Single and Combined Effects of Chlorpyrifos and Glyphosate on the Brain of Common Carp: Based on Biochemical and Molecular Perspective
Abstract
:1. Introduction
2. Results
2.1. Contents of CPF and GLY
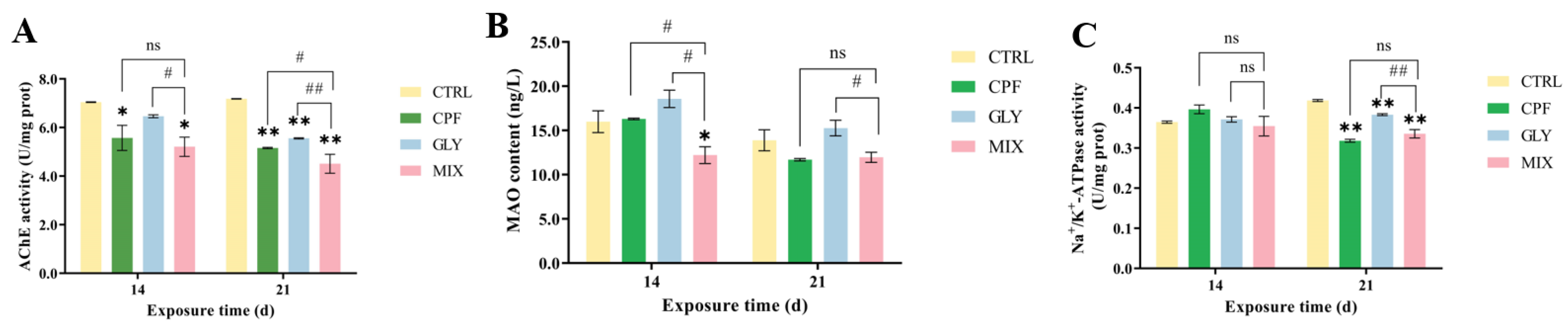
2.2. AChE Activities in the Fish Brain
2.3. Monoamine Oxidase (MAO)
2.4. Na+/K+-ATPase
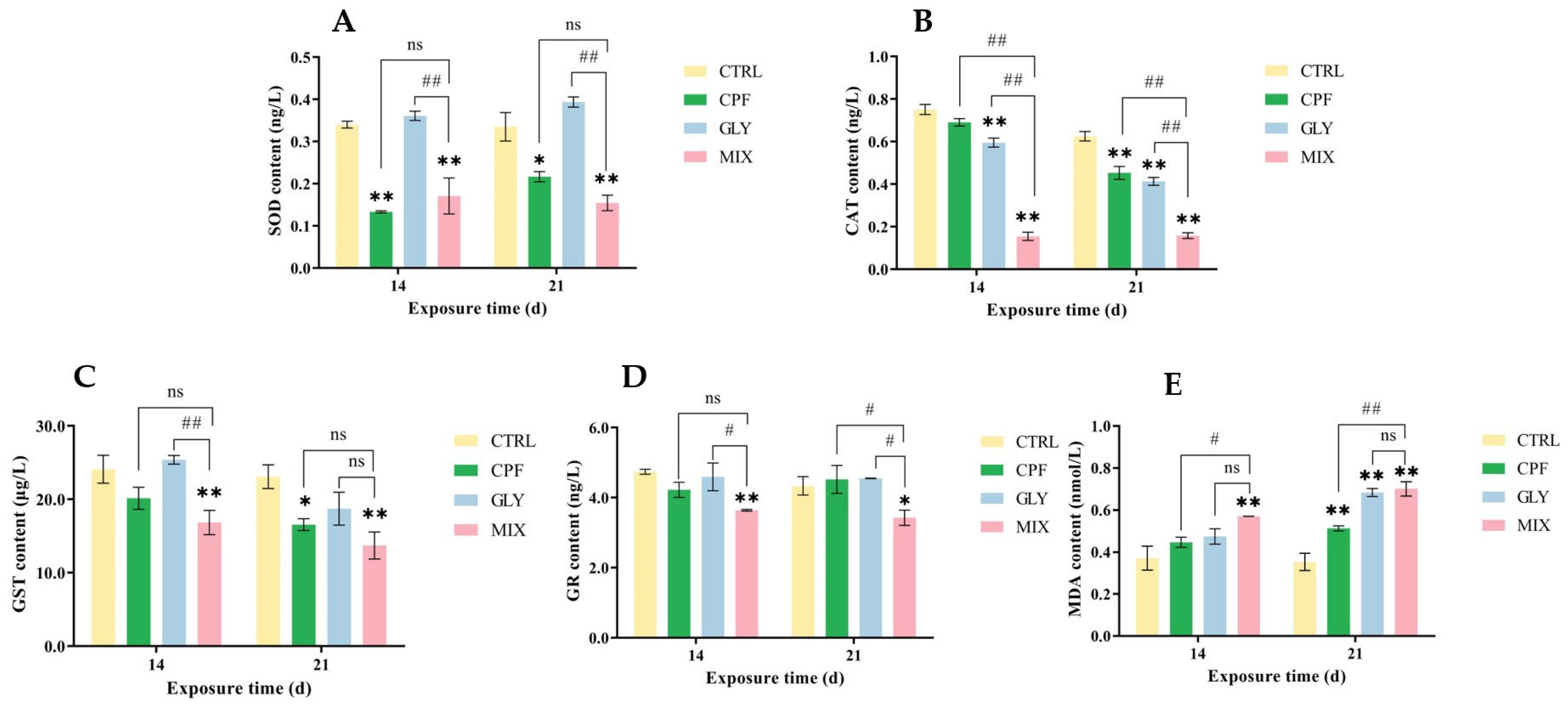
2.5. Oxidative Stress-Related Indicators in the Brain
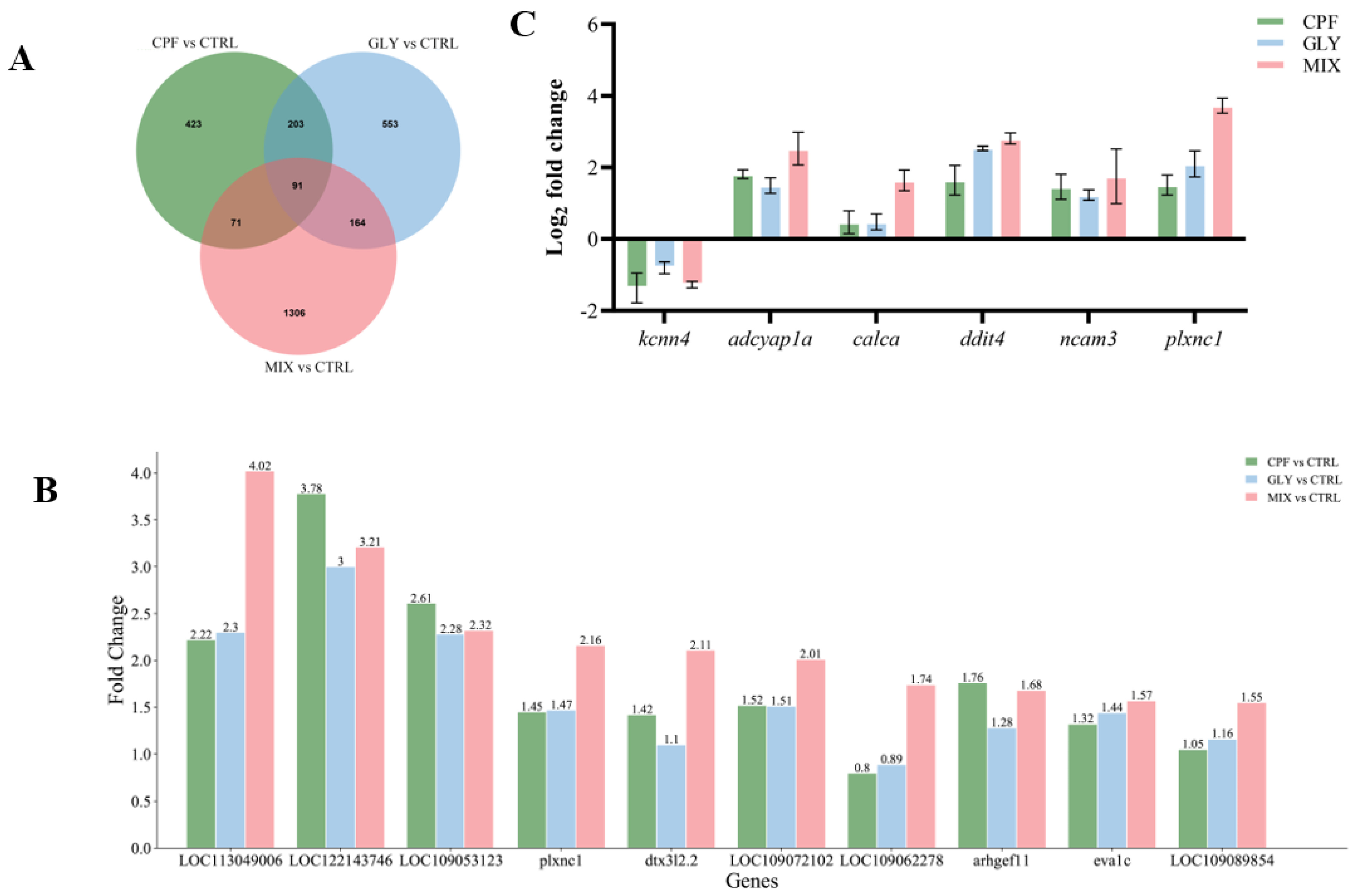
2.6. RNA-seq Data
2.7. DEGs and qPCR Validation
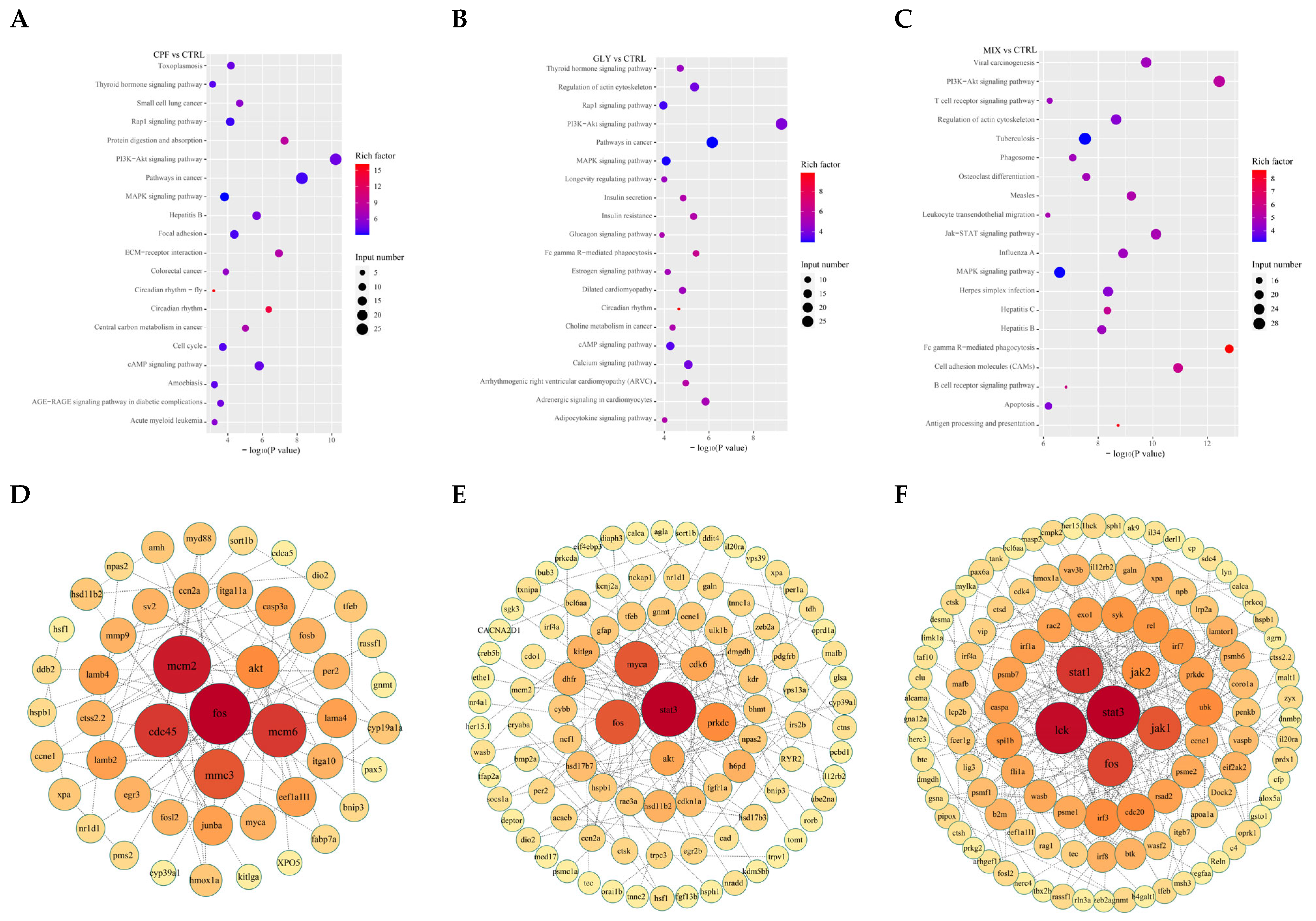
2.8. GO Classification and KEGG Enrichment Analysis of DEGs
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Fish and Experimental Design
4.3. Determination of CPF and GLY Contents in Carp Brain
4.4. Biochemical Assays
4.5. Transcriptomic Analysis
4.6. Real-Time Quantitative PCR (qPCR)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chormare, R.; Kumar, M.A. Environmental health and risk assessment metrics with special mention to biotransfer, bioaccumulation and biomagnification of environmental pollutants. Chemosphere 2022, 302, 134836. [Google Scholar] [CrossRef]
- Naccarato, A.; Tassone, A.; Cavaliere, F.; Elliani, R.; Pirrone, N.; Sprovieri, F.; Tagarelli, A.; Giglio, A. Agrochemical treatments as a source of heavy metals and rare earth elements in agricultural soils and bioaccumulation in ground beetles. Sci. Total Environ. 2020, 749, 141438. [Google Scholar] [CrossRef] [PubMed]
- Eaton, D.L.; Daroff, R.B.; Autrup, H.; Bridges, J.; Buffler, P.; Costa, L.G.; Coyle, J.; McKhann, G.; Mobley, W.C.; Nadel, L.; et al. Review of the toxicology of chlorpyrifos with an emphasis on human exposure and neurodevelopment. Crit. Rev. Toxicol. 2008, 38 (Suppl. 2), 1–125. [Google Scholar] [CrossRef] [PubMed]
- Bhende, R.S.; Jhariya, U.; Srivastava, S.; Bombaywala, S.; Das, S.; Dafale, N.A. Environmental distribution, metabolic fate, and degradation mechanism of chlorpyrifos: Recent and Future Perspectives. Appl. Biochem. Biotechnol. 2022, 194, 2301–2335. [Google Scholar] [CrossRef]
- Wan, Y.; Tran, T.M.; Nguyen, V.T.; Wang, A.; Wang, J.; Kannan, K. Neonicotinoids, fipronil, chlorpyrifos, carbendazim, chlorotriazines, chlorophenoxy herbicides, bentazon, and selected pesticide transformation products in surface water and drinking water from northern Vietnam. Sci. Total Environ. 2021, 750, 141507. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Díaz, J.A.; González-Márquez, L.C.; Longoria-Espinoza, R.M.; Ahumada-Cervantes, R.; Leyva-Morales, J.B.; Rodríguez-Gallegos, H.B. Chlorpyrifos and dimethoate in water and sediments of agricultural drainage ditches in Northern sinaloa, Mexico. Bull Environ. Contam. Toxicol. 2021, 106, 839–843. [Google Scholar] [CrossRef] [PubMed]
- Akoto, O.; Azuure, A.A.; Adotey, K.D. Pesticide residues in water, sediment and fish from Tono Reservoir and their health risk implications. Springerplus 2016, 5, 1849. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.K.; Saha, K.; Bandopadhyay, S.; Ghosh, A.; Mukherjee, M.; Raut, A.; Raman, R.K.; Suresh, V.R.; Mohanty, S.K. Status of pesticide residues in water, sediment, and fishes of Chilika Lake, India. Environ. Monit. Assess 2020, 192, 122. [Google Scholar] [CrossRef]
- Hatami, M.; Banaee, M.; Nematdoost Haghi, B. Sub-lethal toxicity of chlorpyrifos alone and in combination with polyethylene glycol to common carp (Cyprinus carpio). Chemosphere 2019, 219, 981–988. [Google Scholar] [CrossRef]
- Pallotta, M.M.; Ronca, R.; Carotenuto, R.; Porreca, I.; Turano, M.; Ambrosino, C.; Capriglione, T. Specific effects of chronic dietary exposure to chlorpyrifos on brain gene expression-A mouse study. Int. J. Mol. Sci. 2017, 18, 11. [Google Scholar] [CrossRef]
- Colombo, A.; Orsi, F.; Bonfanti, P. Exposure to the organophosphorus pesticide chlorpyrifos inhibits acetylcholinesterase activity and affects muscular integrity in Xenopus laevis larvae. Chemosphere 2005, 61, 1665–1671. [Google Scholar] [CrossRef] [PubMed]
- Renick, V.C.; Weinersmith, K.; Vidal-Dorsch, D.E.; Anderson, T.W. Effects of a pesticide and a parasite on neurological, endocrine, and behavioral responses of an estuarine fish. Aquat. Toxicol. 2016, 170, 335–343. [Google Scholar] [CrossRef]
- Casida, J.E. Organophosphorus Xenobiotic Toxicology. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Shangguan, Y.; Zhu, Y.; Sultan, Y.; Feng, Y.; Zhang, B.; Liu, Y.; Ma, J.; Li, X. Negative impacts of microcystin-LR and glyphosate on zebrafish intestine: Linked with gut microbiota and microRNAs? Environ. Pollut. 2021, 286, 117685. [Google Scholar] [CrossRef] [PubMed]
- Costas-Ferreira, C.; Duran, R.; Faro, L.R.F. Toxic effects of glyphosate on the nervous system: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Lopes, F.M.; Sandrini, J.Z.; Souza, M.M. Toxicity induced by glyphosate and glyphosate-based herbicides in the zebrafish hepatocyte cell line (ZF-L). Ecotoxicol. Environ. Saf. 2018, 162, 201–207. [Google Scholar] [CrossRef]
- Ma, J.; Zhu, J.; Wang, W.; Ruan, P.; Rajeshkumar, S.; Li, X. Biochemical and molecular impacts of glyphosate-based herbicide on the gills of common carp. Environ. Pollut. 2019, 252 Pt B, 1288–1300. [Google Scholar] [CrossRef] [PubMed]
- Carles, L.; Gardon, H.; Joseph, L.; Sanchis, J.; Farre, M.; Artigas, J. Meta-analysis of glyphosate contamination in surface waters and dissipation by biofilms. Environ. Int. 2019, 124, 284–293. [Google Scholar] [CrossRef]
- Geng, Y.; Jiang, L.; Zhang, D.; Liu, B.; Zhang, J.; Cheng, H.; Wang, L.; Peng, Y.; Wang, Y.; Zhao, Y.; et al. Glyphosate, aminomethylphosphonic acid, and glufosinate ammonium in agricultural groundwater and surface water in China from 2017 to 2018: Occurrence, main drivers, and environmental risk assessment. Sci. Total Environ. 2021, 769, 144396. [Google Scholar] [CrossRef]
- Rendon-von Osten, J.; Dzul-Caamal, R. Glyphosate residues in groundwater, drinking water and urine of subsistence farmers from intensive agriculture localities: A survey in Hopelchen, Campeche, Mexico. Int. J. Environ. Res. Public Health 2017, 14, 6. [Google Scholar] [CrossRef]
- Fan, J.; Gen, J.; Wang, X. Determination of Glyphosate content in Taihu Water Body by Ion Chromatography. In Proceedings of the 6th National Conference on Environmental Chemistry and Environmental Scientific Instrument and Analytical Instrument Exhibition, Shanghai, China, 2011; pp. 222–223. (In Chinese). [Google Scholar]
- Székács, A.; Darvas, B. Re-registration Challenges of Glyphosate in the European Union. Front. Environ. Sci. 2018, 6, 00078. [Google Scholar] [CrossRef]
- Mortl, M.; Nemeth, G.; Juracsek, J.; Darvas, B.; Kamp, L.; Rubio, F.; Szekacs, A. Determination of glyphosate residues in Hungarian water samples by immunoassay. Microchem. J. 2013, 107, 143–151. [Google Scholar] [CrossRef]
- Elizalde-Velazquez, A.; Martinez-Rodriguez, H.; Galar-Martinez, M.; Dublan-Garcia, O.; Islas-Flores, H.; Rodriguez-Flores, J.; Castaneda-Penalvo, G.; Lizcano-Sanz, I.; Gomez-Olivan, L.M. Effect of amoxicillin exposure on brain, gill, liver, and kidney of common carp (Cyprinus carpio): The role of amoxicilloic acid. Environ. Toxicol. 2017, 32, 1102–1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, X.; Liu, F.; FangWang; Dong, J.; Zhao, P. Difenoconazole causes cardiotoxicity in common carp (Cyprinus carpio): Involvement of oxidative stress, inflammation, apoptosis and autophagy. Chemosphere 2022, 306, 135562. [Google Scholar] [CrossRef] [PubMed]
- Mieiro, C.L.; Pacheco, M.; Pereira, M.E.; Duarte, A.C. Mercury distribution in key tissues of fish (Liza aurata) inhabiting a contaminated estuary-implications for human and ecosystem health risk assessment. J. Environ. Monit. 2009, 11, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Samanta, P.; Pal, S.; Mukherjee, A.K.; Ghosh, A.R. Biochemical effects of glyphosate based herbicide, Excel Mera 71 on enzyme activities of acetylcholinesterase (AChE), lipid peroxidation (LPO), catalase (CAT), glutathione-S-transferase (GST) and protein content on teleostean fishes. Ecotoxicol. Environ. Saf. 2014, 107, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Li, P.; He, S.W.; Xing, S.Y.; Cao, Z.H.; Zhao, X.L.; Sun, C.; Li, Z.H. Assessing the ecotoxicity of combined exposure to triphenyltin and norfloxacin at environmental levels: A case study of immunotoxicity and metabolic regulation in carp (Cyprinus carpio). Chemosphere 2023, 313, 137381. [Google Scholar] [CrossRef]
- Winstone, J.K.; Pathak, K.V.; Winslow, W.; Piras, I.S.; White, J.; Sharma, R.; Huentelman, M.J.; Pirrotte, P.; Velazquez, R. Glyphosate infiltrates the brain and increases pro-inflammatory cytokine TNFalpha: Implications for neurodegenerative disorders. J. Neuroinflamm. 2022, 19, 193. [Google Scholar] [CrossRef]
- Deepika, D.; Kumar, S.; Bravo, N.; Esplugas, R.; Capodiferro, M.; Sharma, R.P.; Schuhmacher, M.; Grimalt, J.O.; Blanco, J.; Kumar, V. Chlorpyrifos, permethrin and cyfluthrin effect on cell survival, permeability, and tight junction in an in-vitro model of the human blood-brain barrier (BBB). Neurotoxicology 2022, 93, 152–162. [Google Scholar] [CrossRef]
- Basha, D.C.; Rani, M.U.; Devi, C.B.; Kumar, M.R.; Reddy, G.R. Perinatal lead exposure alters postnatal cholinergic and aminergic system in rat brain: Reversal effect of calcium co-administration. Int. J. Dev. Neurosci. 2012, 30, 343–350. [Google Scholar] [CrossRef]
- Jokanovic, M. Medical treatment of acute poisoning with organophosphorus and carbamate pesticides. Toxicol. Lett. 2009, 190, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Pulkrabkova, L.; Svobodova, B.; Konecny, J.; Kobrlova, T.; Muckova, L.; Janousek, J.; Pejchal, J.; Korabecny, J.; Soukup, O. Neurotoxicity evoked by organophosphates and available countermeasures. Arch. Toxicol. 2023, 97, 39–72. [Google Scholar] [CrossRef] [PubMed]
- Sarasamma, S.; Audira, G.; Juniardi, S.; Sampurna, B.P.; Liang, S.T.; Hao, E.; Lai, Y.H.; Hsiao, C.D. Zinc chloride exposure inhibits brain acetylcholine levels, produces neurotoxic signatures, and diminishes memory and motor activities in adult zebrafish. Int. J. Mol. Sci. 2018, 19, 10. [Google Scholar] [CrossRef] [PubMed]
- Abou-Donia, M.B.; Siracuse, B.; Gupta, N.; Sobel Sokol, A. Sarin (GB, O-isopropyl methylphosphonofluoridate) neurotoxicity: Critical review. Crit. Rev. Toxicol. 2016, 46, 845–875. [Google Scholar] [CrossRef] [PubMed]
- Adedara, I.A.; Owoeye, O.; Awogbindin, I.O.; Ajayi, B.O.; Rocha, J.B.T.; Farombi, E.O. Diphenyl diselenide abrogates brain oxidative injury and neurobehavioural deficits associated with pesticide chlorpyrifos exposure in rats. Chem. Biol. Interact. 2018, 296, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Cattani, D.; de Liz Oliveira Cavalli, V.L.; Heinz Rieg, C.E.; Domingues, J.T.; Dal-Cim, T.; Tasca, C.I.; Mena Barreto Silva, F.R.; Zamoner, A. Mechanisms underlying the neurotoxicity induced by glyphosate-based herbicide in immature rat hippocampus: Involvement of glutamate excitotoxicity. Toxicology 2014, 320, 34–45. [Google Scholar] [CrossRef]
- Nagatsu, T. Progress in monoamine oxidase (MAO) research in relation to genetic engineering. Neurotoxicology 2004, 25, 11–20. [Google Scholar] [CrossRef]
- Tabassum, H.; Khan, J.; Salman, M.; Raisuddin, S.; Parvez, S. Propiconazole induced toxicological alterations in brain of freshwater fish Channa punctata Bloch. Ecol. Indic. 2016, 62, 242–248. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Albreht, A. Kinetics, mechanism, and inhibition of monoamine oxidase. J. Neural. Transm. 2018, 125, 1659–1683. [Google Scholar] [CrossRef]
- Reddy, G.R.; Devi, B.C.; Chetty, C.S. Developmental lead neurotoxicity: Alterations in brain cholinergic system. Neurotoxicology 2007, 28, 402–407. [Google Scholar] [CrossRef]
- Souza, L.C.; Wilhelm, E.A.; Bortolatto, C.F.; Nogueira, C.W.; Boeira, S.P.; Jesse, C.R. The protective effect of melatonin against brain oxidative stress and hyperlocomotion in a rat model of mania induced by ouabain. Behav. Brain Res. 2014, 271, 316–324. [Google Scholar] [CrossRef] [PubMed]
- Serafini, S.; de Freitas Souza, C.; Baldissera, M.D.; Baldisserotto, B.; Segat, J.C.; Baretta, D.; Zanella, R.; Schafer da Silva, A. Fish exposed to water contaminated with eprinomectin show inhibition of the activities of AChE and Na(+)/K(+)-ATPase in the brain, and changes in natural behavior. Chemosphere 2019, 223, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Watson, T.A.; Beamish, F.W. The effects of zinc on branchial adenosine triphosphatase enzymes in vitro from rainbow trout, Salmo gairdneri. Comp. Biochem. Physiol. C Comp. Pharmacol. 1981, 68, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Mishra, O.P.; Delivoria-Papadopoulos, M.; Cahillane, G.; Wagerle, L.C. Lipid peroxidation as the mechanism of modification of the affinity of the Na+, K+-ATPase active sites for ATP, K+, Na+, and strophanthidin in vitro. Neurochem. Res. 1989, 14, 845–851. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lee, W.; Bian, J.S. Recent advances in the study of Na(+)/K(+)-ATPase in neurodegenerative Diseases. Cells 2022, 11, 4075. [Google Scholar] [CrossRef] [PubMed]
- Viani, P.; Cervato, G.; Fiorilli, A.; Cestaro, B. Age-related differences in synaptosomal peroxidative damage and membrane properties. J. Neurochem. 1991, 56, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Fang, C.; Jin, C.; Bao, Z.; Yang, G.; Jin, Y. Catechin from green tea had the potential to decrease the chlorpyrifos induced oxidative stress in larval zebrafish (Danio rerio). Pestic. Biochem. Physiol. 2022, 182, 105028. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shangguan, Y.; Zhu, P.; Sultan, Y.; Feng, Y.; Li, X.; Ma, J. Developmental toxicity of glyphosate on embryo-larval zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 2022, 236, 113493. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Yang, J.; Gong, Y.; Cai, J.; Zheng, Y.; Zhang, Z. Chlorpyrifos induces apoptosis and autophagy in common carp lymphocytes by influencing the TCR gamma-dependent PI3K/AKT/JNK pathway. Fish Shellfish Immunol. 2020, 99, 587–593. [Google Scholar] [CrossRef]
- Ubaid Ur Rahman, H.; Asghar, W.; Nazir, W.; Sandhu, M.A.; Ahmed, A.; Khalid, N. A comprehensive review on chlorpyrifos toxicity with special reference to endocrine disruption: Evidence of mechanisms, exposures and mitigation strategies. Sci. Total Environ. 2021, 755 Pt 2, 142649. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Tang, G.; Xiong, C.; Han, S.; Yang, C.; He, K.; Liu, Q.; Luo, J.; Luo, W.; Wang, Y.; et al. Chronic chlorpyrifos exposure induces oxidative stress, apoptosis and immune dysfunction in largemouth bass (Micropterus salmoides). Environ. Pollut. 2021, 282, 117010. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.; Calderon, F.; Wen, Z.; Kim, H.Y. Docosahexaenoic acid: A positive modulator of Akt signaling in neuronal survival. Proc. Natl. Acad. Sci. USA 2005, 102, 10858–10863. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Mendoza, F.; Jauregui-Huerta, F.; Aguilar-Delgadillo, A.; García-Estrada, J.; Luquin, S. Immediate early gene c-fos in the brain: Focus on Glial Cells. Brain Sci. 2022, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Peillex, C.; Pelletier, M. The impact and toxicity of glyphosate and glyphosate-based herbicides on health and immunity. J. Immunotoxicol. 2020, 17, 163–174. [Google Scholar] [CrossRef] [PubMed]
- De Maria, M.; Kroll, K.J.; Yu, F.; Nouri, M.Z.; Silva-Sanchez, C.; Perez, J.G.; Moraga Amador, D.A.; Zhang, Y.; Walsh, M.T.; Denslow, N.D. Endocrine, immune and renal toxicity in male largemouth bass after chronic exposure to glyphosate and Rodeo®. Aquat. Toxicol. 2022, 246, 106142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Dong, Z.; Liu, F.; Pan, E.; He, N.; Ma, F.; Wang, G.; Wang, Y.; Dong, J. Avermectin induces carp neurotoxicity by mediating blood-brain barrier dysfunction, oxidative stress, inflammation, and apoptosis through PI3K/Akt and NF-kappaB pathways. Ecotoxicol. Environ. Saf. 2022, 243, 113961. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Resendiz, K.J.G.; Ortiz-Lazareno, P.C.; Covantes-Rosales, C.E.; Trujillo-Lepe, A.M.; Toledo-Ibarra, G.A.; Ventura-Ramon, G.H.; Giron-Perez, M.I. Effect of diazinon, an organophosphate pesticide, on signal transduction and death induction in mononuclear cells of Nile tilapia fish (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 89, 12–17. [Google Scholar] [CrossRef]
- Yuan, J.; Amin, P.; Ofengeim, D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 2019, 20, 19–33. [Google Scholar] [CrossRef]
- Arrázola, M.S.; Saquel, C.; Catalán, R.J.; Barrientos, S.A.; Hernandez, D.E.; Martínez, N.W.; Catenaccio, A.; Court, F.A. Axonal degeneration is mediated by necroptosis activation. J. Neurosci. 2019, 39, 3832–3844. [Google Scholar] [CrossRef]
- Wang, X.; Xing, H.; Jiang, Y.; Wu, H.; Sun, G.; Xu, Q.; Xu, S. Accumulation, histopathological effects and response of biochemical markers in the spleens and head kidneys of common carp exposed to atrazine and chlorpyrifos. Food Chem. Toxicol. 2013, 62, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Shiogiri, N.S.; Paulino, M.G.; Carraschi, S.P.; Baraldi, F.G.; da Cruz, C.; Fernandes, M.N. Acute exposure of a glyphosate-based herbicide affects the gills and liver of the Neotropical fish, Piaractus mesopotamicus. Environ. Toxicol. Pharmacol. 2012, 34, 388–396. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Ding, W.; Liu, W.; Li, L.; Zhu, G.; Ma, J. Single and Combined Effects of Chlorpyrifos and Glyphosate on the Brain of Common Carp: Based on Biochemical and Molecular Perspective. Int. J. Mol. Sci. 2023, 24, 12934. https://doi.org/10.3390/ijms241612934
Zhang D, Ding W, Liu W, Li L, Zhu G, Ma J. Single and Combined Effects of Chlorpyrifos and Glyphosate on the Brain of Common Carp: Based on Biochemical and Molecular Perspective. International Journal of Molecular Sciences. 2023; 24(16):12934. https://doi.org/10.3390/ijms241612934
Chicago/Turabian StyleZhang, Dongfang, Weikai Ding, Wei Liu, Liuying Li, Gongming Zhu, and Junguo Ma. 2023. "Single and Combined Effects of Chlorpyrifos and Glyphosate on the Brain of Common Carp: Based on Biochemical and Molecular Perspective" International Journal of Molecular Sciences 24, no. 16: 12934. https://doi.org/10.3390/ijms241612934
APA StyleZhang, D., Ding, W., Liu, W., Li, L., Zhu, G., & Ma, J. (2023). Single and Combined Effects of Chlorpyrifos and Glyphosate on the Brain of Common Carp: Based on Biochemical and Molecular Perspective. International Journal of Molecular Sciences, 24(16), 12934. https://doi.org/10.3390/ijms241612934