Alteration of Mitochondrial Transcript Expression in Arabidopsis thaliana Using a Custom-Made Library of Pentatricopeptide Repeat Proteins
Abstract
:1. Introduction
2. Results
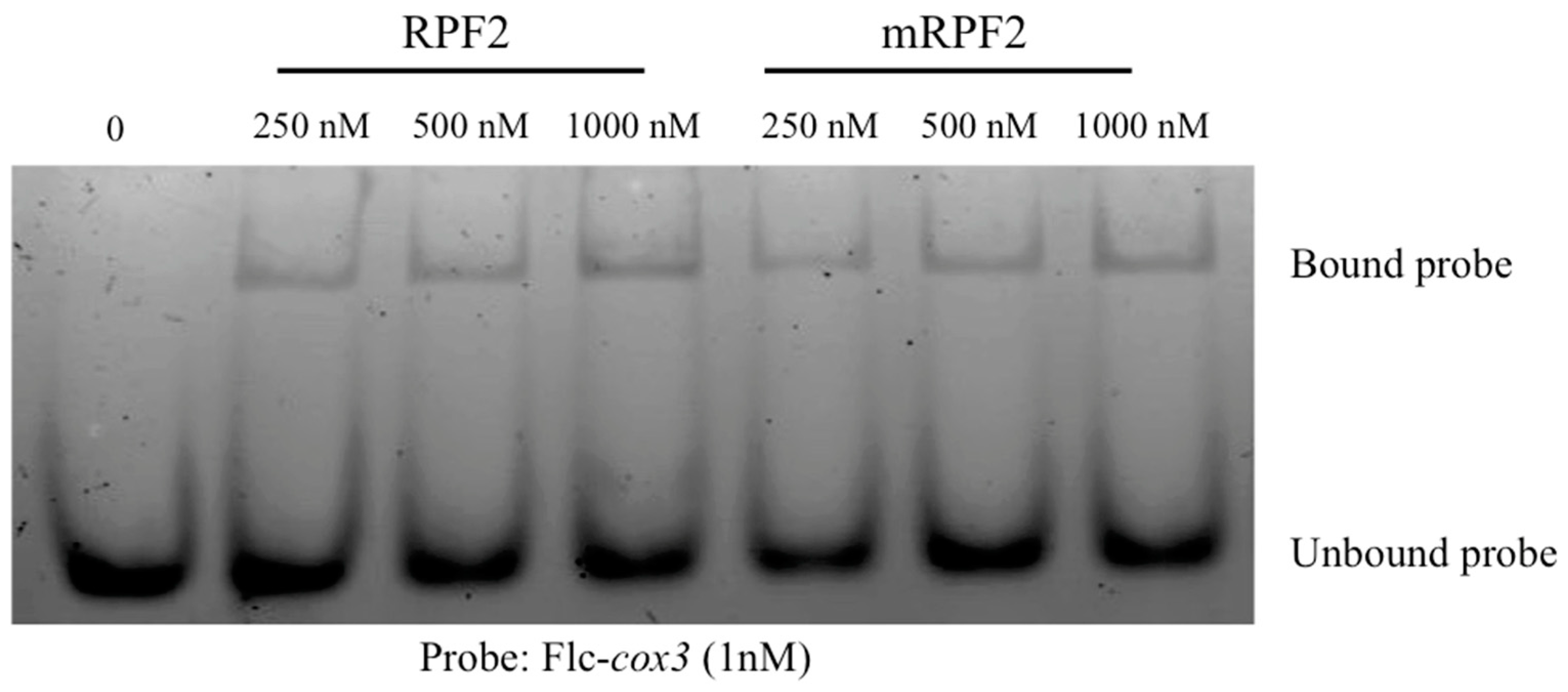
2.1. Creating a Library of RPF2 Proteins
2.2. Genotype and Phenotype Analysis
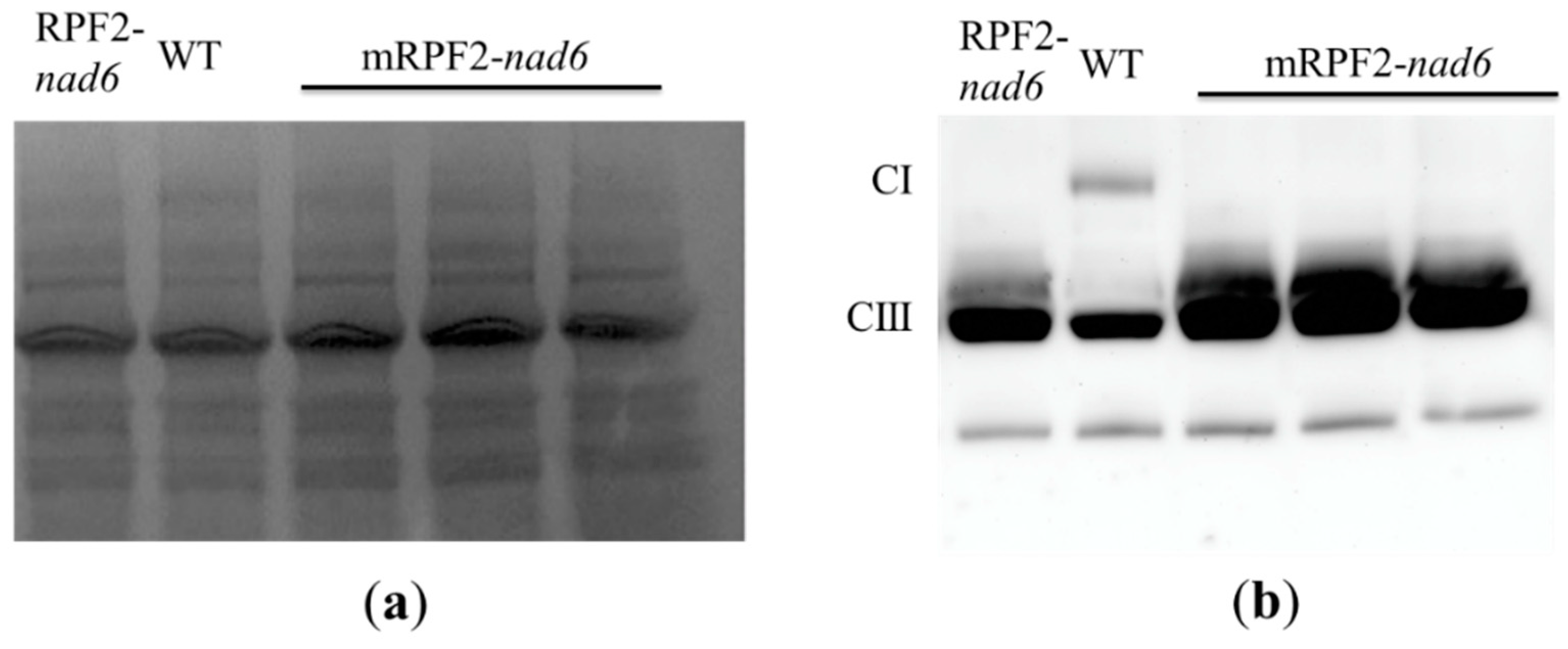
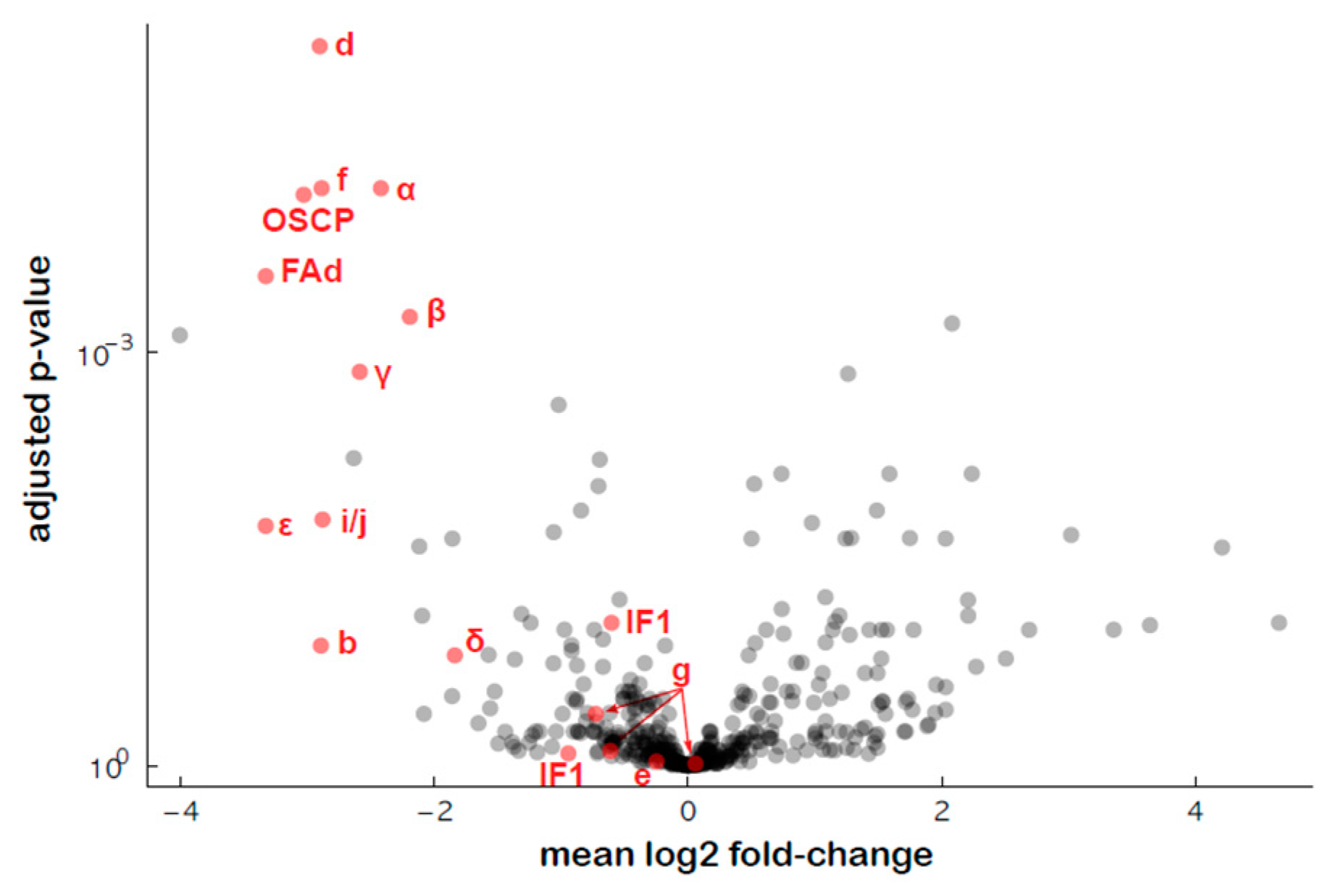
2.3. Analysis of the Mitochondrial Respiratory Complexes
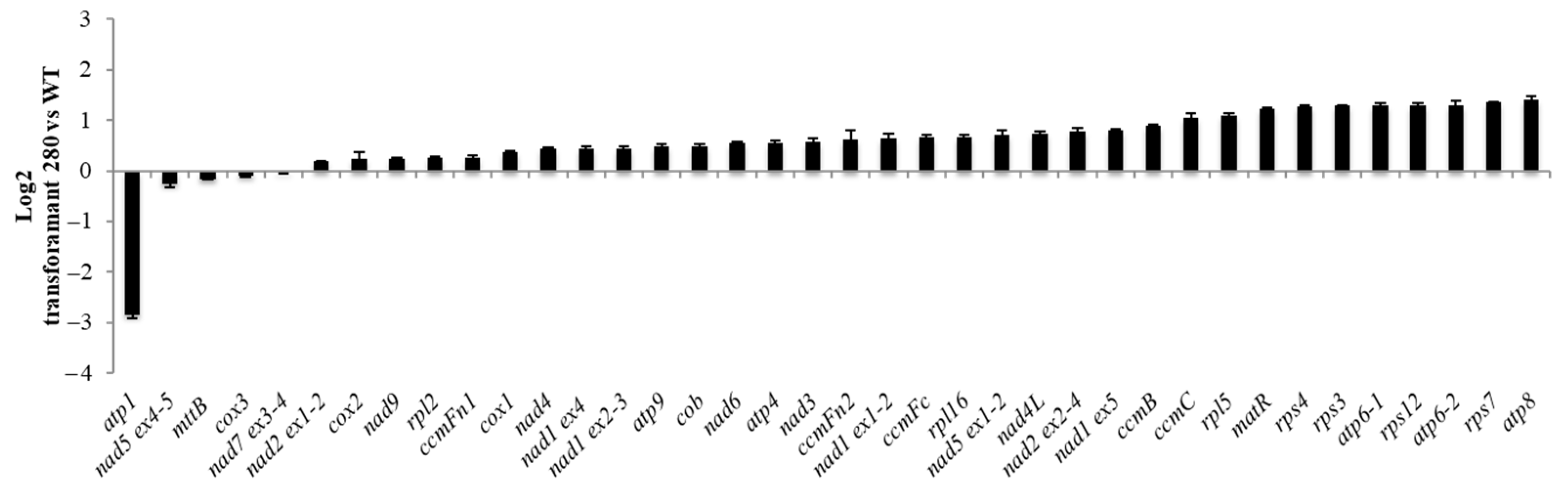
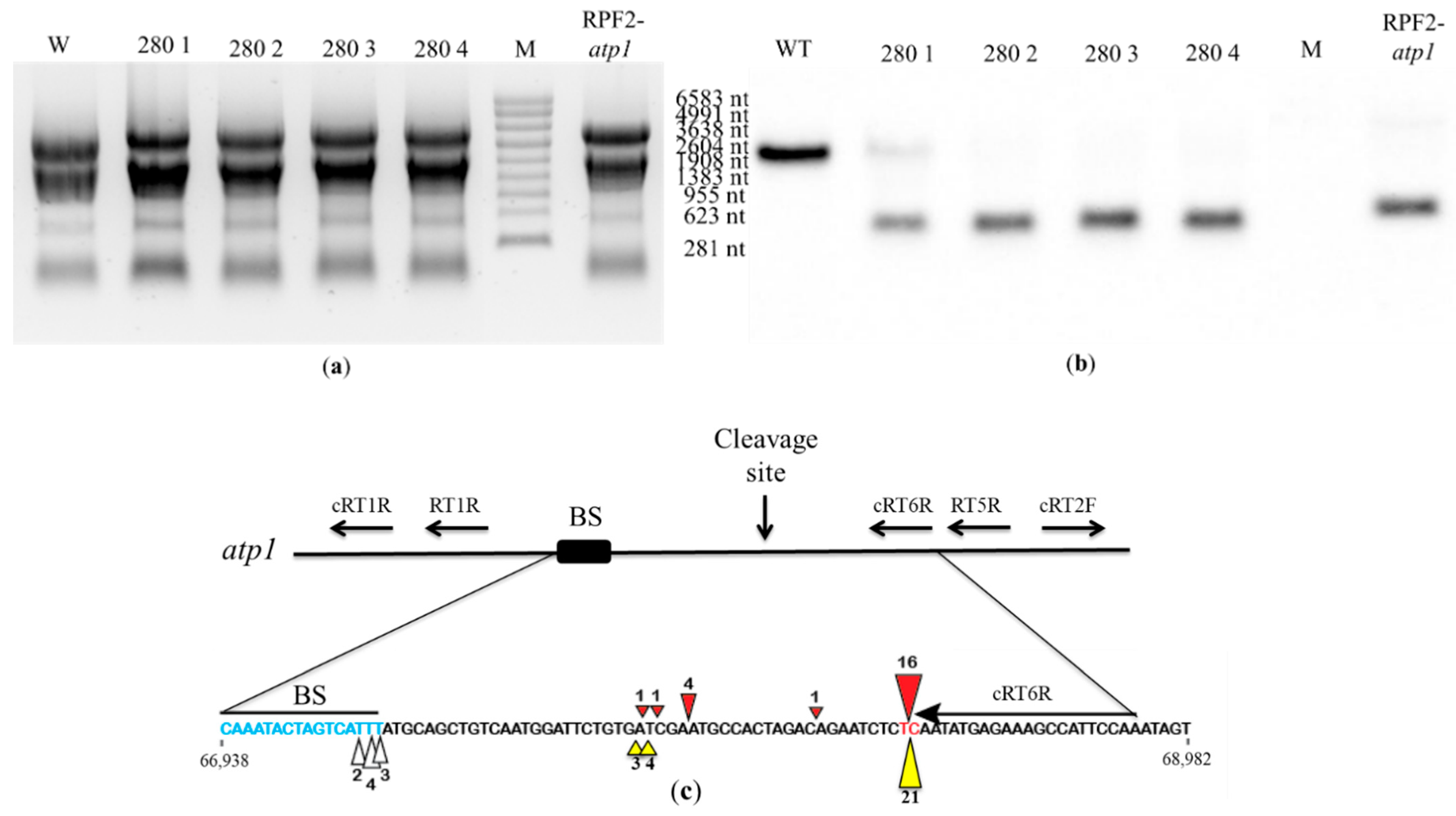
2.4. Further Characterisation of Transformant #280
3. Discussion
4. Materials and Methods
4.1. Plant Growth
4.2. Construction of the PPR Library
4.3. Genotypic Analysis
4.4. Phenotypic Analysis
4.5. High-Throughput BN-PAGE and Western Blotting
4.6. RNA Extraction and qPCR
4.7. Proteomics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bonen, L. Cis-and trans-splicing of group II introns in plant mitochondria. Mitochondrion 2008, 8, 26–34. [Google Scholar] [CrossRef]
- Klodmann, J.; Braun, H.-P. Proteomic approach to characterize mitochondrial complex I from plants. Phytochemistry 2011, 72, 1071–1080. [Google Scholar] [CrossRef]
- Meyer, E.H.; Solheim, C.; Tanz, S.K.; Bonnard, G.; Millar, A.H. Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 2011, 286, 26081–26092. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E.H.; Taylor, N.L.; Millar, A.H. Resolving and identifying protein components of plant mitochondrial respiratory complexes using three dimensions of gel electrophoresis. J. Proteome Res. 2008, 7, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Peters, K.; Belt, K.; Braun, H.-P. 3D gel map of Arabidopsis complex I. Front. Plant Sci. 2013, 4, 153. [Google Scholar] [CrossRef]
- Kubo, T.; Newton, K.J. Angiosperm mitochondrial genomes and mutations. Mitochondrion 2008, 8, 5–14. [Google Scholar] [CrossRef]
- Millar, A.H.; Eubel, H.; Jänsch, L.; Kruft, V.; Heazlewood, J.L.; Braun, H.-P. Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol. Biol. 2004, 56, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Heazlewood, J.L.; Whelan, J.; Millar, A.H. The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 2003, 540, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.F.; Raven, J.A. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J. Mol. Evol. 1996, 42, 482–492. [Google Scholar] [CrossRef]
- Gagliardi, D.; Binder, S. Expression of the plant mitochondrial genome. Plant Mitochondria 2007, 31, 50–95. [Google Scholar]
- Zmudjak, M.; Colas des Francs-Small, C.; Keren, I.; Shaya, F.; Belausov, E.; Small, I.; Ostersetzer-Biran, O. m CSF 1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in A rabidopsis. New Phytol. 2013, 199, 379–394. [Google Scholar] [CrossRef]
- Colas des Francs-Small, C.; Kroeger, T.; Zmudjak, M.; Ostersetzer-Biran, O.; Rahimi, N.; Small, I.; Barkan, A. A PORR domain protein required for rpl2 and ccmFC intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012, 69, 996–1005. [Google Scholar] [CrossRef]
- Hsu, Y.-W.; Wang, H.-J.; Hsieh, M.-H.; Hsieh, H.-L.; Jauh, G.-Y. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 2014, 9, e112360. [Google Scholar]
- Small, I.D.; Peeters, N. The PPR motif–a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 45–47. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Hammani, K.; Giegé, P. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014, 19, 380–389. [Google Scholar] [PubMed]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef] [PubMed]
- Kindgren, P.; Yap, A.; Bond, C.S.; Small, I. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 2015, 27, 403–416. [Google Scholar] [CrossRef]
- Okuda, K.; Shoki, H.; Arai, M.; Shikanai, T.; Small, I.; Nakamura, T. Quantitative analysis of motifs contributing to the interaction between PLS-subfamily members and their target RNA sequences in plastid RNA editing. Plant J. 2014, 80, 870–882. [Google Scholar]
- Shen, C.; Zhang, D.; Guan, Z.; Liu, Y.; Yang, Z.; Yang, Y.; Wang, X.; Wang, Q.; Zhang, Q.; Fan, S. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016, 7, 11285. [Google Scholar]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16 (Suppl. S1), S154–S169. [Google Scholar]
- Gaborieau, L.; Brown, G.G.; Mireau, H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci. 2016, 7, 1816. [Google Scholar] [PubMed]
- Dahan, J.; Mireau, H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 2013, 10, 1469–1476. [Google Scholar] [PubMed]
- Bentolila, S.; Alfonso, A.A.; Hanson, M.R. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 2002, 99, 10887–10892. [Google Scholar]
- Gillman, J.D.; Bentolila, S.; Hanson, M.R. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007, 49, 217–227. [Google Scholar]
- Jonietz, C.; Forner, J.; Hölzle, A.; Thuss, S.; Binder, S. RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 2010, 22, 443–453. [Google Scholar]
- Hölzle, A.; Jonietz, C.; Törjek, O.; Altmann, T.; Binder, S.; Forner, J. A RESTORER OF FERTILITY-like PPR gene is required for 5′-end processing of the nad4 mRNA in mitochondria of Arabidopsis thaliana. Plant J. 2011, 65, 737–744. [Google Scholar]
- Fujii, S.; Bond, C.S.; Small, I.D. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA 2011, 108, 1723–1728. [Google Scholar] [PubMed]
- Colas des Francs-Small, C.; Vincis Pereira Sanglard, L.; Small, I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 2018, 1, 166. [Google Scholar]
- Yang, F.; Vincis Pereira Sanglard, L.; Lee, C.-P.; Stroeher, E.; Singh, S.; Oh, G.G.K.; Millar, A.H.; Small, I.D.; Colas des Francs-Small, C. Knockdown of mitochondrial atp1 mRNA by a custom-designed pentatricopeptide repeat protein alters F1Fo ATP synthase. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kobayashi, T.; Yagi, Y.; Nakamura, T. Comprehensive Prediction of Target RNA Editing Sites for PLS-Class PPR Proteins in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 862–874. [Google Scholar] [PubMed]
- Vincis Pereira Sanglard, L.; des Francs-Small, C.C. High-Throughput BN-PAGE for Mitochondrial Respiratory Complexes. In Plant Mitochondria; Springer: Berlin/Heidelberg, Germany, 2022; pp. 111–119. [Google Scholar]
- Tzfira, T.; Li, J.; Lacroix, B.; Citovsky, V. Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 2004, 20, 375–383. [Google Scholar] [CrossRef]
- Tzfira, T.; Citovsky, V. Agrobacterium: From Biology to Biotechnology; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Arnal, N.; Quadrado, M.; Simon, M.; Mireau, H. A restorer-of-fertility like pentatricopeptide repeat gene directs ribonucleolytic processing within the coding sequence of rps3-rpl16 and orf240a mitochondrial transcripts in Arabidopsis thaliana. Plant J. 2014, 78, 134–145. [Google Scholar] [PubMed]
- Fujii, S.; Suzuki, T.; Giegé, P.; Higashiyama, T.; Koizuka, N.; Shikanai, T. The Restorer-of-fertility-like 2 pentatricopeptide repeat protein and RN ase P are required for the processing of mitochondrial orf291 RNA in Arabidopsis. Plant J. 2016, 86, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Stoll, K.; Jonietz, C.; Binder, S. In Arabidopsis thaliana two co-adapted cyto-nuclear systems correlate with distinct ccmC transcript sizes. Plant J. 2015, 81, 247–257. [Google Scholar]
- Stoll, K.; Jonietz, C.; Schleicher, S.; Colas des Francs-Small, C.; Small, I.; Binder, S. In Arabidopsis thaliana distinct alleles encoding mitochondrial RNA PROCESSING FACTOR 4 support the generation of additional 5′ termini of ccmB transcripts. Plant Mol. Biol. 2017, 93, 659–668. [Google Scholar]
- Nakazawa, M.; Matsui, M. Selection of hygromycin-resistant Arabidopsis seedlings. Biotechniques 2003, 34, 28–30. [Google Scholar]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar]
- Royan, S.; Gutmann, B.; Colas des Francs-Small, C.; Honkanen, S.; Schmidberger, J.; Soet, A.; Sun, Y.K.; Vincis Pereira Sanglard, L.; Bond, C.S.; Small, I. A synthetic RNA editing factor edits its target site in chloroplasts and bacteria. Commun. Biol. 2021, 4, 545. [Google Scholar]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, 85. [Google Scholar]
- Bernath-Levin, K.; Schmidberger, J.; Honkanen, S.; Gutmann, B.; Sun, Y.K.; Pullakhandam, A.; Colas des Francs-Small, C.; Bond, C.S.; Small, I. Cofactor-independent RNA editing by a synthetic S-type PPR protein. Synth. Biol. 2022, 7, ysab034. [Google Scholar] [CrossRef] [PubMed]
- McDowell, R.; Small, I.; Bond, C.S. Synthetic PPR proteins as tools for sequence-specific targeting of RNA. Methods 2022, 208, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., III; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Bui, M.; Liu, Z. Simple allele-discriminating PCR for cost-effective and rapid genotyping and mapping. Plant Methods 2009, 5, 1. [Google Scholar] [CrossRef]
- Delannoy, E.; de Longevialle, A.F.; Colas des Francs-Small, C. Mitochondrial RNA transcript analysis assay of Arabidopsis leaf tissues. Bio. Protoc. 2015, 5, 1620e. [Google Scholar] [CrossRef]
- Colas des Francs-Small, C.; de Longevialle, A.F.; Li, Y.; Lowe, E.; Tanz, S.K.; Smith, C.; Bevan, M.W.; Small, I. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 2014, 165, 1409–1416. [Google Scholar] [CrossRef]
- Petereit, J.; Duncan, O.; Murcha, M.W.; Fenske, R.; Cincu, E.; Cahn, J.; Pružinská, A.; Ivanova, A.; Kollipara, L.; Wortelkamp, S. Mitochondrial CLPP2 assists coordination and homeostasis of respiratory complexes. Plant Physiol. 2020, 184, 148–164. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; Garcia-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincis Pereira Sanglard, L.; Small, I.D.; Colas des Francs-Small, C. Alteration of Mitochondrial Transcript Expression in Arabidopsis thaliana Using a Custom-Made Library of Pentatricopeptide Repeat Proteins. Int. J. Mol. Sci. 2023, 24, 13233. https://doi.org/10.3390/ijms241713233
Vincis Pereira Sanglard L, Small ID, Colas des Francs-Small C. Alteration of Mitochondrial Transcript Expression in Arabidopsis thaliana Using a Custom-Made Library of Pentatricopeptide Repeat Proteins. International Journal of Molecular Sciences. 2023; 24(17):13233. https://doi.org/10.3390/ijms241713233
Chicago/Turabian StyleVincis Pereira Sanglard, Lilian, Ian D. Small, and Catherine Colas des Francs-Small. 2023. "Alteration of Mitochondrial Transcript Expression in Arabidopsis thaliana Using a Custom-Made Library of Pentatricopeptide Repeat Proteins" International Journal of Molecular Sciences 24, no. 17: 13233. https://doi.org/10.3390/ijms241713233




