
| MCHK_1792 | DNA-binding protein HU | 9.18 | 0.63 | 0.000009 | ||
| MCHK_2180 | hfq | RNA chaperone Hfq\ | 11.57 | 0.63 | 0.000042 | 0.40 |
| MCHK_3965 | Transcriptional regulator | 23.35 | 0.27 | 0.000081 | ||
| Unknown function proteins | ||||||
| MCHK_2160 | Uncharacterized protein | 13.09 | 0.67 | 0.001235 | ||
| MCHK_3617 | Uncharacterized protein | 7.02 | 0.67 | 0.003551 | ||
| MCHK_1009 | Uncharacterized protein | 14.09 | 0.67 | 0.000005 | ||
| MCHK_3463 | Uncharacterized protein | 11.22 | 0.66 | 0.000455 | ||
| MCHK_5458 | Uncharacterized protein | 10.60 | 0.64 | 0.000965 | ||
| MCHK_3109 | Uncharacterized protein | 11.20 | 0.64 | 0.000074 | ||
| MCHK_6262 | Uncharacterized protein | 14.25 | 0.64 | 0.000203 | ||
| MCHK_1978 | Uncharacterized protein | 15.74 | 0.64 | 0.000033 | ||
| MCHK_4545 | Uncharacterized protein | 17.29 | 0.64 | 0.000246 | ||
| MCHK_6162 | Uncharacterized protein | 7.90 | 0.64 | 0.000537 | ||
| MCHK_0805 | Uncharacterized protein | 37.47 | 0.63 | 0.000066 | ||
| MCHK_1574 | Uncharacterized protein | 24.45 | 0.63 | 0.000404 | ||
| MCHK_5383 | Uncharacterized protein | 23.59 | 0.62 | 0.000388 | ||
| MCHK_6128 | Uncharacterized protein | 18.16 | 0.61 | 0.000374 | ||
| MCHK_5147 | Uncharacterized protein | 6.78 | 0.61 | 0.000204 | ||
| MCHK_1260 | Uncharacterized protein | 7.60 | 0.60 | 0.000010 | ||
| MCHK_5627 | Uncharacterized protein | 18.69 | 0.59 | 0.001819 | ||
| MCHK_5063 | Uncharacterized protein | 11.40 | 0.57 | 0.000147 | ||
| MCHK_12690 * | Uncharacterized protein | 9.32 | 0.47 | 0.000240 | ||
| Strains | Description | Reference, Source, Sequence |
|---|---|---|
| M. huakuii 7653R | Wild type, Nod+ on Astragalus sinicus | [48] |
| M. huakuii HKhypE | 7653R hypE:pk19mob, Strr Neor | This study |
| M. huakuii HKhypE(pBBRhypE) | HKhypE carrying pBBRhypE; Strr Neor Gmr | This study |
| DH5α | F− lacZDM15 recA1 hsdR17 supE44 D(lacZYA argF) | This study |
| Plasmids | ||
| pK19mob | pUC19 derivative lacZ mob Kmr | [49] |
| pRK2013 | Helper plasmid for mobilizing plasmids Kmr | [50] |
| pKhypE | hypEfor/hypErev PCR product in pK19mob, Kmr | This study |
| pBBR1MCS-5 | lacPOZ′ mob, broad host range, Gmr | [51] |
| pBBRhypE | ||
| Primer * | ||
| hypEfor | Sense primer for hypE mutation | TTTAAGCTTATCGAGGAAGGCATGAAGG |
| hypErev | Antisense primer for hypE mutation | TTTTCTAGACTGCATGGTCACGCGCCCCG |
| hypEmap | Mapping PCR primer for hypE mutation | GCCAAGCCGCTCTATCTGTC |
| pK19A | pK19mob mapping primer | ATCAGATCTTGATCCCCTGC |
| pK19B | pK19mob mapping primer | GCACGAGGGAGCTTCCAGGG |
| chypEfor | Sense PCR primer for complementation of hypE mutant | TTTGGATCCGGTGATCATGGTCATGCGAA |
| chypErev | Antisense PCR primer for complementation of hypE mutant | TTTTCTAGACAGTATGGCGGCGTCAAGAA |
| M13-F | Sense primer for LacZ | CGCCAGGGTTTTCCCAGTCACGAC |
| M13-R | Antisense primer for LacZ | CACACAGGAAACAGCTATGAC |
| QhypE_F | Sense primer for qRT-PCR of hypE | TGAAAGACCTGATCGACGAC |
| QhypE_R | Antisense primer for qRT-PCR of hypE | CAAGCCGGTCGCCATGTTTT |
| QgyrB_F | Sense primer for qRT-PCR of gyrB | TTCGACCAGAATTCCTACAA |
| QgyrB_R | Antisense primer for qRT-PCR of gyrB | GCTCATTTCGAAGATCTGGC |
| MCHK_11255F | Sense primer for qRT-PCR of MCHK_11255 | GCCTCTCACTCGTCACTGAC |
| MCHK_11255R | Antisense primer for qRT-PCR of MCHK_11255 | GCCGAAATGGGCATTGAGGT |
| MCHK_0751F | Sense primer for qRT-PCR of MCHK_0751 | AAGATGATCATCGCCCCGGT |
| MCHK_0751R | Antisense primer for qRT-PCR of MCHK_0751 | CCAGCGTCGAGAAGGTGAGG |
| MCHK_5188F | Sense primer for qRT-PCR of MCHK_5188 | GGGGACGAGCACCTATGTGA |
| MCHK_5188R | Antisense primer for qRT-PCR of MCHK_5188 | AAAATGATCTGAGTCAGCAC |
| MCHK_2180F | Sense primer for qRT-PCR of MCHK_2180 | GATGATGTTTTCCCAGGTCA |
| MCHK_2180R | Antisense primer for qRT-PCR of MCHK_2180 | TGCCCATCAACGTGCATGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, S.; Su, M.; Chen, X.; Hu, A.; Yu, F.; Zou, Q.; Cheng, G. Proteomic and Mutant Analysis of Hydrogenase Maturation Protein Gene hypE in Symbiotic Nitrogen Fixation of Mesorhizobium huakuii. Int. J. Mol. Sci. 2023, 24, 12534. https://doi.org/10.3390/ijms241612534
Long S, Su M, Chen X, Hu A, Yu F, Zou Q, Cheng G. Proteomic and Mutant Analysis of Hydrogenase Maturation Protein Gene hypE in Symbiotic Nitrogen Fixation of Mesorhizobium huakuii. International Journal of Molecular Sciences. 2023; 24(16):12534. https://doi.org/10.3390/ijms241612534
Chicago/Turabian StyleLong, Songhua, Min Su, Xiaohong Chen, Aiqi Hu, Fuyan Yu, Qian Zou, and Guojun Cheng. 2023. "Proteomic and Mutant Analysis of Hydrogenase Maturation Protein Gene hypE in Symbiotic Nitrogen Fixation of Mesorhizobium huakuii" International Journal of Molecular Sciences 24, no. 16: 12534. https://doi.org/10.3390/ijms241612534
APA StyleLong, S., Su, M., Chen, X., Hu, A., Yu, F., Zou, Q., & Cheng, G. (2023). Proteomic and Mutant Analysis of Hydrogenase Maturation Protein Gene hypE in Symbiotic Nitrogen Fixation of Mesorhizobium huakuii. International Journal of Molecular Sciences, 24(16), 12534. https://doi.org/10.3390/ijms241612534




