Expression Profiling along the Murine Intestine: Different Mucosal Protection Systems and Alterations in Tff1-Deficient Animals
Abstract
:1. Introduction
2. Results
2.1. Expression Profiling of the Murine Intestine (RT-PCR Analysis)
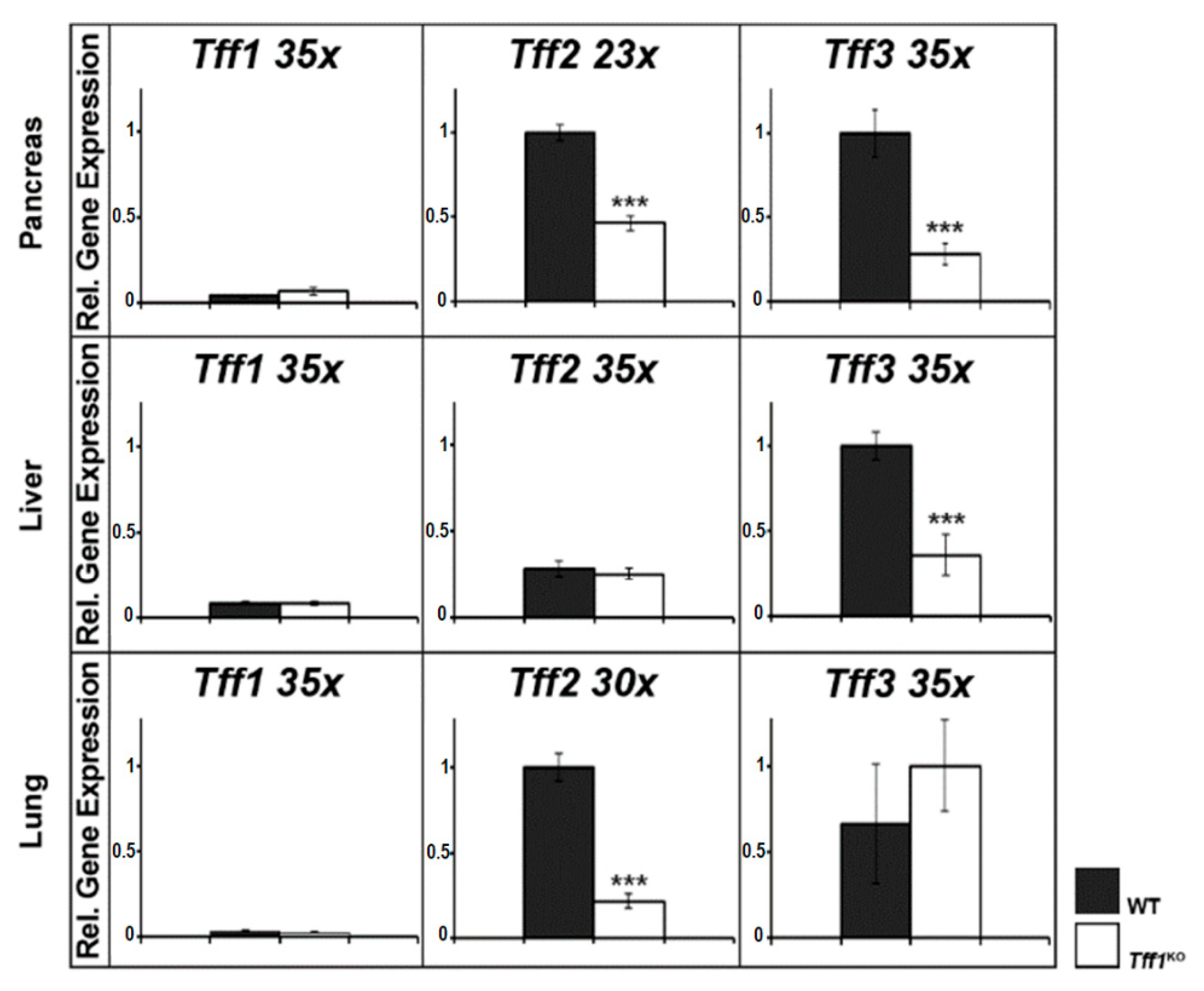
2.2. Expression Profiling of Tffs in the Murine Pancreas, Liver, and Lung (RT-PCR Analysis)
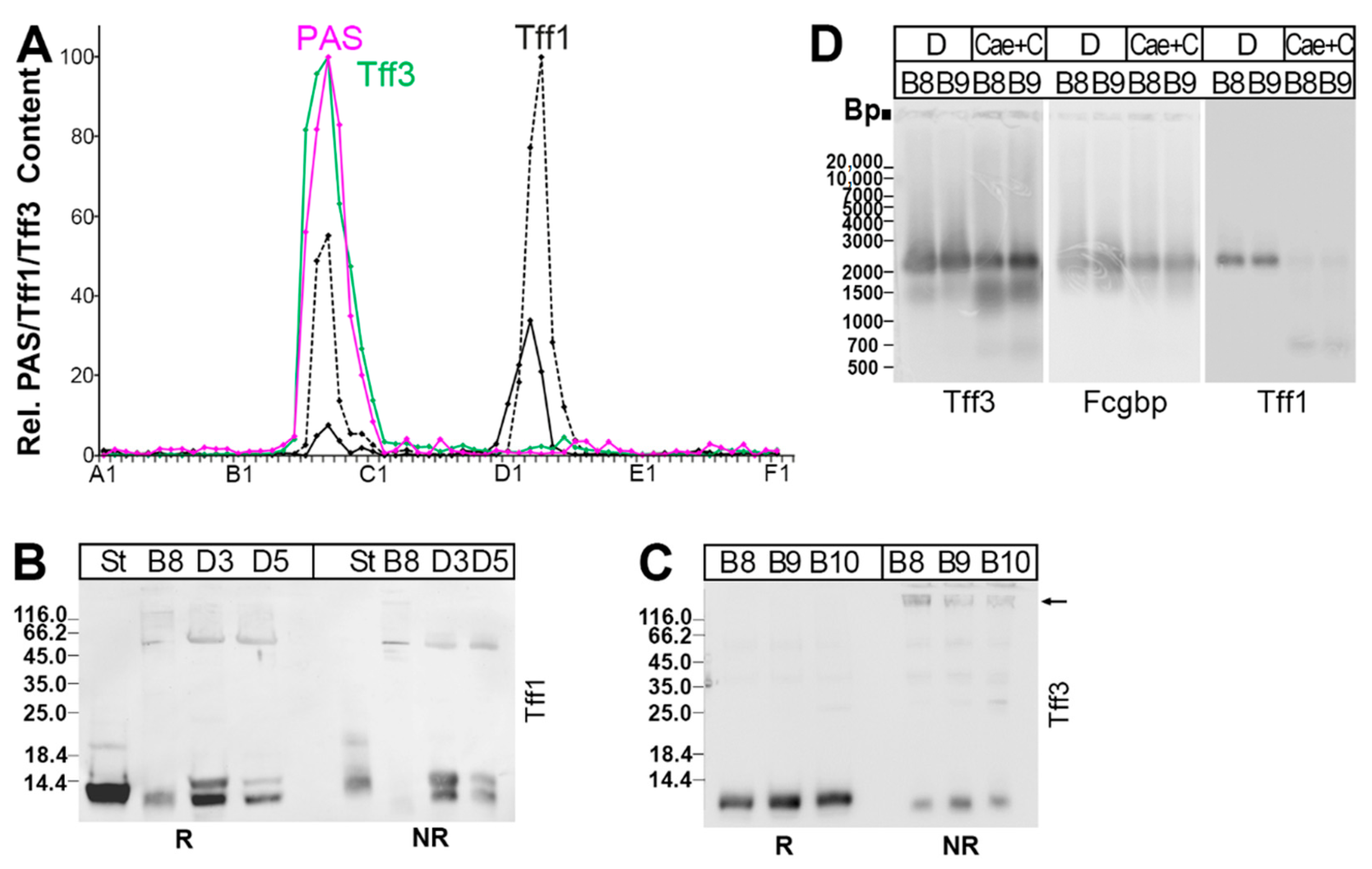
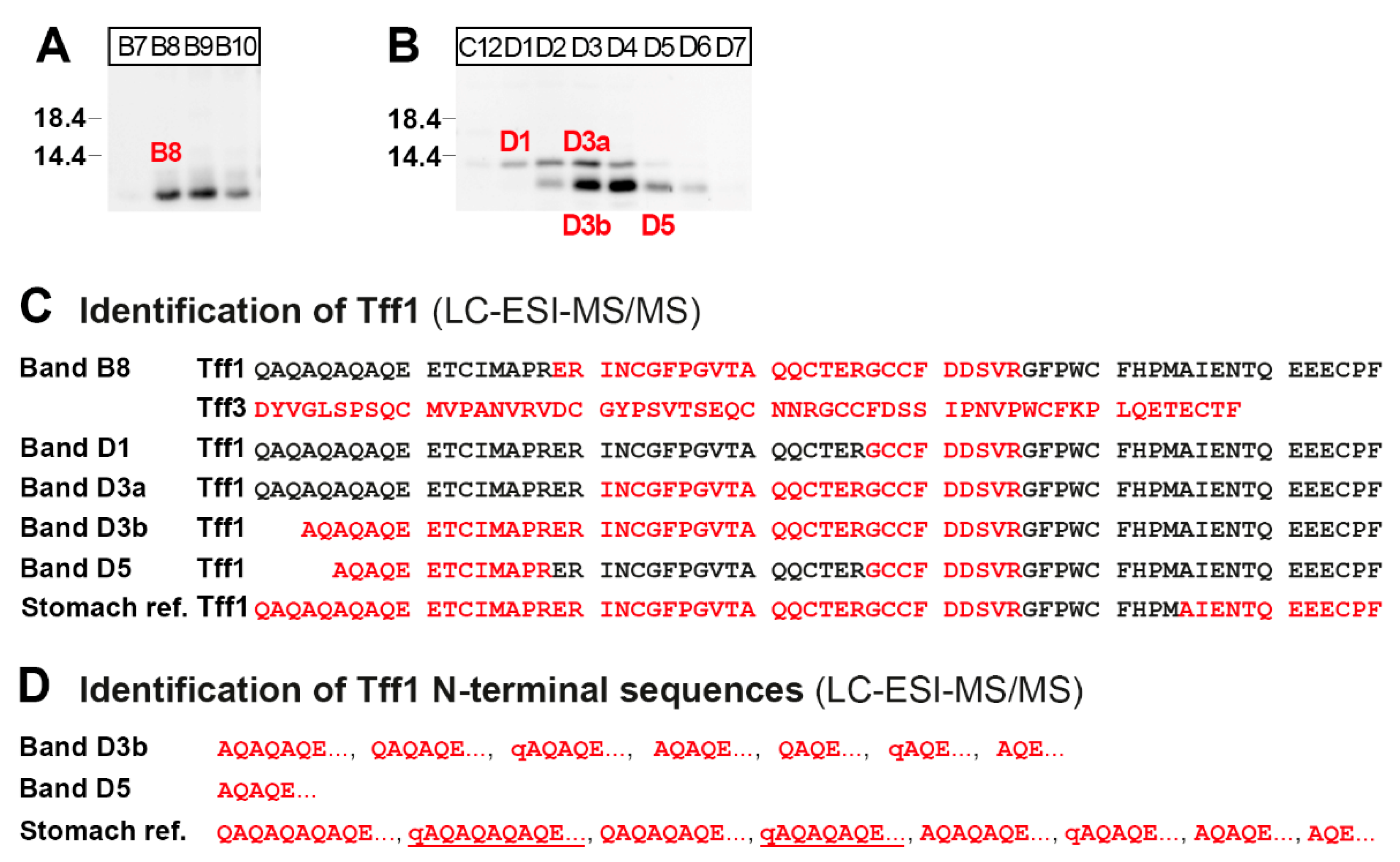
2.3. Protein Analysis of the Murine Duodenum
2.4. Protein Analysis of the Murine Large Intestine
3. Discussion
3.1. Expression Profiling along the Murine Intestinal Tract: Indications for Different Mucosal Protection Systems
3.1.1. Basic Protection of the Entire Intestinal Tract by Goblet Cell Products (Muc2, Tff3-Fcgbp)
3.1.2. Specific Protection of the Proximal Duodenum and the Colon by the Tff2/Muc6 Complex
3.1.3. Specific Protection of the Duodenum by Tff1 and Gastrokines
3.1.4. Specific Protection of the Proximal Duodenum and the Colon by Epithelial Fucosylation
3.1.5. Specific Protection of the Proximal Duodenum and Particularly the Colon by ROS-Generating Enzymes
3.1.6. Summary of the Different Mucosal Protection Systems along the Murine Intestine
3.2. Transcriptional Changes in the Intestine of Tff1KO Mice
3.3. Tff Expression Is Cross-Regulated in the Pancreas, Liver, and Lung of Tff1KO Mice
4. Materials and Methods
4.1. Animals
4.2. RNA Extraction, PCR Analysis
4.3. Extraction of Proteins, Protein Purification via SEC
4.4. SDS-PAGE, AgGE, and Western Blot Analysis
4.5. Identification of Proteins via Bottom-Up Proteomics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGC | Antral gland cell |
| AgGE | Agarose gel electrophoresis |
| FCGBP | IgG Fc binding protein |
| GI | Gastrointestinal |
| GKN | Gastrokine |
| MNC | Mucous neck cell |
| SEC | Size exclusion chromatography |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| TFF | Trefoil factor family |
References
- Clevers, H.; Batlle, E. SnapShot: The intestinal crypt. Cell 2013, 152, 1198–1198.e2. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [Green Version]
- Garrett, W.S.; Gordon, J.I.; Glimcher, L.H. Homeostasis and inflammation in the intestine. Cell 2010, 140, 859–870. [Google Scholar] [CrossRef] [Green Version]
- Ermund, A.; Schütte, A.; Johansson, M.E.V.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilchez-Vargas, R.; Salm, F.; Znalesniak, E.B.; Haupenthal, K.; Schanze, D.; Zenker, M.; Link, A.; Hoffmann, W. Profiling of the bacterial microbiota along the murine alimentary tract. Int. J. Mol. Sci. 2022, 23, 1783. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Larsson, J.M.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4659–4665. [Google Scholar] [CrossRef]
- Gustafsson, J.K.; Johansson, M.E.V. The role of goblet cells and mucus in intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 785–803. [Google Scholar] [CrossRef]
- Holmén Larsson, J.M.; Thomsson, K.A.; Rodríguez-Piñeiro, A.M.; Karlsson, H.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. III. Gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G357–G363. [Google Scholar] [CrossRef] [Green Version]
- Albert, T.K.; Laubinger, W.; Müller, S.; Hanisch, F.-G.; Kalinski, T.; Meyer, F.; Hoffmann, W. Human intestinal TFF3 forms disulfide-linked heteromers with the mucus-associated FCGBP protein and is released by hydrogen sulfide. J. Proteome Res. 2010, 9, 3108–3117. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Piñeiro, A.M.; Bergström, J.H.; Ermund, A.; Gustafsson, J.K.; Schütte, A.; Johansson, M.E.V.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. II. Gastrointestinal mucus proteome reveals Muc2 and Muc5ac accompanied by a set of core proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G348–G356. [Google Scholar] [CrossRef]
- Lang, T.; Klasson, S.; Larsson, E.; Johansson, M.E.V.; Hansson, G.C.; Samuelsson, T. Searching the Evolutionary Origin of Epithelial Mucus Protein Components-Mucins and FCGBP. Mol. Biol. Evol. 2016, 33, 1921–1936. [Google Scholar] [CrossRef]
- Nyström, E.E.L.; Birchenough, G.M.H.; van der Post, S.; Arike, L.; Gruber, A.D.; Hansson, G.C.; Johansson, M.E.V. Calcium-activated Chloride Channel Regulator 1 (CLCA1) Controls Mucus Expansion in Colon by Proteolytic Activity. EBioMedicine 2018, 33, 134–143. [Google Scholar] [CrossRef]
- Hoffmann, W. Salivary Trefoil Factor Family (TFF) Peptides and Their Roles in Oral and Esophageal Protection: Therapeutic Potential. Int. J. Mol. Sci. 2021, 22, 12221. [Google Scholar] [CrossRef]
- Weste, J.; Houben, T.; Harder, S.; Schlüter, H.; Lücke, E.; Schreiber, J.; Hoffmann, W. Different Molecular Forms of TFF3 in the Human Respiratory Tract: Heterodimerization with IgG Fc Binding Protein (FCGBP) and Proteolytic Cleavage in Bronchial Secretions. Int. J. Mol. Sci. 2022, 23, 15359. [Google Scholar] [CrossRef]
- Park, S.W.; Zhen, G.; Verhaeghe, C.; Nakagami, Y.; Nguyenvu, L.T.; Barczak, A.J.; Killeen, N.; Erle, D.J. The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. USA 2009, 106, 6950–6955. [Google Scholar] [CrossRef]
- Bergström, J.H.; Berg, K.A.; Rodríguez-Piñeiro, A.M.; Stecher, B.; Johansson, M.E.V.; Hansson, G.C. AGR2, an endoplasmic reticulum protein, is secreted into the gastrointestinal mucus. PLoS ONE 2014, 9, e104186. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, U.; Duku, M.; Katoh, M.; Jörns, J.; Krause, W.J. Histochemical similarities of mucins produced by Brunner’s glands and pyloric glands: A comparative study. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2004, 278, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J. Dual Roles of Gastric Gland Mucin-specific O-glycans in Prevention of Gastric Cancer. Acta Histochem. Cytochem. 2014, 47, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oinuma, T.; Kawano, J.; Suganuma, T. Glycoconjugate histochemistry of Xenopus laevis fundic gland with special reference to mucous neck cells during development. Anat. Rec. 1991, 230, 502–512. [Google Scholar] [CrossRef]
- Hoffmann, W. TFF2, a MUC6-binding lectin stabilizing the gastric mucus barrier and more. Int. J. Oncol. 2015, 47, 806–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Znalesniak, E.B.; Laskou, A.; Salm, F.; Haupenthal, K.; Harder, S.; Schlüter, H.; Hoffmann, W. The Forms of the Lectin Tff2 Differ in the Murine Stomach and Pancreas: Indications for Different Molecular Functions. Int. J. Mol. Sci. 2023, 24, 7059. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Hayama, M.; Momose, M.; El-Zimaity, H.M.T.; Matsuda, K.; Sano, K.; Maruta, F.; Okumura, N.; Katsuyama, T. Co-localization of TFF2 with gland mucous cell mucin in gastric mucous cells and in extracellular mucous gel adherent to normal and damaged gastric mucosa. Histochem. Cell Biol. 2006, 126, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Belikov, A.V.; Schraven, B.; Simeoni, L. T cells and reactive oxygen species. J. Biomed. Sci. 2015, 22, 85. [Google Scholar] [CrossRef] [PubMed]
- Aviello, G.; Knaus, U.G. NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol. 2018, 11, 1011–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedard, K.; Krause, K.-H. The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Allaoui, A.; Botteaux, A.; Dumont, J.E.; Hoste, C.; De Deken, X. Dual oxidases and hydrogen peroxide in a complex dialogue between host mucosae and bacteria. Trends Mol. Med. 2009, 15, 571–579. [Google Scholar] [CrossRef]
- Sarr, D.; Tóth, E.; Gingerich, A.; Rada, B. Antimicrobial actions of dual oxidases and lactoperoxidase. J. Microbiol. 2018, 56, 373–386. [Google Scholar] [CrossRef]
- Fattman, C.L.; Schaefer, L.M.; Oury, T.D. Extracellular superoxide dismutase in biology and medicine. Free Radic. Biol. Med. 2003, 35, 236–256. [Google Scholar] [CrossRef]
- Menheniott, T.R.; Kurklu, B.; Giraud, A.S. Gastrokines: Stomach-specific proteins with putative homeostatic and tumor suppressor roles. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 304, G109–G121. [Google Scholar] [CrossRef] [Green Version]
- Stappenbeck, T.S. Paneth cell development, differentiation, and function: New molecular cues. Gastroenterology 2009, 137, 30–33. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667–685. [Google Scholar] [CrossRef] [PubMed]
- Braga Emidio, N.; Hoffmann, W.; Brierley, S.M.; Muttenthaler, M. Trefoil factor family: Unresolved questions and clinical perspectives. Trends Biochem. Sci. 2019, 44, 387–390. [Google Scholar] [CrossRef]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides and Their Diverse Molecular Functions in Mucus Barrier Protection and More: Changing the Paradigm. Int. J. Mol. Sci. 2020, 21, 4535. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides. Encyclopedia 2021, 1, 974–987. [Google Scholar] [CrossRef]
- Hauser, F.; Roeben, C.; Hoffmann, W. xP2, a new member of the P-domain peptide family of potential growth factors, is synthesized in Xenopus laevis skin. J. Biol. Chem. 1992, 267, 14451–14455. [Google Scholar] [CrossRef]
- Stürmer, R.; Reising, J.; Hoffmann, W. The TFF peptides xP1 and xP4 appear in distinctive forms in the Xenopus laevis gastric mucosa: Indications for different protective functions. Int. J. Mol. Sci. 2019, 20, 6052. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, O.; Chenard, M.-P.; Masson, R.; Linares, J.; Dierich, A.; LeMeur, M.; Wendling, C.; Tomasetto, C.; Chambon, P.; Rio, M.-C. Gastric mucosa abnormalities and tumorigenesis in mice lacking the pS2 trefoil protein. Science 1996, 274, 259–262. [Google Scholar] [CrossRef]
- Tomasetto, C.; Rio, M.-C. Pleiotropic effects of Trefoil Factor 1 deficiency. Cell. Mol. Life Sci. 2005, 62, 2916–2920. [Google Scholar] [CrossRef]
- Hoffmann, W. Self-Renewal and Cancers of the Gastric Epithelium: An Update and the Role of the Lectin TFF1 as an Antral Tumor Suppressor. Int. J. Mol. Sci. 2022, 23, 5377. [Google Scholar] [CrossRef]
- Znalesniak, E.B.; Salm, F.; Hoffmann, W. Molecular Alterations in the Stomach of Tff1-Deficient Mice: Early Steps in Antral Carcinogenesis. Int. J. Mol. Sci. 2020, 21, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, O.; Wolf, C.; Kédinger, M.; Chenard, M.P.; Tomasetto, C.; Chambon, P.; Rio, M.C. The mouse one P-domain (pS2) and two P-domain (mSP) genes exhibit distinct patterns of expression. J. Cell Biol. 1993, 122, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Playford, R.J.; Marchbank, T.; Goodlad, R.A.; Chinery, R.A.; Poulsom, R.; Hanby, A.M.; Wright, N.A. Transgenic mice that overexpress the human trefoil peptide pS2 have an increased resistance to intestinal damage. Proc. Natl. Acad. Sci. USA 1996, 93, 2137–2142. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, K.; Hans, W.; Van Huysse, J.; Neirynck, S.; Demetter, P.; Remaut, E.; Rottiers, P.; Steidler, L. Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology 2004, 127, 502–513. [Google Scholar] [CrossRef]
- Houben, T.; Harder, S.; Schlüter, H.; Kalbacher, H.; Hoffmann, W. Different forms of TFF3 in the human saliva: Heterodimerization with IgG Fc binding protein (FCGBP). Int. J. Mol. Sci. 2019, 20, 5000. [Google Scholar] [CrossRef] [Green Version]
- Larsson, L.I.; Madsen, O.D.; Serup, P.; Jonsson, J.; Edlund, H. Pancreatic-duodenal homeobox 1 -role in gastric endocrine patterning. Mech. Dev. 1996, 60, 175–184. [Google Scholar] [CrossRef]
- Hoffmann, W. Trefoil Factor Family (TFF) Peptides and their Different Roles in the Mucosal Innate Immune Defense and More: An Update. Curr. Med. Chem. 2021, 28, 7387–7399. [Google Scholar] [CrossRef]
- Noah, T.K.; Kazanjian, A.; Whitsett, J.; Shroyer, N.F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res. 2010, 316, 452–465. [Google Scholar] [CrossRef] [Green Version]
- Carleton, A. The distribution of Brunner’s glands in the duodenum of mammals. Proc. Zool. Soc. Lond. 1935, 105, 385–390. [Google Scholar] [CrossRef]
- Judd, L.M.; Chalinor, H.V.; Walduck, A.; Pavlic, D.I.; Däbritz, J.; Dubeykovskaya, Z.; Wang, T.C.; Menheniott, T.R.; Giraud, A.S. TFF2 deficiency exacerbates weight loss and alters immune cell and cytokine profiles in DSS colitis, and this cannot be rescued by wild-type bone marrow. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G12–G24. [Google Scholar] [CrossRef] [Green Version]
- Poulsen, S.S.; Thulesen, J.; Hartmann, B.; Kissow, H.L.; Nexo, E.; Thim, L. Injected TFF1 and TFF3 bind to TFF2-immunoreactive cells in the gastrointestinal tract in rats. Regul. Pept. 2003, 115, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Nakayama, J.; Momose, M.; Kurihara, M.; Ishihara, K.; Hotta, K.; Katsuyama, T. New monoclonal antibodies against gastric gland mucous cell-type mucins: A comparative immunohistochemical study. Histochem. Cell Biol. 1998, 110, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Ota, H.; Katsuyama, T.; Akamatsu, T.; Ishihara, K.; Kurihara, M.; Hotta, K. Histochemical reactivity of normal, metaplastic, and neoplastic tissues to alpha-linked N-acetylglucosamine residue-specific monoclonal antibody HIK1083. J. Histochem. Cytochem. 1998, 46, 793–801. [Google Scholar] [CrossRef] [Green Version]
- Altmann, G.G. Morphological observations on mucus-secreting nongoblet cells in the deep crypts of the rat ascending colon. Am. J. Anat. 1983, 167, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Komiya, T.; Tanigawa, Y.; Hirohashi, S. Cloning of the gene gob-4, which is expressed in intestinal goblet cells in mice. Biochim. Biophys. Acta 1999, 1444, 434–438. [Google Scholar] [CrossRef]
- Gupta, A.; Wodziak, D.; Tun, M.; Bouley, D.M.; Lowe, A.W. Loss of anterior gradient 2 (Agr2) expression results in hyperplasia and defective lineage maturation in the murine stomach. J. Biol. Chem. 2013, 288, 4321–4333. [Google Scholar] [CrossRef]
- Bouchalova, P.; Sommerova, L.; Potesil, D.; Martisova, A.; Lapcik, P.; Koci, V.; Scherl, A.; Vonka, P.; Planas-Iglesias, J.; Chevet, E.; et al. Characterization of the AGR2 Interactome Uncovers New Players of Protein Disulfide Isomerase Network in Cancer Cells. Mol. Cell. Proteom. 2022, 21, 100188. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Liu, C.Y.; Katada, K.; Thai, M.H.; Hsieh, J.J.; Hansten, B.J.; Waddell, A.; Rosen, M.J.; Frey, M.R. Deep Crypt Secretory Cell Differentiation in the Colonic Epithelium Is Regulated by Sprouty2 and Interleukin 13. Cell. Mol. Gastroenterol. Hepatol. 2023, 15, 971–984. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Sasaki, N.; Sachs, N.; Wiebrands, K.; Ellenbroek, S.I.J.; Fumagalli, A.; Lyubimova, A.; Begthel, H.; van den Born, M.; van Es, J.H.; Karthaus, W.R.; et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad. Sci. USA 2016, 113, E5399–E5407. [Google Scholar] [CrossRef]
- Heuer, J.; Heuer, F.; Stürmer, R.; Harder, S.; Schlüter, H.; Braga Emidio, N.; Muttenthaler, M.; Jechorek, D.; Meyer, F.; Hoffmann, W. The Tumor Suppressor TFF1 Occurs in Different Forms and Interacts with Multiple Partners in the Human Gastric Mucus Barrier: Indications for Diverse Protective Functions. Int. J. Mol. Sci. 2020, 21, 2508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purwaha, P.; Silva, L.P.; Hawke, D.H.; Weinstein, J.N.; Lorenzi, P.L. An artifact in LC-MS/MS measurement of glutamine and glutamic acid: In-source cyclization to pyroglutamic acid. Anal. Chem. 2014, 86, 5633–5637. [Google Scholar] [CrossRef] [PubMed]
- Menheniott, T.R.; Peterson, A.J.; O’Connor, L.; Lee, K.S.; Kalantzis, A.; Kondova, I.; Bontrop, R.E.; Bell, K.M.; Giraud, A.S. A novel gastrokine, Gkn3, marks gastric atrophy and shows evidence of adaptive gene loss in humans. Gastroenterology 2010, 138, 1823–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, Y.; Obata, T.; Kunisawa, J.; Sato, S.; Ivanov, I.I.; Lamichhane, A.; Takeyama, N.; Kamioka, M.; Sakamoto, M.; Matsuki, T.; et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science 2014, 345, 1254009. [Google Scholar] [CrossRef] [PubMed]
- Nanthakumar, N.N.; Meng, D.; Newburg, D.S. Fucosylated TLR4 mediates communication between mutualist fucotrophic microbiota and mammalian gut mucosa. Front. Med. 2023, 10, 1070734. [Google Scholar] [CrossRef] [PubMed]
- Bry, L.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A model of host-microbial interactions in an open mammalian ecosystem. Science 1996, 273, 1380–1383. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Xu, J.; Falk, P.G.; Midtvedt, T.; Gordon, J.I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 1999, 96, 9833–9838. [Google Scholar] [CrossRef]
- Coyne, M.J.; Reinap, B.; Lee, M.M.; Comstock, L.E. Human symbionts use a host-like pathway for surface fucosylation. Science 2005, 307, 1778–1781. [Google Scholar] [CrossRef]
- Comstock, L.E.; Kasper, D.L. Bacterial glycans: Key mediators of diverse host immune responses. Cell 2006, 126, 847–850. [Google Scholar] [CrossRef] [Green Version]
- Nyström, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, eabb1590. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, D.; Chen, K.-Y.; Cui, M.; Wang, W.; Huang, X.; Awadellah, A.; Li, Q.; Friedman, A.; Xin, W.W.; et al. Fucosylation Deficiency in Mice Leads to Colitis and Adenocarcinoma. Gastroenterology 2017, 152, 193–205. [Google Scholar] [CrossRef] [Green Version]
- El Hassani, R.A.; Benfares, N.; Caillou, B.; Talbot, M.; Sabourin, J.-C.; Belotte, V.; Morand, S.; Gnidehou, S.; Agnandji, D.; Ohayon, R.; et al. Dual oxidase2 is expressed all along the digestive tract. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 288, G933–G942. [Google Scholar] [CrossRef] [PubMed]
- Rigoni, A.; Poulsom, R.; Jeffery, R.; Mehta, S.; Lewis, A.; Yau, C.; Giannoulatou, E.; Feakins, R.; Lindsay, J.O.; Colombo, M.P.; et al. Separation of Dual Oxidase 2 and Lactoperoxidase Expression in Intestinal Crypts and Species Differences May Limit Hydrogen Peroxide Scavenging During Mucosal Healing in Mice and Humans. Inflamm. Bowel Dis. 2018, 24, 136–148. [Google Scholar] [CrossRef]
- Geiszt, M.; Lekstrom, K.; Brenner, S.; Hewitt, S.M.; Dana, R.; Malech, H.L.; Leto, T.L. NAD(P)H oxidase 1, a product of differentiated colon epithelial cells, can partially replace glycoprotein 91phox in the regulated production of superoxide by phagocytes. J. Immunol. 2003, 171, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Toback, F.G.; Walsh-Reitz, M.M.; Musch, M.W.; Chang, E.B.; Del Valle, J.; Ren, H.; Huang, E.; Martin, T.E. Peptide fragments of AMP-18, a novel secreted gastric antrum mucosal protein, are mitogenic and motogenic. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G344–G353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurt-Jones, E.A.; Cao, L.; Sandor, F.; Rogers, A.B.; Whary, M.T.; Nambiar, P.R.; Cerny, A.; Bowen, G.; Yan, J.; Takaishi, S.; et al. Trefoilf factor 2 is expressed in murine gastric and immune cells and controls both gastrointestinal inflammation and systemic immune response. Infect. Immun. 2007, 75, 471–480. [Google Scholar] [CrossRef] [Green Version]
- Bossenmeyer-Pourié, C.; Kannan, R.; Ribieras, S.; Wendling, C.; Stoll, I.; Thim, L.; Tomasetto, C.; Rio, M.-C. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J. Cell Biol. 2002, 157, 761–770. [Google Scholar] [CrossRef]
- Karam, S.M.; Tomasetto, C.; Rio, M.-C. Trefoil factor 1 is required for the commitment programme of mouse oxyntic epithelial progenitors. Gut 2004, 53, 1408–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karam, S.M.; Tomasetto, C.; Rio, M.-C. Amplification and invasiveness of epithelial progenitors during gastric carcinogenesis in trefoil factor 1 knockout mice. Cell Prolif. 2008, 41, 923–935. [Google Scholar] [CrossRef]
- Maurel, M.; Obacz, J.; Avril, T.; Ding, Y.P.; Papadodima, O.; Treton, X.; Daniel, F.; Pilalis, E.; Hörberg, J.; Hou, W.; et al. Control of anterior GRadient 2 (AGR2) dimerization links endoplasmic reticulum proteostasis to inflammation. EMBO Mol. Med. 2019, 11, e10120. [Google Scholar] [CrossRef]
- Delom, F.; Mohtar, M.A.; Hupp, T.; Fessart, D. The anterior gradient-2 interactome. Am. J. Physiol. Cell Physiol. 2020, 318, C40–C47. [Google Scholar] [CrossRef]
- Torres, L.F.; Karam, S.M.; Wendling, C.; Chenard, M.P.; Kershenobich, D.; Tomasetto, C.; Rio, M.-C. Trefoil factor 1 (TFF1/pS2) deficiency activates the unfolded protein response. Mol. Med. 2002, 8, 273–282. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, W.; Jagla, W. Cell type specific expression of secretory TFF peptides: Colocalization with mucins and synthesis in the brain. Int. Rev. Cytol. 2002, 213, 147–181. [Google Scholar]
- Ribieras, S.; Lefèbvre, O.; Tomasetto, C.; Rio, M.-C. Mouse Trefoil factor genes: Genomic organization, sequences and methylation analyses. Gene 2001, 266, 67–75. [Google Scholar] [CrossRef]
- Hertel, S.C.; Chwieralski, C.E.; Hinz, M.; Rio, M.-C.; Tomasetto, C.; Hoffmann, W. Profiling trefoil factor family (TFF) expression in the mouse: Identification of an antisense TFF1-related transcript in the kidney and liver. Peptides 2004, 25, 755–762. [Google Scholar] [CrossRef]
- Yamaguchi, J.; Mino-Kenudson, M.; Liss, A.S.; Chowdhury, S.; Wang, T.C.; Castillo, C.F.-D.; Lillemoe, K.D.; Warshaw, A.L.; Thayer, S.P. Loss of Trefoil Factor 2 From Pancreatic Duct Glands Promotes Formation of Intraductal Papillary Mucinous Neoplasms in Mice. Gastroenterology 2016, 151, 1232–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hung, L.Y.; Oniskey, T.K.; Sen, D.; Krummel, M.F.; Vaughan, A.E.; Cohen, N.A.; Herbert, D.R. Trefoil Factor 2 Promotes Type 2 Immunity and Lung Repair through Intrinsic Roles in Hematopoietic and Nonhematopoietic Cells. Am. J. Pathol. 2018, 188, 1161–1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozaki, I.; Lunz, J.G., 3rd; Specht, S.; Park, J.I.; Giraud, A.S.; Murase, N.; Demetris, A.J. Regulation and function of trefoil factor family 3 expression in the biliary tree. Am. J. Pathol. 2004, 165, 1907–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.C.; Olver, W.I.; Donnelly, C.J.; May, M.E.; Naggert, J.K.; Shaffer, D.J.; Roopenian, D.C. Searching QTL by gene expression: Analysis of diabesity. BMC Genet. 2005, 6, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillén, N.; Navarro, M.-A.; Arnal, C.; Noone, E.; Arbonés-Mainar, J.M.; Acín, S.; Surra, J.C.; Muniesa, P.; Roche, H.M.; Osada, J. Microarray analysis of hepatic gene expression identifies new genes involved in steatotic liver. Physiol. Genom. 2009, 37, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Bujak, M.; Bujak, I.T.; Sobočanec, S.; Mihalj, M.; Novak, S.; Ćosić, A.; Levak, M.T.; Kopačin, V.; Mihaljević, B.; Balog, T.; et al. Trefoil Factor 3 Deficiency Affects Liver Lipid Metabolism. Cell. Physiol. Biochem. 2018, 47, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Znalesniak, E.B.; Fu, T.; Guttek, K.; Händel, U.; Reinhold, D.; Hoffmann, W. Increased cerebral Tff1 expression in two murine models of neuroinflammation. Cell. Physiol. Biochem. 2016, 39, 2287–2296. [Google Scholar] [CrossRef]
- Fu, T.; Znalesniak, E.B.; Kalinski, T.; Möhle, L.; Biswas, A.; Salm, F.; Dunay, I.R.; Hoffmann, W. TFF peptides play a role in the immune response following oral infection of mice with Toxoplasma gondii. Eur. J. Microbiol. Immunol. 2015, 5, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Kalbacher, H.; Hoffmann, W. TFF1 is differentially expressed in stationary and migratory rat gastric epithelial cells (RGM-1) after in vitro wounding: Influence of TFF1 RNA interference on cell migration. Cell. Physiol. Biochem. 2013, 32, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Znalesniak, E.B.; Fu, T.; Salm, F.; Händel, U.; Hoffmann, W. Transcriptional responses in the murine spleen after Toxoplasma gondii infection: Inflammasome and mucus-associated genes. Int. J. Mol. Sci. 2017, 18, 1245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanisch, F.-G.; Ragge, H.; Kalinski, T.; Meyer, F.; Kalbacher, H.; Hoffmann, W. Human gastric TFF2 peptide contains an N-linked fucosylated N,N′-diacetyllactosediamine (LacdiNAc) oligosaccharide. Glycobiology 2013, 23, 2–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stürmer, R.; Müller, S.; Hanisch, F.-G.; Hoffmann, W. Porcine gastric TFF2 is a mucus constituent and differs from pancreatic TFF2. Cell. Physiol. Biochem. 2014, 33, 895–904. [Google Scholar] [CrossRef]
- Jagla, W.; Wiede, A.; Kölle, S.; Hoffmann, W. Differential expression of the TFF-peptides xP1 and xP4 in the gastrointestinal tract of Xenopus laevis. Cell Tiss. Res. 1998, 291, 13–18. [Google Scholar] [CrossRef]
- Kouznetsova, I.; Laubinger, W.; Kalbacher, H.; Kalinski, T.; Meyer, F.; Roessner, A.; Hoffmann, W. Biosynthesis of gastrokine-2 in the human gastric mucosa: Restricted spatial expression along the antral gland axis and differential interaction with TFF1, TFF2 and mucins. Cell. Physiol. Biochem. 2007, 20, 899–908. [Google Scholar] [CrossRef] [Green Version]
- Probst, J.C.; Skutella, T.; Müller-Schmid, A.; Jirikowski, G.F.; Hoffmann, W. Molecular and cellular analysis of rP1.B in the rat hypothalamus: In situ hybridization and immunohistochemistry of a new P-domain neuropeptide. Mol. Brain Res. 1995, 33, 269–276. [Google Scholar] [CrossRef]
- Wilhelm, B.; Keppler, C.; Henkeler, A.; Schilli-Westermann, M.; Linder, D.; Aumüller, G.; Seitz, J. Identification and Characterization of an IgG Binding Protein in the Secretion of the Rat Coagulating Gland. Biol. Chem. 2002, 383, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Ota, H.; Katsuyama, T. Alternating laminated array of two types of mucin in the human gastric surface mucous layer. Histochem. J. 1992, 24, 86–92. [Google Scholar] [CrossRef] [PubMed]



| Genes Accession No. | Primer No. | Primer Pairs | Nucleotide Positions | Annealing T Size (bp) |
|---|---|---|---|---|
| Agr2 NM_011783.2 | MB2190 MB2191 | GTCTGCAATCCTGCTTCTTGT GTCTTTAGCAGCTTGAGAGCTT | 70–90 570–549 | 60 °C 501 |
| Cdx1 NM_009880.4 | MB2354 MB2355 | GGACGCCCTACGAATGGAT ACCAGATCTTTACCTGCCGC | 489–507 704–685 | 60 °C 216 |
| Cdx2 NM_007673.3 | MB2352 MB2353 | AGCCAAGTGAAAACCAGGACA GATGCTGTTCGTGGGTAGGA | 799–819 1320–1301 | 60 °C 522 |
| Clca1 NM_017474.2 | MB2176 MB2177 | CTTATCACCTGGACAACGCA TGGTCCCTGAGATCAACGAT | 1589–1608 2436–2417 | 60 °C 848 |
| Duox2 NM_001362755.1 | MB2726 MB2727 | GCCTGTCGAGTCTCGTTCAT CCGCAAGAAGGTGATGAGGT | 2934–2953 3383–3364 | 60 °C 450 |
| Fut2 NM_001271993.1 | MB2468 MB2469 | CTCCCCCGGGATCCTTATCT GTGGTAATTCTGCCACGGG | 252–271 699–681 | 60 °C 448 |
| Lgr5 NM_010195.2 | MB2492 MB2493 | GTCTCCTACATCGCCTCTGC AGAAGGGTTGCCTACGAACG | 538–557 1133–1114 | 60 °C 596 |
| Lpo NM_080420.3 | MB3005 MB3006 | GGCTGCCACGGGAGGTCAA TTATAGGGTGGTGTGGGGCA | 11–29 906–887 | 60 °C 896 |
| Mki67 NM_001081117.2 | MB2458 MB2459 | AGAGCTAACTTGCGCTGACTG TCTTGAGGCTCGCCTTGATG | 129–149 618–599 | 60 °C 490 |
| Muc2 NM_023566.4 | MB2178 MB2179 | GGCTCTACAGACAAGCAGAC CATGAAGGTATGGTCAGGGC | 1329–1348 2141–2122 | 60 °C 813 |
| Nox1 NM_172203.2 | MB2883 MB2884 | AAGTTTCTCTCCCGAAGGACC CCCTCAAGAAGGACAGCAGA | 74–94 387–368 | 60 °C 314 |
| Pdia6 NM_027959.4 | MB2991 MB2992 | TGGTCGGACGAGATCTGACA TGAGACGCTGAGGTTCACTG | 806–825 1513–1494 | 60 °C 708 |
| Qsox1 NM_001024945.1 | MB2546 MB2547 | TATAGTGAGGCCCACCCACA GTACATCTAGGGCAGTGGCTC | 1370–1389 1895–1875 | 60 °C 526 |
| Sod1 NM_011434.2 | MB2837 MB2838 | CGGTGAACCAGTTGTGTTGTC GGTCTCCAACATGCCTCTCT | 174–194 349–330 | 60 °C 176 |
| Sod2 NM_013671.3 | MB2839 MB2840 | CTGGACAAACCTGAGCCCTA GTTGTTCCTTGCAATGGGTCC | 510–529 728–708 | 60 °C 219 |
| Sod3 NM_011435.3 | MB2184 MB2185 | CTGCTGCTCGCTCACATAA CGCCTGGAGACATCTATGC | 119–137 1077–1059 | 60 °C 959 |
| Spdef NM_013891.4 | MB2200 MB2201 | AAGATATTGAGACGGCCTGC TGTCTATCTGGGACCTTGGG | 790–809 1528–1509 | 60 °C 739 |
| Zg16 NM_026918.3 | MB2995 MB2996 | CCTCGGCCTCTGCTAATTCC CCTGGATCACAGATTCCCCG | 85–104 339–320 | 60 °C 255 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salm, F.; Znalesniak, E.B.; Laskou, A.; Harder, S.; Schlüter, H.; Hoffmann, W. Expression Profiling along the Murine Intestine: Different Mucosal Protection Systems and Alterations in Tff1-Deficient Animals. Int. J. Mol. Sci. 2023, 24, 12684. https://doi.org/10.3390/ijms241612684
Salm F, Znalesniak EB, Laskou A, Harder S, Schlüter H, Hoffmann W. Expression Profiling along the Murine Intestine: Different Mucosal Protection Systems and Alterations in Tff1-Deficient Animals. International Journal of Molecular Sciences. 2023; 24(16):12684. https://doi.org/10.3390/ijms241612684
Chicago/Turabian StyleSalm, Franz, Eva B. Znalesniak, Aikaterini Laskou, Sönke Harder, Hartmut Schlüter, and Werner Hoffmann. 2023. "Expression Profiling along the Murine Intestine: Different Mucosal Protection Systems and Alterations in Tff1-Deficient Animals" International Journal of Molecular Sciences 24, no. 16: 12684. https://doi.org/10.3390/ijms241612684






