THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC
Abstract
:1. Introduction
2. Results
2.1. Identification of ECM Clusters Based on DEGs Associated with the ECM Scores in OSCC
2.2. Characteristics of Immune Infiltration in Different ECM Clusters of OSCC
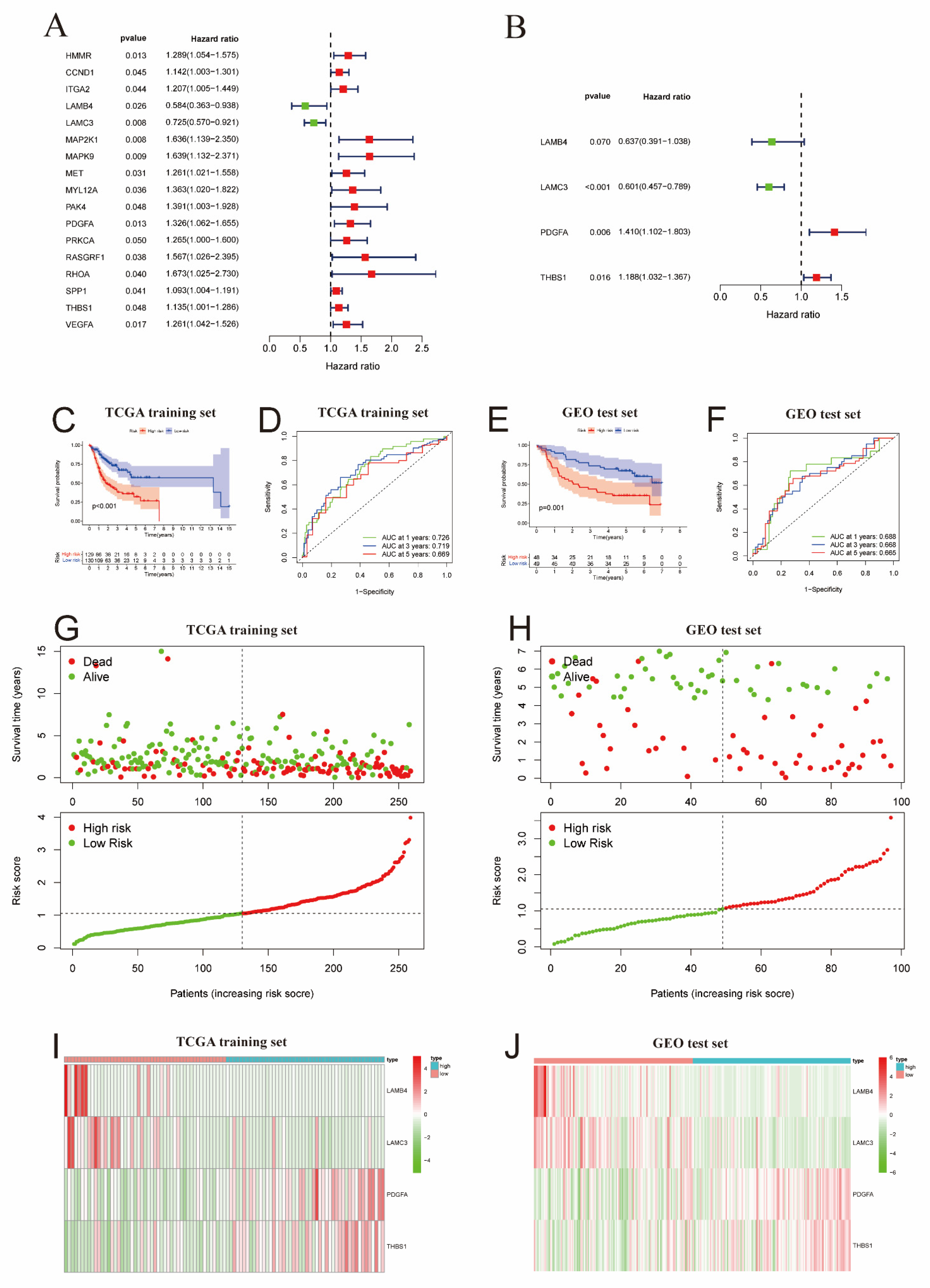
2.3. Development and Verification of the ECM Prognostic Signature for OSCC
2.4. Correlation Analysis of ECM Risk Scores and Immune Infiltration in OSCC
2.5. Ectopic Expression of THBS1 Is Correlated with Clinical Parameters of OSCC
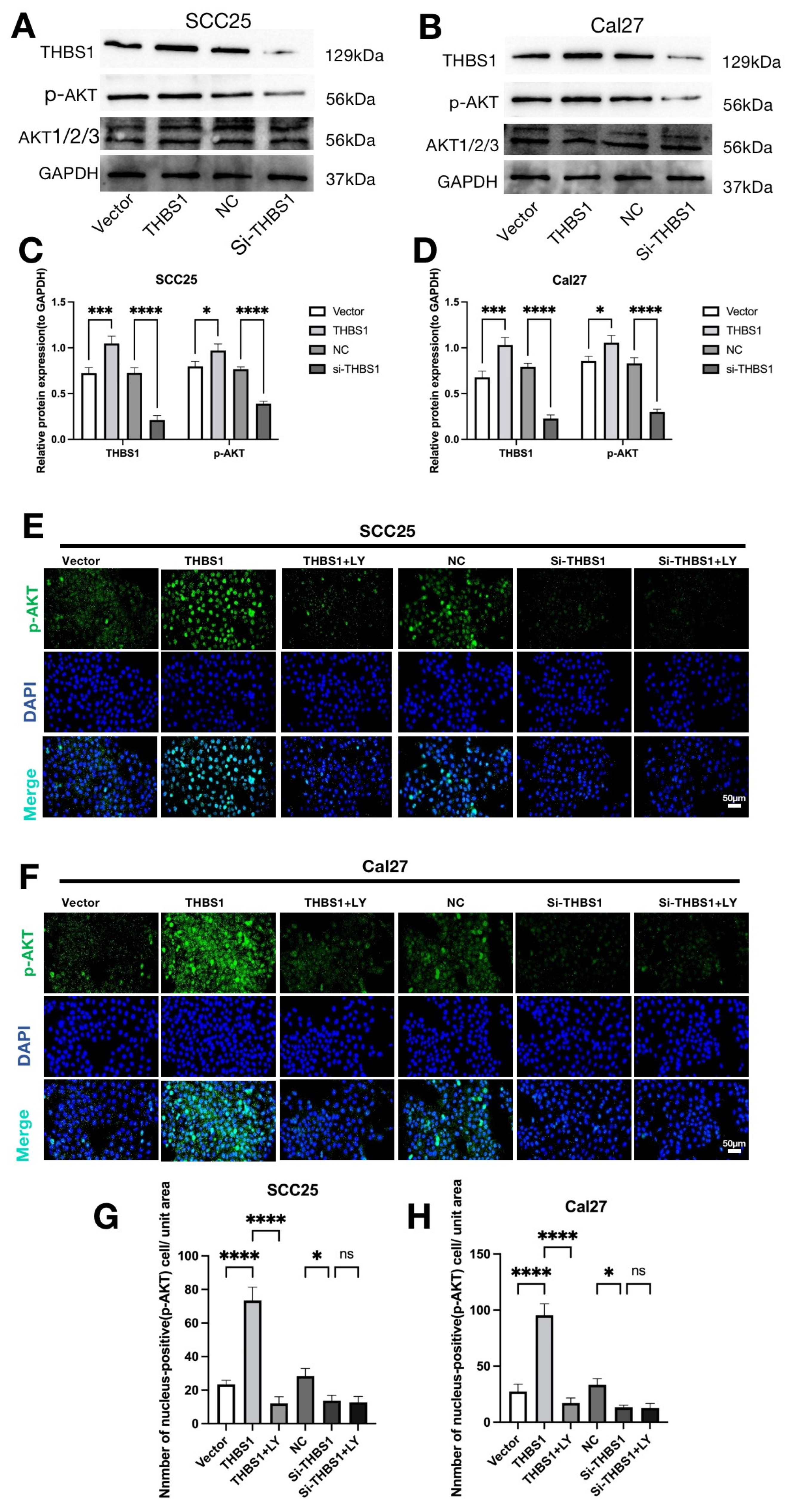
2.6. THBS1 Can Over-Activate the PI3K/AKT Signaling Pathway in OSCC
2.7. THBS1 Mediates Cell Proliferation via the PI3K/AKT Signaling Pathway in OSCC
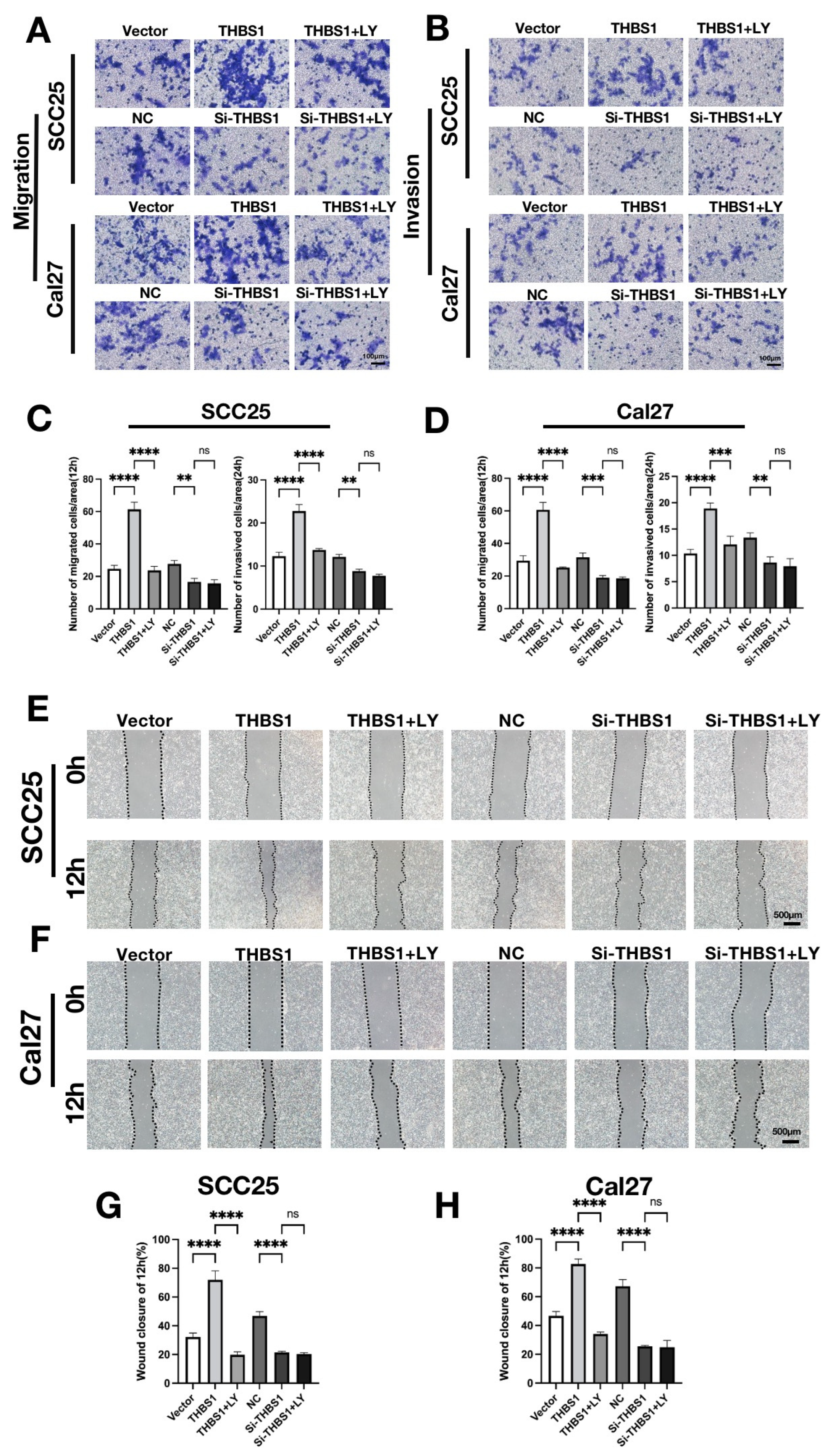
2.8. THBS1 Mediates the Migration and Invasion Behaviors via the PI3K/AKT Signaling Pathway in OSCC
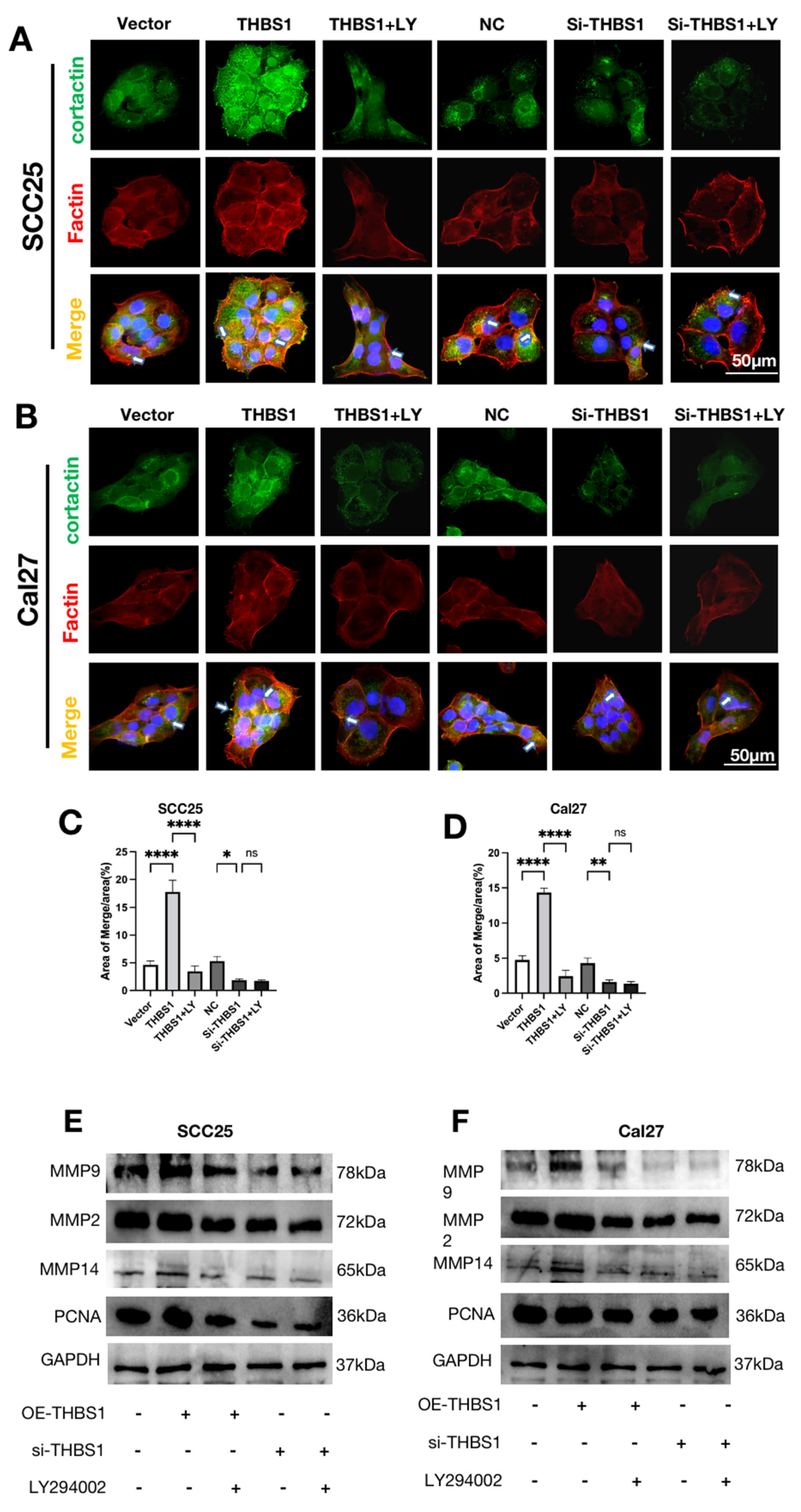
2.9. THBS1 Promotes Invadopodia Formation and ECM Degradation through PI3K/AKT Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Data Collection and Processing
4.2. Identification of ECM-Related DGEs (Differentially Expressed Genes)
4.3. Tumor Sample Clustering
4.4. Analysis of Tumor Gene Mutations
4.5. Development and Validation of ECM-Related Signature for OSCC
4.6. Functional and Pathway Enrichment Analysis
4.7. Immune Infiltration Study of OSCC
4.8. Cell Culture, Cell Transfection, and Reagents
4.9. Transwell Assay
4.10. Western Blotting
4.11. Wound Healing Assay
4.12. Cell Proliferation Assay
4.13. Cell Immunofluorescence Staining Assay
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chai, A.W.Y.; Lim, K.P.; Cheong, S.C. Translational genomics and recent advances in oral squamous cell carcinoma. Semin. Cancer Biol. 2020, 61, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Magnano, S.; Barroeta, P.H.; Duffy, R.; O’Sullivan, J.; Zisterer, D.M. Cisplatin induces autophagy-associated apoptosis in human oral squamous cell carcinoma (OSCC) me-diated in part through reactive oxygen species. Toxicol. Appl. Pharmacol. 2021, 427, 115646. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-P.; Chen, K.-C.; Chen, M.-Y.; Huang, L.-Y.; Su, A.-Y.; Chiang, W.-F.; Huang, W.-T.; Huang, T.-T. Epigenetic Deregulation of Protein Tyrosine Kinase 6 Promotes Carcinogenesis of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 4495. [Google Scholar] [CrossRef] [PubMed]
- Almangush, A.; Leivo, I.; Mäkitie, A.A. Biomarkers for Immunotherapy of Oral Squamous Cell Carcinoma: Current Status and Challenges. Front. Oncol. 2021, 11, 616629. [Google Scholar] [CrossRef]
- Niu, Q.; Sun, Q.; Bai, R.; Zhang, Y.; Zhuang, Z.; Zhang, X.; Xin, T.; Chen, S.; Han, B. Progress of Nanomaterials-Based Photothermal Therapy for Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 10428. [Google Scholar] [CrossRef]
- Rossi, G.R.; Trindade, E.S.; Souza-Fonseca-Guimaraes, F. Tumor Microenvironment-Associated Extracellular Matrix Components Regulate NK Cell Function. Front. Immunol. 2020, 11, 73. [Google Scholar] [CrossRef]
- Mohan, V.; Das, A.; Sagi, I. Emerging roles of ECM remodeling processes in cancer. Semin. Cancer Biol. 2020, 62, 192–200. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef]
- Xu, X.; Wu, Y.; Qian, X.; Wang, Y.; Wang, J.; Li, J.; Li, Y.; Zhang, Z. Nanomedicine Strategies to Circumvent Intratumor Extracellular Matrix Barriers for Cancer Therapy. Adv. Health Mater. 2022, 11, e2101428. [Google Scholar] [CrossRef]
- Perrin, L.; Belova, E.; Bayarmagnai, B.; Tüzel, E.; Gligorijevic, B. Invadopodia enable cooperative invasion and metastasis of breast cancer cells. Commun. Biol. 2022, 5, 758. [Google Scholar] [CrossRef]
- Niland, S.; Riscanevo, A.X.; Eble, J.A. Matrix Metalloproteinases Shape the Tumor Microenvironment in Cancer Pro-gression. Int. J. Mol. Sci. 2021, 23, 146. [Google Scholar] [CrossRef] [PubMed]
- Karamanou, K.; Franchi, M.; Vynios, D.; Brézillon, S. Epithelial-to-mesenchymal transition and invadopodia markers in breast cancer: Lumican a key reg-ulator. Semin. Cancer Biol. 2020, 62, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yu, X.; Sun, S.; Zhang, X.; Yang, W.; Zhang, J.; Zhang, X.; Jiang, Z. Increased expression of MMP-2 and MMP-9 indicates poor prognosis in glioma recurrence. Biomed. Pharmacother. 2019, 118, 109369. [Google Scholar] [CrossRef] [PubMed]
- Martins, J.M.A.; Rabelo-Santos, S.H.; Westin, M.C.D.A.; Zeferino, L.C. Tumoral and stromal expression of MMP-2, MMP-9, MMP-14, TIMP-1, TIMP-2, and VEGF-A in cervical cancer patient survival: A competing risk analysis. BMC Cancer 2020, 20, 660. [Google Scholar] [CrossRef]
- Ye, Y.; Kuang, X.; Xie, Z.; Liang, L.; Zhang, Z.; Zhang, Y.; Ma, F.; Gao, Q.; Chang, R.; Lee, H.H.; et al. Small-molecule MMP2/MMP9 inhibitor SB-3CT modulates tumor immune surveillance by regulating PD-L1. Genome Med. 2020, 12, 83. [Google Scholar] [CrossRef]
- Chen, Z.; Wei, H.; Zhao, X.; Xin, X.; Peng, L.; Ning, Y.; Wang, Y.; Lan, Y.; Zhang, Q. Metformin treatment alleviates polycystic ovary syndrome by decreasing the expression of MMP-2 and MMP-9 via H19/miR-29b-3p and AKT/mTOR/autophagy signaling pathways. J. Cell Physiol. 2019, 234, 19964–19976. [Google Scholar] [CrossRef]
- Salik, B.; Smyth, M.J.; Nakamura, K. Targeting immune checkpoints in hematological malignancies. J. Hematol. Oncol. 2020, 13, 111. [Google Scholar] [CrossRef]
- Gonzalez-Molina, J.; Moyano-Galceran, L.; Single, A.; Gultekin, O.; Alsalhi, S.; Lehti, K. Chemotherapy as a regulator of extracellular matrix-cell communication: Implications in therapy resistance. Semin. Cancer Biol. 2022, 86 Pt 3, 224–236. [Google Scholar] [CrossRef]
- Gao, S.; Yang, X.; Xu, J.; Qiu, N.; Zhai, G. Nanotechnology for Boosting Cancer Immunotherapy and Remodeling Tumor Microenvironment: The Hori-zons in Cancer Treatment. ACS Nano 2021, 15, 12567–12603. [Google Scholar] [CrossRef]
- Tu, J.; Xu, H.; Ma, L.; Li, C.; Qin, W.; Chen, X.; Yi, M.; Sun, L.; Liu, B.; Yuan, X. Nintedanib enhances the efficacy of PD-L1 blockade by upregulating MHC-I and PD-L1 expression in tumor cells. Theranostics 2022, 12, 747–766. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Liu, J.; Dang, P.; Hu, S.; Yuan, W.; Sun, Z.; Liu, Y.; Wang, C. Roles of tumor-associated macrophages in anti-PD-1/PD-L1 immunotherapy for solid cancers. Mol. Cancer 2023, 22, 58. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Acerbi, I.; Cassereau, L.; Dean, I.; Shi, Q.; Au, A.; Park, C.; Chen, Y.Y.; Liphardt, J.; Hwang, E.S.; Weaver, V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015, 7, 1120–1134. [Google Scholar] [CrossRef]
- Cox, T.R. The matrix in cancer. Nat. Rev. Cancer 2021, 21, 217–238. [Google Scholar] [CrossRef]
- Parker, A.L.; Cox, T.R. The Role of the ECM in Lung Cancer Dormancy and Outgrowth. Front. Oncol. 2020, 10, 1766. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Zhang, S.-Y.; Ren, X.-Y.; Wang, C.-Y.; Chen, X.-J.; Cao, R.-Y.; Liu, Q.; Pan, X.; Zhou, J.-Y.; Zhang, W.-L.; Tang, X.-R.; et al. Comprehensive Characterization of Immune Landscape Based on Epithelial-Mesenchymal Transition Signature in OSCC: Implication for Prognosis and Immunotherapy. Front. Oncol. 2021, 11, 587862. [Google Scholar] [CrossRef]
- Kandimalla, R.; Tomihara, H.; Banwait, J.K.; Yamamura, K.; Singh, G.; Baba, H.; Goel, A. A 15-Gene Immune, Stromal, and Proliferation Gene Signature that Significantly Associates with Poor Survival in Patients with Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020, 26, 3641–3648. [Google Scholar] [CrossRef]
- Fakih, M.; Ouyang, C.; Wang, C.; Tu, T.Y.; Gozo, M.C.; Cho, M.; Sy, M.; Longmate, J.A.; Lee, P.P. Immune overdrive signature in colorectal tumor subset predicts poor clinical outcome. J. Clin. Investig. 2019, 129, 4464–4476. [Google Scholar] [CrossRef]
- Colbeck, E.J.; Jones, E.; Hindley, J.P.; Smart, K.; Schulz, R.; Browne, M.; Cutting, S.; Williams, A.; Parry, L.; Godkin, A.; et al. Treg Depletion Licenses T Cell–Driven HEV Neogenesis and Promotes Tumor Destruction. Cancer Immunol. Res. 2017, 5, 1005–1015. [Google Scholar] [CrossRef]
- Peske, J.D.; Woods, A.B.; Engelhard, V.H. Control of CD8 T-Cell Infiltration into Tumors by Vasculature and Microenvironment. Adv. Cancer Res. 2015, 128, 263–307. [Google Scholar]
- Salmon, H.; Franciszkiewicz, K.; Damotte, D.; Dieu-Nosjean, M.-C.; Validire, P.; Trautmann, A.; Mami-Chouaib, F.; Donnadieu, E. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J. Clin. Investig. 2012, 122, 899–910. [Google Scholar] [CrossRef]
- Hammoud, M.K.; Dietze, R.; Pesek, J.; Finkernagel, F.; Unger, A.; Bieringer, T.; Nist, A.; Stiewe, T.; Bhagwat, A.M.; Nockher, W.A.; et al. Arachidonic acid, a clinically adverse mediator in the ovarian cancer microenvironment, impairs JAK-STAT signaling in macrophages by perturbing lipid raft structures. Mol. Oncol. 2022, 16, 3146–3166. [Google Scholar] [CrossRef] [PubMed]
- Lauson, C.B.N.; Tiberti, S.; Corsetto, P.A.; Conte, F.; Tyagi, P.; Machwirth, M.; Ebert, S.; Loffreda, A.; Scheller, L.; Sheta, D.; et al. Linoleic acid potentiates CD8+ T cell metabolic fitness and antitumor immunity. Cell Metab. 2023, 35, 633–650.e9. [Google Scholar] [CrossRef]
- Mittal, B.; Tulsyan, S.; Kumar, S.; Mittal, R.D.; Agarwal, G. Cytochrome P450 in Cancer Susceptibility and Treatment. Adv. Clin. Chem. 2015, 71, 77–139. [Google Scholar] [CrossRef]
- Guo, Z.; Johnson, V.; Barrera, J.; Porras, M.; Hinojosa, D.; Hernández, I.; McGarrah, P.; Potter, D.A. Targeting cytochrome P450-dependent cancer cell mitochondria: Cancer associated CYPs and where to find them. Cancer Metastasis Rev. 2018, 37, 409–423. [Google Scholar] [CrossRef]
- Waxman, D.J.; Chen, L.; Hecht, J.E.D.; Jounaidi, Y. Cytochrome P450-based cancer gene therapy: Recent advances and future prospects. Drug Metab. Rev. 1999, 31, 503–522. [Google Scholar] [CrossRef]
- Bartoschek, M.; Pietras, K. PDGF family function and prognostic value in tumor biology. Biochem. Biophys. Res. Commun. 2018, 503, 984–990. [Google Scholar] [CrossRef]
- Sahraei, M.; Roy, L.D.; Curry, J.M.; Teresa, T.L.; Nath, S.; Besmer, D.; Kidiyoor, A.; Dalia, R.; Gendler, S.J.; Mukherjee, P. MUC1 regulates PDGFA expression during pancreatic cancer progression. Oncogene 2012, 31, 4935–4945. [Google Scholar] [CrossRef]
- Ding, Z.; Ke, R.; Zhang, Y.; Fan, Y.; Fan, J. FOXE1 inhibits cell proliferation, migration and invasion of papillary thyroid cancer by regulating PDGFA. Mol. Cell. Endocrinol. 2019, 493, 110420. [Google Scholar] [CrossRef]
- Cui, H.; Wei, W.; Qian, M.; Tian, R.; Fu, X.; Li, H.; Nan, G.; Yang, T.; Lin, P.; Chen, X.; et al. PDGFA-associated protein 1 is a novel target of c-Myc and contributes to colorectal cancer initiation and progression. Cancer Commun. 2022, 42, 750–767. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Yun, J.H.; Choi, C.; Cho, S.; Kim, S.J.; Kim, J.H. Autocrine DUSP28 signaling mediates pancreatic cancer malignancy via regulation of PDGF-A. Sci. Rep. 2017, 7, 12760. [Google Scholar] [CrossRef]
- Pal, S.K.; Nguyen, C.T.K.; Morita, K.-I.; Miki, Y.; Kayamori, K.; Yamaguchi, A.; Sakamoto, K. THBS1 is induced by TGFB1 in the cancer stroma and promotes invasion of oral squamous cell carcinoma. J. Oral. Pathol. Med. 2016, 45, 730–739. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Zhang, J.; Chen, W.; Chen, W. M1-like tumor-associated macrophages activated by exosome-transferred THBS1 promote malignant migra-tion in oral squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 143. [Google Scholar] [CrossRef]
- You, Y.; Tian, Z.; Du, Z.; Wu, K.; Xu, G.; Dai, M.; Wang, Y.A.; Xiao, M. M1-like tumor-associated macrophages cascade a mesenchymal/stem-like phenotype of oral squamous cell carcinoma via the IL6/Stat3/THBS1 feedback loop. J. Exp. Clin. Cancer Res. 2022, 41, 10. [Google Scholar] [CrossRef]
- Ji, F.H.; Qiu, X.G. THBS1, a fatty acid-related metabolic gene, can promote the development of laryngeal cancer. Sci. Rep. 2022, 12, 18809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, Q.; Cui, Q.; Xu, Y. MiR-3612 targeting THBS1 suppresses nasopharyngeal carcinoma progression by PI3K/AKT signaling pathway. Hum. Exp. Toxicol. 2023, 42, 9603271221150248. [Google Scholar] [CrossRef]
- Xu, Y.; Sui, L.; Qiu, B.; Yin, X.; Liu, J.; Zhang, X. ANXA4 promotes trophoblast invasion via the PI3K/Akt/eNOS pathway in preeclampsia. Am. J. Physiol. Physiol. 2019, 316, C481–C491. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, L.; Lu, S.; Li, W.; Dong, W. KIFC3 Promotes Proliferation, Migration, and Invasion in Colorectal Cancer via PI3K/AKT/mTOR Signaling Pathway. Front. Genet. 2022, 13, 848926. [Google Scholar] [CrossRef]
- Chai, S.; Wen, Z.; Zhang, R.; Bai, Y.; Liu, J.; Li, J.; Kongling, W.; Chen, W.; Wang, F.; Gao, L. CCL25/CCR9 interaction promotes the malignant behavior of salivary adenoid cystic carcinoma via the PI3K/AKT signaling pathway. PeerJ 2022, 10, e13844. [Google Scholar] [CrossRef]
- Luengo-Gil, G.; González-Billalabeitia, E.; Chaves-Benito, A.; Martínez, E.G.; Garre, E.G.; Vicente, V.; de la Peña, F.A. Effects of conventional neoadjuvant chemotherapy for breast cancer on tumor angiogenesis. Breast Cancer Res. Treat. 2015, 151, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.F.; Johansen, J.; Bjørnevoll, I.; Sylvander, A.E.; Steinsbekk, K.S.; Sætrom, P.; Sandvik, A.K.; Drabløs, F.; Sjursen, W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Fam. Cancer 2015, 14, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.R.; An, C.H.; Yoo, N.J.; Lee, S.H. Laminin gene LAMB4 is somatically mutated and expressionally altered in gastric and colorectal cancers. APMIS 2015, 123, 65–71. [Google Scholar] [CrossRef]
- He, Y.; Deng, Y.; Peng, P.; Wang, N.; Wang, J.; Ding, Z.; Chen, X.; Zhou, X. A novel messenger RNA and long noncoding RNA signature associated with the progression of nonmuscle invasive bladder cancer. J. Cell. Biochem. 2019, 120, 8101–8109. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.G.; Naven, M.; Harris, R.; Colley, J.; West, H.; Li, N.; Liu, Y.; Adams, R.; Maughan, T.S.; Nichols, L.; et al. Exome Resequencing Identifies Potential Tumor-Suppressor Genes that Predispose to Colorectal Cancer. Hum. Mutat. 2013, 34, 1026–1034. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, X.; Xie, W.; Li, Z.; Liu, W.; Liu, A. Identification of a basement membrane-related gene signature for predicting prognosis and estimating the tumor immune microenvironment in breast cancer. Front. Endocrinol. 2022, 13, 1065530. [Google Scholar] [CrossRef]
- Ishikawa, S.; Sugimoto, M.; Konta, T.; Kitabatake, K.; Ueda, S.; Edamatsu, K.; Okuyama, N.; Yusa, K.; Iino, M. Salivary Metabolomics for Prognosis of Oral Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 789248. [Google Scholar] [CrossRef]
- Yang, Q.; Cheng, C.; Zhu, R.; Guo, F.; Lai, R.; Liu, X.; Li, M. A N6-methyladenosine-related long noncoding RNAs model for predicting prognosis in oral squamous cell carcinoma: Association with immune cell infiltration and tumor metastasis. Oral. Oncol. 2022, 127, 105771. [Google Scholar] [CrossRef]
- Zhou, J.-Y.; Wang, W.-J.; Zhang, C.-Y.; Ling, Y.-Y.; Hong, X.-J.; Su, Q.; Li, W.-G.; Mao, Z.-W.; Cheng, B.; Tan, C.-P.; et al. Ru(II)-modified TiO2 nanoparticles for hypoxia-adaptive photo-immunotherapy of oral squamous cell carcinoma. Biomaterials 2022, 289, 121757. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Variable selection with stepwise and best subset approaches. Ann. Transl. Med. 2016, 4, 136. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Chen, Z.; Chen, W.; Yuan, L.; Liu, B. The prognostic value of circular RNA regulatory genes in competitive endogenous RNA network in gastric cancer. Cancer Gene Ther. 2021, 28, 1175–1187. [Google Scholar] [CrossRef]
- Xie, T.; Guo, X.; Wu, D.; Li, S.; Lu, Y.; Wang, X.; Chen, L. PAFAH1B3 Expression Is Correlated With Gastric Cancer Cell Proliferation and Immune Infiltration. Front. Oncol. 2021, 11, 591545. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Z.; Zhang, Y.; Wang, X.; Wu, Y.; Mao, J.; Li, Q.; Gong, S. THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC. Int. J. Mol. Sci. 2023, 24, 13312. https://doi.org/10.3390/ijms241713312
Wen Z, Zhang Y, Wang X, Wu Y, Mao J, Li Q, Gong S. THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC. International Journal of Molecular Sciences. 2023; 24(17):13312. https://doi.org/10.3390/ijms241713312
Chicago/Turabian StyleWen, Zhihao, Yuxiao Zhang, Xiangyao Wang, Yaxin Wu, Jing Mao, Qilin Li, and Shiqiang Gong. 2023. "THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC" International Journal of Molecular Sciences 24, no. 17: 13312. https://doi.org/10.3390/ijms241713312
APA StyleWen, Z., Zhang, Y., Wang, X., Wu, Y., Mao, J., Li, Q., & Gong, S. (2023). THBS1-Mediated Degradation of Collagen via the PI3K/AKT Pathway Facilitates the Metastasis and Poor Prognosis of OSCC. International Journal of Molecular Sciences, 24(17), 13312. https://doi.org/10.3390/ijms241713312






