The Future of Precision Oncology
Abstract
1. Introduction
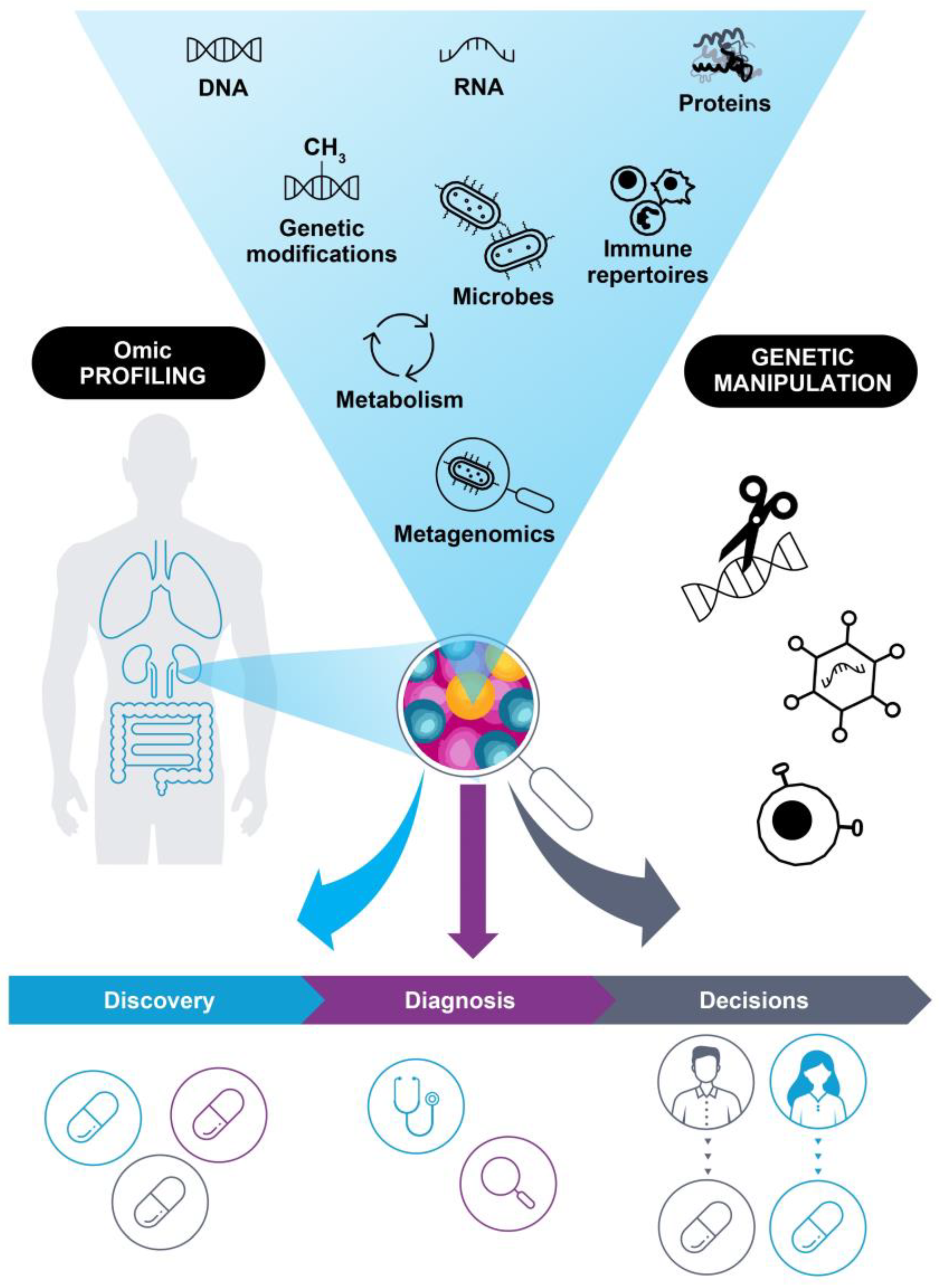
2. The Current State of Molecular Profiling in Precision Oncology
2.1. Landmark Discoveries in Precision Oncology
2.2. Comprehensive Diagnostics for Patients with Cancer
2.3. Increasing Information to Guide Treatment Decisions
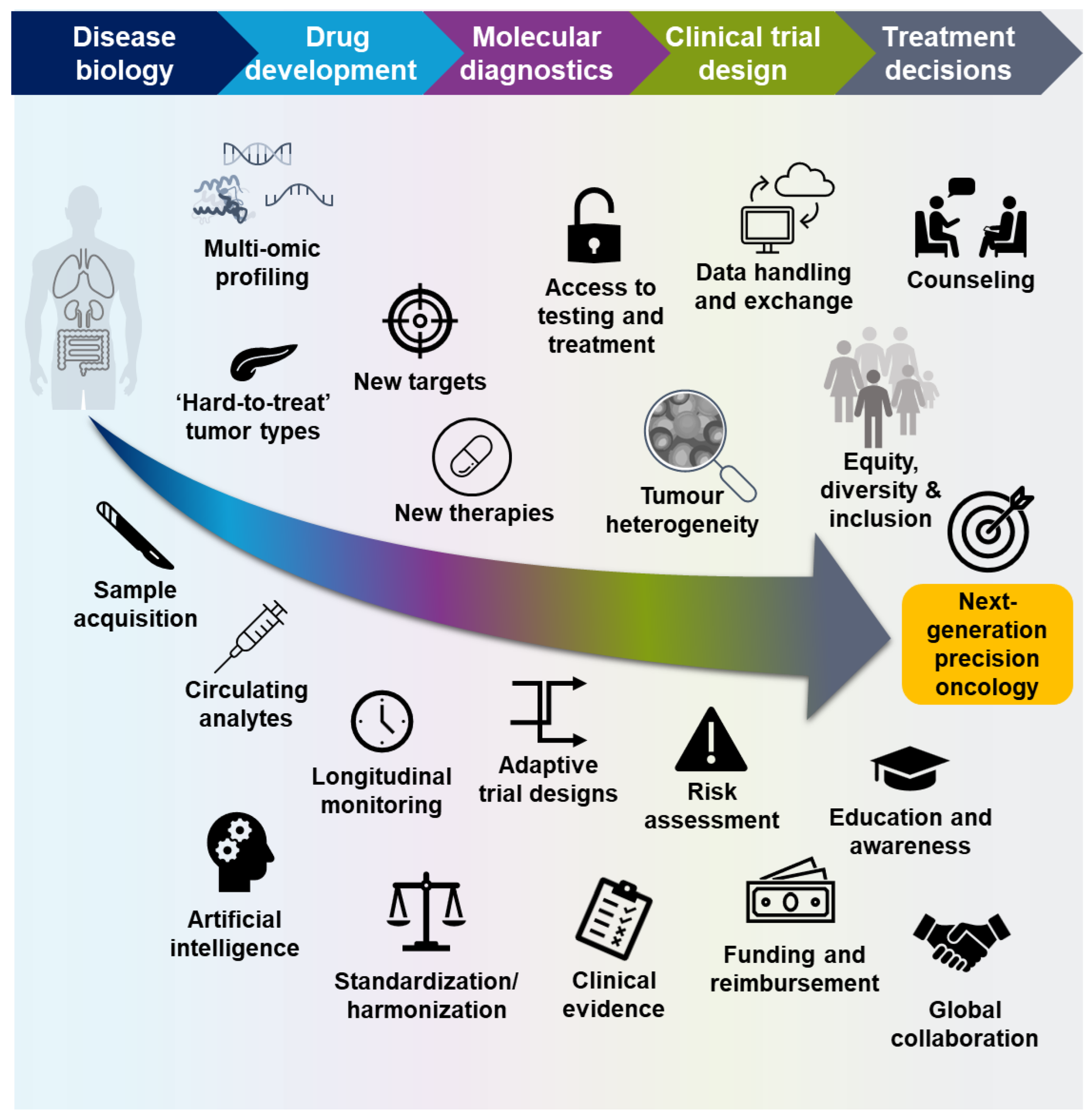
3. Future Directions for Molecular Profiling of Patients with Cancer
3.1. Multi-Omic Profiling in the Characterization of Disease Biology
3.2. Novel Approaches to Drug Development
3.3. Clinical Molecular Diagnostics
3.4. Novel Approaches to Clinical Trials
3.5. Guiding Treatment Decisions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nature. Milestones in Cancer. Available online: https://www.nature.com/immersive/d42859-020-00083-8/index.html (accessed on 27 June 2023).
- Shin, S.H.; Bode, A.M.; Dong, Z. Addressing the challenges of applying precision oncology. NPJ Precis. Oncol. 2017, 1, 28. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, H.; Balling, R.; Beerenwinkel, N.; Kohlbacher, O.; Kumar, S.; Lengauer, T.; Maathuis, M.H.; Moreau, Y.; Murphy, S.A.; Przytycka, T.M.; et al. From hype to reality: Data science enabling personalized medicine. BMC Med. 2018, 16, 150. [Google Scholar] [CrossRef] [PubMed]
- Schwartzberg, L.; Kim, E.S.; Liu, D.; Schrag, D. Precision oncology: Who, how, what, when, and when not? In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2017; Volume 37, pp. 160–169. [Google Scholar] [CrossRef]
- D’Adamo, G.L.; Widdop, J.T.; Giles, E.M. The future is now? Clinical and translational aspects of “omics” technologies. Immunol. Cell Biol. 2021, 99, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Yang, J.; Chen, L.; Wu, T. Editorial: Applications of Metagenomics in Studying Human Cancer. Front. Genet. 2021, 12, 760141. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Hsu, M.H.; Tu, S.J.; Yen, J.C.; Lee, Y.T.; Fang, H.Y.; Chang, J.G. Metatranscriptomic analysis of human lung metagenomes from patients with lung cancer. Genes 2021, 12, 1458. [Google Scholar] [CrossRef]
- Villanueva, L.; Álvarez-Errico, D.; Esteller, M. The Contribution of Epigenetics to Cancer Immunotherapy. Trends Immunol. 2020, 41, 676–691. [Google Scholar] [CrossRef]
- Bulaklak, K.; Gersbach, C.A. The once and future gene therapy. Nat. Commun. 2020, 11, 5820. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Hu, C.; Zhang, D.; Shao, Z.; Jin, Q.; Yang, J.; Xie, H.; Liu, B.; Hu, M.; et al. Synthetic lethality-based identification of targets for anticancer drugs in the human signaling network. Sci. Rep. 2018, 8, 8440. [Google Scholar] [CrossRef]
- Smetana, J.; Brož, P. National genome initiatives in Europe and the United Kingdom in the era of whole-genome sequencing: A comprehensive review. Genes 2022, 13, 556. [Google Scholar] [CrossRef]
- Berlanga, P.; Pierron, G.; Lacroix, L.; Chicard, M.; de Beaumais, T.A.; Marchais, A.; Harttrampf, A.C.; Iddir, Y.; Larive, A.; Soriano Fernandez, A.; et al. The European MAPPYACTS Trial: Precision medicine program in pediatric and adolescent patients with recurrent malignancies. Cancer Discov. 2022, 12, 1266–1281. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018, 173, 371–385.e18. [Google Scholar] [CrossRef]
- Senga, S.S.; Grose, R.P. Hallmarks of cancer-the new testament. Open Biol. 2021, 11, 200358. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Gruber, S.B.; Wu, Z.; Schmidt, E.M.; Zawistowski, M.; Moser, S.E.; Blanc, V.M.; Brummett, C.M.; Kheterpal, S.; Abecasis, G.R.; et al. Association of polygenic risk scores for multiple cancers in a phenome-wide study: Results from The Michigan Genomics Initiative. Am. J. Hum. Genet. 2018, 102, 1048–1061. [Google Scholar] [CrossRef]
- Grant, R.C.; Denroche, R.E.; Borgida, A.; Virtanen, C.; Cook, N.; Smith, A.L.; Connor, A.A.; Wilson, J.M.; Peterson, G.; Roberts, N.J.; et al. Exome-wide association study of pancreatic cancer risk. Gastroenterology 2018, 154, 719–722.e3. [Google Scholar] [CrossRef]
- The 100,000 Genomes Project Pilot Investigators. 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N. Engl. J. Med. 2021, 385, 1868–1880. [Google Scholar] [CrossRef]
- Lee, J.S. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016, 49, 607–611. [Google Scholar] [CrossRef]
- Jensen, E.V.; Block, G.E.; Smith, S.; Kyser, K.; DeSombre, E.R. Estrogen receptors and breast cancer response to adrenalectomy. Natl. Cancer Inst. Monogr. 1971, 34, 55–70. [Google Scholar]
- Cohen, M.H.; Williams, G.A.; Sridhara, R.; Chen, G.; McGuinn, W.D., Jr.; Morse, D.; Abraham, S.; Rahman, A.; Liang, C.; Lostritto, R.; et al. United States Food and Drug Administration Drug approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin. Cancer Res. 2004, 10, 1212–1218. [Google Scholar] [CrossRef]
- Ricciuti, B.; Genova, C.; Crinò, L.; Libra, M.; Leonardi, G.C. Antitumor activity of larotrectinib in tumors harboring NTRK gene fusions: A short review on the current evidence. Onco. Targets Ther. 2019, 12, 3171–3179. [Google Scholar] [CrossRef]
- Cunningham, D.; Humblet, Y.; Siena, S.; Khayat, D.; Bleiberg, H.; Santoro, A.; Bets, D.; Mueser, M.; Harstrick, A.; Verslype, C.; et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N. Engl. J. Med. 2004, 351, 337–345. [Google Scholar] [CrossRef]
- Skoulidis, F.; Li, B.T.; Dy, G.K.; Price, T.J.; Falchook, G.S.; Wolf, J.; Italiano, A.; Schuler, M.; Borghaei, H.; Barlesi, F.; et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021, 384, 2371–2381. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef]
- Eiermann, W. Trastuzumab combined with chemotherapy for the treatment of HER2-positive metastatic breast cancer: Pivotal trial data. Ann. Oncol. 2001, 12 (Suppl. S1), S57–S62. [Google Scholar] [CrossRef]
- O’Brien, S.G.; Guilhot, F.; Larson, R.A.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef]
- Shih, C.; Weinberg, R.A. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell 1982, 29, 161–169. [Google Scholar] [CrossRef]
- Reinartz, J.; Bruyns, E.; Lin, J.Z.; Burcham, T.; Brenner, S.; Bowen, B.; Kramer, M.; Woychik, R. Massively parallel signature sequencing (MPSS) as a tool for in-depth quantitative gene expression profiling in all organisms. Brief. Funct. Genom. 2002, 1, 95–104. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351. [Google Scholar] [CrossRef]
- Vieira, A.F.; Schmitt, F. An update on breast cancer multigene prognostic tests—Emergent clinical biomarkers. Front. Med. 2018, 5, 248. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Nucleic Acid Based Tests. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/nucleic-acid-based-tests#human (accessed on 27 June 2023).
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- US Food and Drug Administration. Considerations for Design, Development, and Analytical Validation of Next Generation Sequencing (NGS)—Based In Vitro Diagnostics (IVDs) Intended to Aid in the Diagnosis of Suspected Germline Diseases. Available online: https://www.fda.gov/media/99030/download (accessed on 27 June 2023).
- Colomer, R.; Mondejar, R.; Romero-Laorden, N.; Alfranca, A.; Sanchez-Madrid, F.; Quintela-Fandino, M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine 2020, 25, 100487. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhou, Y.; Shi, J. Extrachromosomal circular DNA: A new potential role in cancer progression. J. Transl. Med. 2021, 19, 257. [Google Scholar] [CrossRef]
- Dai, J.; Su, Y.; Zhong, S.; Cong, L.; Liu, B.; Yang, J.; Tao, Y.; He, Z.; Chen, C.; Jiang, Y. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct. Target. Ther. 2020, 5, 145. [Google Scholar] [CrossRef]
- Russo, A.; Incorvaia, L.; Del Re, M.; Malapelle, U.; Capoluongo, E.; Gristina, V.; Castiglia, M.; Danesi, R.; Fassan, M.; Giuffre, G.; et al. The molecular profiling of solid tumors by liquid biopsy: A position paper of the AIOM-SIAPEC-IAP-SIBioC-SIC-SIF Italian Scientific Societies. ESMO Open 2021, 6, 100164. [Google Scholar] [CrossRef]
- Openshaw, M.R.; Mohamed, A.A.; Ottolini, B.; Fernandez-Garcia, D.; Richards, C.J.; Page, K.; Guttery, D.S.; Thomas, A.L.; Shaw, J.A. Longitudinal monitoring of circulating tumour DNA improves prognostication and relapse detection in gastroesophageal adenocarcinoma. Br. J. Cancer 2020, 123, 1271–1279. [Google Scholar] [CrossRef] [PubMed]
- Buttitta, F.; Felicioni, L.; Lorito, A.D.; Cortellini, A.; Irtelli, L.; Brocco, D.; Marino, P.D.; Traisci, D.; D’Ostilio, N.; Paolo, A.D.; et al. Early prediction of resistance to tyrosine kinase inhibitors by plasma monitoring of EGFR mutations in NSCLC: A new algorithm for patient selection and personalized treatment. Oncotarget 2020, 11, 982–991. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and plasma EGFR mutation analysis in the FLAURA trial: Osimertinib versus comparator EGFR tyrosine kinase inhibitor as first-line treatment in patients with EGFR-mutated advanced non-small cell lung cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef] [PubMed]
- Jamal-Hanjani, M.; Hackshaw, A.; Ngai, Y.; Shaw, J.; Dive, C.; Quezada, S.; Middleton, G.; de Bruin, E.; Le Quesne, J.; Shafi, S.; et al. Tracking genomic cancer evolution for precision medicine: The lung TRACERx study. PLoS Biol. 2014, 12, e1001906. [Google Scholar] [CrossRef]
- Prince, E.A.; Sanzari, J.K.; Pandya, D.; Huron, D.; Edwards, R. Analytical concordance of PD-L1 assays utilizing antibodies from FDA-approved diagnostics in advanced cancers: A systematic literature review. JCO Precis. Oncol. 2021, 5, 953–973. [Google Scholar] [CrossRef]
- Büttner, R.; Longshore, J.W.; López-Ríos, F.; Merkelbach-Bruse, S.; Normanno, N.; Rouleau, E.; Penault-Llorca, F. Implementing TMB measurement in clinical practice: Considerations on assay requirements. ESMO Open 2019, 4, e000442. [Google Scholar] [CrossRef]
- Valla, V.; Alzabin, S.; Koukoura, A.; Lewis, A.; Nielsen, A.A.; Vassiliadis, E. Companion diagnostics: State of the art and new regulations. Biomark Insights 2021, 16, 11772719211047763. [Google Scholar] [CrossRef]
- Phillips, K.A.; Douglas, M.P.; Wordsworth, S.; Buchanan, J.; Marshall, D.A. Availability and funding of clinical genomic sequencing globally. BMJ Glob. Health 2021, 6, e004415. [Google Scholar] [CrossRef]
- NHS England. National Genomic Test Directory. Available online: https://www.england.nhs.uk/publication/national-genomic-test-directories/ (accessed on 27 June 2023).
- Oberg, J.A.; Glade Bender, J.L.; Sulis, M.L.; Pendrick, D.; Sireci, A.N.; Hsiao, S.J.; Turk, A.T.; Dela Cruz, F.S.; Hibshoosh, H.; Remotti, H.; et al. Implementation of next generation sequencing into pediatric hematology-oncology practice: Moving beyond actionable alterations. Genome Med. 2016, 8, 133. [Google Scholar] [CrossRef]
- Harris, M.H.; DuBois, S.G.; Glade Bender, J.L.; Kim, A.; Crompton, B.D.; Parker, E.; Dumont, I.P.; Hong, A.L.; Guo, D.; Church, A.; et al. Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: The individualized cancer therapy (iCat) study. JAMA Oncol. 2016, 2, 608–615. [Google Scholar] [CrossRef]
- Chang, W.; Brohl, A.S.; Patidar, R.; Sindiri, S.; Shern, J.F.; Wei, J.S.; Song, Y.K.; Yohe, M.E.; Gryder, B.; Zhang, S.; et al. Multidimensional clinomics for precision therapy of children and adolescent young adults with relapsed and refractory cancer: A report from the Center for Cancer Research. Clin. Cancer Res. 2016, 22, 3810–3820. [Google Scholar] [CrossRef]
- Izquierdo, E.; Yuan, L.; George, S.; Hubank, M.; Jones, C.; Proszek, P.; Shipley, J.; Gatz, S.A.; Stinson, C.; Moore, A.S.; et al. Development of a targeted sequencing approach to identify prognostic, predictive and diagnostic markers in paediatric solid tumours. Oncotarget 2017, 8, 112036–112050. [Google Scholar] [CrossRef]
- Harttrampf, A.C.; Lacroix, L.; Deloger, M.; Deschamps, F.; Puget, S.; Auger, N.; Vielh, P.; Varlet, P.; Balogh, Z.; Abbou, S.; et al. Molecular screening for cancer treatment optimization (MOSCATO-01) in pediatric patients: A single-institutional prospective molecular stratification trial. Clin. Cancer Res. 2017, 23, 6101–6112. [Google Scholar] [CrossRef]
- Surrey, L.F.; MacFarland, S.P.; Chang, F.; Cao, K.; Rathi, K.S.; Akgumus, G.T.; Gallo, D.; Lin, F.; Gleason, A.; Raman, P.; et al. Clinical utility of custom-designed NGS panel testing in pediatric tumors. Genome Med. 2019, 11, 32. [Google Scholar] [CrossRef]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Wong, M.; Mayoh, C.; Lau, L.M.S.; Khuong-Quang, D.A.; Pinese, M.; Kumar, A.; Barahona, P.; Wilkie, E.E.; Sullivan, P.; Bowen-James, R.; et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 2020, 26, 1742–1753. [Google Scholar] [CrossRef]
- Parsons, D.W.; Janeway, K.A.; Patton, D.R.; Winter, C.L.; Coffey, B.; Williams, P.M.; Roy-Chowdhuri, S.; Tsongalis, G.J.; Routbort, M.; Ramirez, N.C.; et al. Actionable tumor alterations and treatment protocol enrollment of pediatric and young adult patients with refractory cancers in The National Cancer Institute-Children’s Oncology Group pediatric MATCH trial. J. Clin. Oncol. 2022, 40, 2224–2234. [Google Scholar] [CrossRef]
- Chabanon, R.M.; Pedrero, M.; Lefebvre, C.; Marabelle, A.; Soria, J.C.; Postel-Vinay, S. Mutational landscape and sensitivity to immune checkpoint blockers. Clin. Cancer Res. 2016, 22, 4309–4321. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Peeters, M.; Siena, S.; Humblet, Y.; Hendlisz, A.; Neyns, B.; Canon, J.L.; Van Laethem, J.L.; Maurel, J.; Richardson, G.; et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J. Clin. Oncol. 2007, 25, 1658–1664. [Google Scholar] [CrossRef]
- Zhu, A.X.; Macarulla, T.; Javle, M.M.; Kelley, R.K.; Lubner, S.J.; Adeva, J.; Cleary, J.M.; Catenacci, D.V.T.; Borad, M.J.; Bridgewater, J.A.; et al. Final overall survival efficacy results of ivosidenib for patients with advanced cholangiocarcinoma with IDH1 mutation: The phase 3 randomized clinical ClarIDHy trial. JAMA Oncol. 2021, 7, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.W.; Jin, J. The value of companion diagnostics in oncology drug development. Expert. Rev. Mol. Diagn. 2022, 22, 591–593. [Google Scholar] [CrossRef] [PubMed]
- The IQVIA Institute. Supporting Precision Oncology: Targeted Therapies, Immuno-Oncology, and Predictive Biomarker-Based Medicines. Available online: https://www.iqvia.com/insights/the-iqvia-institute/reports/supporting-precision-oncology (accessed on 27 June 2023).
- Smith, J.C.; Sheltzer, J.M. Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 2022, 38, 110569. [Google Scholar] [CrossRef] [PubMed]
- García-Figueiras, R.; Baleato-González, S.; Padhani, A.R.; Luna-Alcalá, A.; Vallejo-Casas, J.A.; Sala, E.; Vilanova, J.C.; Koh, D.M.; Herranz-Carnero, M.; Vargas, H.A. How clinical imaging can assess cancer biology. Insights Imaging 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef]
- Cercek, A.; Lumish, M.; Sinopoli, J.; Weiss, J.; Shia, J.; Lamendola-Essel, M.; El Dika, I.H.; Segal, N.; Shcherba, M.; Sugarman, R.; et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N. Engl. J. Med. 2022, 386, 2363–2376. [Google Scholar] [CrossRef]
- Bilkey, G.A.; Burns, B.L.; Coles, E.P.; Mahede, T.; Baynam, G.; Nowak, K.J. Optimizing precision medicine for public health. Front. Public Health 2019, 7, 42. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Campbell, B.B.; Light, N.; Fabrizio, D.; Zatzman, M.; Fuligni, F.; de Borja, R.; Davidson, S.; Edwards, M.; Elvin, J.A.; Hodel, K.P.; et al. Comprehensive analysis of hypermutation in human cancer. Cell 2017, 171, 1042–1056. [Google Scholar] [CrossRef]
- Golia D’Augè, T.; Cuccu, I.; Santangelo, G.; Muzii, L.; Giannini, A.; Bogani, G.; Di Donato, V. Novel insights into molecular mechanisms of endometrial diseases. Biomolecules 2023, 13, 499. [Google Scholar] [CrossRef]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Cuccu, I.; D’Oria, O.; Sgamba, L.; De Angelis, E.; Golia D’Augè, T.; Turetta, C.; Di Dio, C.; Scudo, M.; Bogani, G.; Di Donato, V.; et al. Role of genomic and molecular biology in the modulation of the treatment of endometrial cancer: Narrative review and perspectives. Healthcare 2023, 11, 571. [Google Scholar] [CrossRef]
- Ossandon, M.R.; Agrawal, L.; Bernhard, E.J.; Conley, B.A.; Dey, S.M.; Divi, R.L.; Guan, P.; Lively, T.G.; McKee, T.C.; Sorg, B.S.; et al. Circulating tumor DNA assays in clinical cancer research. JNCI J. Natl. Cancer Inst. 2018, 110, 929–934. [Google Scholar] [CrossRef]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour heterogeneity in the clinic. Nature 2013, 501, 355–364. [Google Scholar] [CrossRef]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Leroux, C.; Konstantinidou, G. Targeted therapies for pancreatic cancer: Overview of current treatments and new opportunities for personalized oncology. Cancers 2021, 13, 799. [Google Scholar] [CrossRef]
- Torres, C.; Grippo, P.J. Pancreatic cancer subtypes: A roadmap for precision medicine. Ann. Med. 2018, 50, 277–287. [Google Scholar] [CrossRef]
- Lin, W.; Noel, P.; Borazanci, E.H.; Lee, J.; Amini, A.; Han, I.W.; Heo, J.S.; Jameson, G.S.; Fraser, C.; Steinbach, M.; et al. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med. 2020, 12, 80. [Google Scholar] [CrossRef]
- Grünwald, B.T.; Devisme, A.; Andrieux, G.; Vyas, F.; Aliar, K.; McCloskey, C.W.; Macklin, A.; Jang, G.H.; Denroche, R.; Romero, J.M.; et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 2021, 184, 5577–5592.e18. [Google Scholar] [CrossRef]
- Ligorio, M.; Sil, S.; Malagon-Lopez, J.; Nieman, L.T.; Misale, S.; Di Pilato, M.; Ebright, R.Y.; Karabacak, M.N.; Kulkarni, A.S.; Liu, A.; et al. Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell 2019, 178, 160–175.e27. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, S.; Winter, P.S.; Navia, A.W.; Williams, H.L.; DenAdel, A.; Lowder, K.E.; Galvez-Reyes, J.; Kalekar, R.L.; Mulugeta, N.; Kapner, K.S.; et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021, 184, 6119–6137.e26. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.J.; Nowak, J.A.; Camarda, N.D.; Moffitt, R.A.; Ghazani, A.A.; Hazar-Rethinam, M.; Raghavan, S.; Kim, J.; Brais, L.K.; Ragon, D.; et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018, 8, 1096–1111. [Google Scholar] [CrossRef] [PubMed]
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Wang, Q.; Li, M.; Guo, H.; Liu, W.; Wang, F.; Tian, X.; Yang, Y. Single-cell RNA-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression. EBioMedicine 2021, 66, 103315. [Google Scholar] [CrossRef]
- Menezes, S.; Okail, M.H.; Jalil, S.M.A.; Kocher, H.M.; Cameron, A.J.M. Cancer-associated fibroblasts in pancreatic cancer: New subtypes, new markers, new targets. J. Pathol. 2022, 257, 526–544. [Google Scholar] [CrossRef]
- Kwon, Y.W.; Jo, H.S.; Bae, S.; Seo, Y.; Song, P.; Song, M.; Yoon, J.H. Application of proteomics in cancer: Recent trends and approaches for biomarkers discovery. Front. Med. 2021, 8, 747333. [Google Scholar] [CrossRef]
- Casado, P.; Cutillas, P.R. Proteomic characterization of acute myeloid leukemia for precision medicine. Mol. Cell Proteom. 2023, 22, 100517. [Google Scholar] [CrossRef]
- Higgins, L.; Gerdes, H.; Cutillas, P.R. Principles of phosphoproteomics and applications in cancer research. Biochem. J. 2023, 480, 403–420. [Google Scholar] [CrossRef]
- Casado, P.; Rio-Machin, A.; Miettinen, J.J.; Bewicke-Copley, F.; Rouault-Pierre, K.; Krizsan, S.; Parsons, A.; Rajeeve, V.; Miraki-Moud, F.; Taussig, D.C.; et al. Integrative phosphoproteomics defines two biologically distinct groups of KMT2A rearranged acute myeloid leukaemia with different drug response phenotypes. Signal Transduct. Target. Ther. 2023, 8, 80. [Google Scholar] [CrossRef]
- Dudani, J.S.; Warren, A.D.; Bhatia, S.N. Harnessing protease activity to improve cancer care. Annu. Rev. Cancer Biol. 2018, 2, 353–376. [Google Scholar] [CrossRef]
- Gerdes, H.; Casado, P.; Dokal, A.; Hijazi, M.; Akhtar, N.; Osuntola, R.; Rajeeve, V.; Fitzgibbon, J.; Travers, J.; Britton, D.; et al. Drug ranking using machine learning systematically predicts the efficacy of anti-cancer drugs. Nat. Commun. 2021, 12, 1850. [Google Scholar] [CrossRef]
- Macklin, A.; Khan, S.; Kislinger, T. Recent advances in mass spectrometry based clinical proteomics: Applications to cancer research. Clin. Proteom. 2020, 17, 17. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Wolf, S.; Buettner, F.; Alexe, G.; Häupl, B.; Comoglio, F.; Schneider, C.; Doebele, C.; Fuhrmann, D.C.; Wagner, S.; et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell 2022, 40, 301–317.e12. [Google Scholar] [CrossRef]
- Tierney, C.; Bazou, D.; Majumder, M.M.; Anttila, P.; Silvennoinen, R.; Heckman, C.A.; Dowling, P.; O’Gorman, P. Next generation proteomics with drug sensitivity screening identifies sub-clones informing therapeutic and drug development strategies for multiple myeloma patients. Sci. Rep. 2021, 11, 12866. [Google Scholar] [CrossRef]
- Dupont, C.A.; Riegel, K.; Pompaiah, M.; Juhl, H.; Rajalingam, K. Druggable genome and precision medicine in cancer: Current challenges. FEBS J. 2021, 288, 6142–6158. [Google Scholar] [CrossRef]
- Oprea, T.I.; Bologa, C.G.; Brunak, S.; Campbell, A.; Gan, G.N.; Gaulton, A.; Gomez, S.M.; Guha, R.; Hersey, A.; Holmes, J.; et al. Unexplored therapeutic opportunities in the human genome. Nat. Rev. Drug. Discov. 2018, 17, 377. [Google Scholar] [CrossRef]
- Coleman, N.; Rodon, J. Taking aim at the undruggable. In American Society of Clinical Oncology Educational Book; American Society of Clinical Oncology: Alexandria, VA, USA, 2021; pp. e145–e152. [Google Scholar] [CrossRef]
- Neijssen, J.; Cardoso, R.M.F.; Chevalier, K.M.; Wiegman, L.; Valerius, T.; Anderson, G.M.; Moores, S.L.; Schuurman, J.; Parren, P.; Strohl, W.R.; et al. Discovery of amivantamab (JNJ-61186372), a bispecific antibody targeting EGFR and MET. J. Biol. Chem. 2021, 296, 100641. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Huang, C.S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Mudd, G.E.; Scott, H.; Chen, L.; van Rietschoten, K.; Ivanova-Berndt, G.; Dzionek, K.; Brown, A.; Watcham, S.; White, L.; Park, P.U.; et al. Discovery of BT8009: A nectin-4 targeting bicycle toxin conjugate for the treatment of cancer. J. Med. Chem. 2022, 65, 14337–14347. [Google Scholar] [CrossRef]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, E.; Segura-Cabrera, A.; Pacini, C.; Picco, G.; Behan, F.M.; Jaaks, P.; Coker, E.A.; van der Meer, D.; Barthorpe, A.; Lightfoot, H.; et al. Drug mechanism-of-action discovery through the integration of pharmacological and CRISPR screens. Mol. Syst. Biol. 2020, 16, e9405. [Google Scholar] [CrossRef] [PubMed]
- AstraZeneca UK Ltd. LYNPARZA. Summary of Product Characteristics. Available online: https://www.medicines.org.uk/emc/product/9204/smpc/print (accessed on 27 June 2023).
- Clovis Oncology UK Ltd. RUBRACA®. Summary of Product Characteristics. Available online: https://www.medicines.org.uk/emc/product/10027/smpc/print (accessed on 27 June 2023).
- GlaxoSmithKline UK. ZEJULA. Summary of Product Characteristics. Available online: https://www.medicines.org.uk/emc/product/8828/smpc/print (accessed on 27 June 2023).
- Lord, C.J.; Quinn, N.; Ryan, C.J. Integrative analysis of large-scale loss-of-function screens identifies robust cancer-associated genetic interactions. eLife 2020, 9, e58925. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Boucher, J.I.; Paulsen, J.; Matuszewski, S.; Eide, C.A.; Ou, J.; Eickelberg, G.; Press, R.D.; Zhu, L.J.; Druker, B.J.; et al. CRISPR-Cas9–mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc. Natl. Acad. Sci. USA 2017, 114, 11751–11756. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yan, Z. Systematic prediction of drug resistance caused by transporter genes in cancer cells. Sci. Rep. 2021, 11, 7400. [Google Scholar] [CrossRef]
- Zhang, Z.; Rohweder, P.J.; Ongpipattanakul, C.; Basu, K.; Bohn, M.F.; Dugan, E.J.; Steri, V.; Hann, B.; Shokat, K.M.; Craik, C.S. A covalent inhibitor of K-Ras(G12C) induces MHC class I presentation of haptenated peptide neoepitopes targetable by immunotherapy. Cancer Cell 2022, 40, 1060–1069.e7. [Google Scholar] [CrossRef]
- Choi, J.; Manzano, A.; Dong, W.; Bellone, S.; Bonazzoli, E.; Zammataro, L.; Yao, X.; Deshpande, A.; Zaidi, S.; Guglielmi, A.; et al. Integrated mutational landscape analysis of uterine leiomyosarcomas. Proc. Natl. Acad. Sci. USA 2021, 118, e2025182118. [Google Scholar] [CrossRef]
- Prendergast, S.C.; Strobl, A.C.; Cross, W.; Pillay, N.; Strauss, S.J.; Ye, H.; Lindsay, D.; Tirabosco, R.; Chalker, J.; Mahamdallie, S.S.; et al. Sarcoma and the 100,000 Genomes Project: Our experience and changes to practice. J. Pathol. Clin. Res. 2020, 6, 297–307. [Google Scholar] [CrossRef]
- Schipper, L.J.; Monkhorst, K.; Samsom, K.G.; Bosch, L.J.W.; Snaebjornsson, P.; van Boven, H.; Roepman, P.; van der Kolk, L.E.; van Houdt, W.J.; van der Graaf, W.T.A.; et al. Clinical impact of prospective whole genome sequencing in sarcoma patients. Cancers 2022, 14, 436. [Google Scholar] [CrossRef]
- Chen, X.; Gole, J.; Gore, A.; He, Q.; Lu, M.; Min, J.; Yuan, Z.; Yang, X.; Jiang, Y.; Zhang, T.; et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat. Commun. 2020, 11, 3475. [Google Scholar] [CrossRef]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Ruhen, O.; Lak, N.S.M.; Stutterheim, J.; Danielli, S.G.; Chicard, M.; Iddir, Y.; Saint-Charles, A.; Di Paolo, V.; Tombolan, L.; Gatz, S.A.; et al. Molecular characterization of circulating tumor DNA in pediatric rhabdomyosarcoma: A feasibility study. JCO Precis. Oncol. 2022, 6, e2100534. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Bifulco, C.; Palmieri, G.; Peters, S.; Sidiropoulos, N. Preanalytic variables and tissue stewardship for reliable next-generation sequencing (NGS) clinical analysis. J. Mol. Diagn. 2019, 21, 756–767. [Google Scholar] [CrossRef]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef]
- Friends of Cancer Research. Regulatory Advancements for Patients: 2021 Scientific Report. Available online: https://friendsofcancerresearch.org/wp-content/uploads/2021-Friends-of-Cancer-Research-Scientific-Report.pdf (accessed on 27 June 2023).
- International Quality Network for Pathology. Mission. Available online: http://www.iqnpath.org/mission/ (accessed on 27 June 2023).
- Quality in Pathology (QuIP). QuIP: Biomarker Information. Available online: https://www.qualityinpathology.com/en_GB/biomarker-information (accessed on 27 June 2023).
- BloodPAC. Annual Report: Improving Patient Outcomes through Collaboration. Available online: https://static1.squarespace.com/static/5f9346ec7a77064e62dee0ad/t/613faa8d0487ba1ca41f8b6b/1631562384617/BP-Annual2020.pdf (accessed on 27 June 2023).
- Cancer Treatment and Monitoring through Identification of Circulating Tumor Cells and Tumor Related Nucleic Acids in Blood. Available online: https://cordis.europa.eu/project/rcn/203725/en (accessed on 27 June 2023).
- Horgan, D.; Curigliano, G.; Riess, O.; Hofman, P.; Buttner, R.; Conte, P.; Cufer, T.; Gallagher, W.M.; Georges, N.; Kerr, K.; et al. Identifying the steps required to effectively implement next-generation sequencing in oncology at a national level in Europe. J. Pers. Med. 2022, 12, 72. [Google Scholar] [CrossRef]
- Slembrouck, L.; Darrigues, L.; Laurent, C.; Mittempergher, L.; Delahaye, L.J.; Vanden Bempt, I.; Vander Borght, S.; Vliegen, L.; Sintubin, P.; Raynal, V.; et al. Decentralization of next-generation RNA sequencing-based MammaPrint® and BluePrint® kit at University Hospitals Leuven and Curie Institute Paris. Transl. Oncol. 2019, 12, 1557–1565. [Google Scholar] [CrossRef]
- Deak, K.L.; Jackson, J.B.; Valkenburg, K.C.; Keefer, L.A.; Robinson Gerding, K.M.; Angiuoli, S.V.; Datto, M.B.; McCall, S.J. Next-generation sequencing concordance analysis of comprehensive solid tumor profiling between a centralized specialty laboratory and the decentralized personal genome diagnostics elio tissue complete kitted solution. J. Mol. Diagn. 2021, 23, 1324–1333. [Google Scholar] [CrossRef]
- Shen, Q.; Lu, D.; Schelin, M.E.C.; Jöud, A.; Cao, Y.; Adami, H.-O.; Cnattingius, S.; Fall, K.; Valdimarsdóttir, U.; Fang, F. Injuries before and after diagnosis of cancer: Nationwide register based study. BMJ 2016, 354, i4218. [Google Scholar] [CrossRef]
- Noor, N.M.; Love, S.B.; Isaacs, T.; Kaplan, R.; Parmar, M.K.B.; Sydes, M.R. Uptake of the multi-arm multi-stage (MAMS) adaptive platform approach: A trial-registry review of late-phase randomised clinical trials. BMJ Open 2022, 12, e055615. [Google Scholar] [CrossRef]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef]
- Fors, M.; González, P. Current status of Bayesian clinical trials for oncology, 2020. Contemp. Clin. Trials Commun. 2020, 20, 100658. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Arida, J.A.; Donovan, H.S. The application of crowdsourcing approaches to cancer research: A systematic review. Cancer Med. 2017, 6, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Ballester, P.J.; Carmona, J. Artificial intelligence for the next generation of precision oncology. NPJ Precis. Oncol. 2021, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Blum, A.; Wang, P.; Zenklusen, J.C. SnapShot: TCGA-analyzed tumors. Cell 2018, 173, 530. [Google Scholar] [CrossRef]
- Vincent, B.G.; Szustakowski, J.D.; Doshi, P.; Mason, M.; Guinney, J.; Carbone, D.P. Pursuing better biomarkers for immunotherapy response in cancer through a crowdsourced data challenge. JCO Precis. Oncol. 2021, 5, 51–54. [Google Scholar] [CrossRef]
- Guinney, J.; Wang, T.; Laajala, T.D.; Winner, K.K.; Bare, J.C.; Neto, E.C.; Khan, S.A.; Peddinti, G.; Airola, A.; Pahikkala, T.; et al. Prediction of overall survival for patients with metastatic castration-resistant prostate cancer: Development of a prognostic model through a crowdsourced challenge with open clinical trial data. Lancet Oncol. 2017, 18, 132–142. [Google Scholar] [CrossRef]
- Ciardiello, F.; Adams, R.; Tabernero, J.; Seufferlein, T.; Taieb, J.; Moiseyenko, V.; Ma, B.; Lopez, G.; Vansteenkiste, J.F.; Esser, R.; et al. Awareness, understanding, and adoption of precision medicine to deliver personalized treatment for patients with cancer: A multinational survey comparison of physicians and patients. Oncologist 2016, 21, 292–300. [Google Scholar] [CrossRef]
- Miga, K.H.; Wang, T. The need for a human pangenome reference sequence. Annu. Rev. Genom. Hum. Genet. 2021, 22, 81–102. [Google Scholar] [CrossRef]
- Oyer, R.A.; Hurley, P.; Boehmer, L.; Bruinooge, S.S.; Levit, K.; Barrett, N.; Benson, A.; Bernick, L.A.; Byatt, L.; Charlot, M.; et al. Increasing racial and ethnic diversity in cancer clinical trials: An American Society of Clinical Oncology and Association of Community Cancer Centers joint research statement. J. Clin. Oncol. 2022, 40, 2163–2171. [Google Scholar] [CrossRef]
- Strobl, M.A.R.; West, J.; Viossat, Y.; Damaghi, M.; Robertson-Tessi, M.; Brown, J.S.; Gatenby, R.A.; Maini, P.K.; Anderson, A.R.A. Turnover modulates the need for a cost of resistance in adaptive therapy. Cancer Res. 2021, 81, 1135–1147. [Google Scholar] [CrossRef]
- Trapani, D.; Franzoi, M.A.; Burstein, H.J.; Carey, L.A.; Delaloge, S.; Harbeck, N.; Hayes, D.F.; Kalinsky, K.; Pusztai, L.; Regan, M.M.; et al. Risk-adapted modulation through de-intensification of cancer treatments: An ESMO classification. Ann. Oncol. 2022, 33, 702–712. [Google Scholar] [CrossRef]
- Sanz-Garcia, E.; Zhao, E.; Bratman, S.V.; Siu, L.L. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: Current research, opportunities, and challenges. Sci. Adv. 2022, 8, eabi8618. [Google Scholar] [CrossRef]
- Plana, D.; Palmer, A.C.; Sorger, P.K. Independent drug action in combination therapy: Implications for precision oncology. Cancer Discov. 2022, 12, 606–624. [Google Scholar] [CrossRef]
- Kimmelman, J.; Tannock, I. The paradox of precision medicine. Nat. Rev. Clin. Oncol. 2018, 15, 341–342. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rulten, S.L.; Grose, R.P.; Gatz, S.A.; Jones, J.L.; Cameron, A.J.M. The Future of Precision Oncology. Int. J. Mol. Sci. 2023, 24, 12613. https://doi.org/10.3390/ijms241612613
Rulten SL, Grose RP, Gatz SA, Jones JL, Cameron AJM. The Future of Precision Oncology. International Journal of Molecular Sciences. 2023; 24(16):12613. https://doi.org/10.3390/ijms241612613
Chicago/Turabian StyleRulten, Stuart L., Richard P. Grose, Susanne A. Gatz, J. Louise Jones, and Angus J. M. Cameron. 2023. "The Future of Precision Oncology" International Journal of Molecular Sciences 24, no. 16: 12613. https://doi.org/10.3390/ijms241612613
APA StyleRulten, S. L., Grose, R. P., Gatz, S. A., Jones, J. L., & Cameron, A. J. M. (2023). The Future of Precision Oncology. International Journal of Molecular Sciences, 24(16), 12613. https://doi.org/10.3390/ijms241612613






