The Regulation of Ferroptosis by Noncoding RNAs
Abstract
1. Introduction
2. The Core Mechanism and Regulators of Ferroptosis
2.1. Iron Metabolism
2.2. Lipid Metabolism
2.3. Antioxidant Defense Systems
2.4. Transcription Factors Regulating Ferroptosis
3. The Classification and Function of ncRNAs in Ferroptosis
3.1. MiRNAs in Ferroptosis
3.2. LncRNAs in Ferroptosis
3.3. CircRNAs in Ferroptosis
3.4. PiRNAs in Ferroptosis
3.5. Structural ncRNAs in Ferroptosis
4. ncRNAs Regulating Ferroptosis
4.1. NcRNAs Regulating Ferroptosis through Iron Metabolism
4.2. NcRNAs Regulating Ferroptosis through Lipid Metabolism
4.3. NcRNAs Regulating Ferroptosis through Antioxidant Defense
4.4. NcRNAs Regulating Ferroptosis through Other Ferroptosis Regulators
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Estevao, D.; da Cruz-Ribeiro, M.; Cardoso, A.P.; Costa, A.M.; Oliveira, M.J.; Duarte, T.L.; da Cruz, T.B. Iron metabolism in colorectal cancer: A balancing act. Cell Oncol. 2023. [Google Scholar] [CrossRef]
- Li, Y.; Huang, X.; Wang, J.; Huang, R.; Wan, D. Regulation of Iron Homeostasis and Related Diseases. Mediat. Inflamm. 2020, 2020, 6062094. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascon, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Li, J.; Cao, F.; Yin, H.L.; Huang, Z.J.; Lin, Z.T.; Mao, N.; Sun, B.; Wang, G. Ferroptosis: Past, present and future. Cell Death Dis. 2020, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Shaghaghi, Z.; Motieian, S.; Alvandi, M.; Yazdi, A.; Asadzadeh, B.; Farzipour, S.; Abbasi, S. Ferroptosis Inhibitors as Potential New Therapeutic Targets for Cardiovascular Disease. Mini Rev. Med. Chem. 2022, 22, 2271–2286. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Hirschhorn, T.; Stockwell, B.R. The development of the concept of ferroptosis. Free Radic. Biol. Med. 2019, 133, 130–143. [Google Scholar] [CrossRef]
- Hu, W.; Liang, K.; Zhu, H.; Zhao, C.; Hu, H.; Yin, S. Ferroptosis and Its Role in Chronic Diseases. Cells 2022, 11, 2040. [Google Scholar] [CrossRef]
- Lei, G.; Zhuang, L.; Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer 2022, 22, 381–396. [Google Scholar] [CrossRef]
- Sun, Y.; Xia, X.; Basnet, D.; Zheng, J.C.; Huang, J.; Liu, J. Mechanisms of Ferroptosis and Emerging Links to the Pathology of Neurodegenerative Diseases. Front. Aging Neurosci. 2022, 14, 904152. [Google Scholar] [CrossRef]
- Van Coillie, S.; Van San, E.; Goetschalckx, I.; Wiernicki, B.; Mukhopadhyay, B.; Tonnus, W.; Choi, S.M.; Roelandt, R.; Dumitrascu, C.; Lamberts, L.; et al. Targeting ferroptosis protects against experimental (multi)organ dysfunction and death. Nat. Commun. 2022, 13, 1046. [Google Scholar] [CrossRef]
- Zhou, L.; Han, S.; Guo, J.; Qiu, T.; Zhou, J.; Shen, L. Ferroptosis-A New Dawn in the Treatment of Organ Ischemia-Reperfusion Injury. Cells 2022, 11, 3653. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P. The ENCODE (ENCyclopedia of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Seal, R.L.; Chen, L.L.; Griffiths-Jones, S.; Lowe, T.M.; Mathews, M.B.; O’Reilly, D.; Pierce, A.J.; Stadler, P.F.; Ulitsky, I.; Wolin, S.L.; et al. A guide to naming human non-coding RNA genes. EMBO J. 2020, 39, e103777. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 16, 20190027. [Google Scholar] [CrossRef]
- Frazer, D.M.; Anderson, G.J. The regulation of iron transport. BioFactors 2014, 40, 206–214. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Kang, R.; Tang, D. Iron Metabolism in Ferroptosis. Front. Cell Dev. Biol. 2020, 8, 590226. [Google Scholar] [CrossRef]
- Knutson, M.D. Steap proteins: Implications for iron and copper metabolism. Nutr. Rev. 2007, 65, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Xie, M.; Kang, R.; Fan, Y.; Niu, X.; Wang, H.; Cao, L.; Tang, D. HSPB1 as a novel regulator of ferroptotic cancer cell death. Oncogene 2015, 34, 5617–5625. [Google Scholar] [CrossRef]
- West, A.R.; Oates, P.S. Mechanisms of heme iron absorption: Current questions and controversies. World J. Gastroenterol. 2008, 14, 4101–4110. [Google Scholar] [CrossRef]
- Chiang, S.K.; Chen, S.E.; Chang, L.C. A Dual Role of Heme Oxygenase-1 in Cancer Cells. Int. J. Mol. Sci. 2019, 20, 39. [Google Scholar] [CrossRef]
- Consoli, V.; Sorrenti, V.; Pittala, V.; Greish, K.; D’Amico, A.G.; Romeo, G.; Intagliata, S.; Salerno, L.; Vanella, L. Heme Oxygenase Modulation Drives Ferroptosis in TNBC Cells. Int. J. Mol. Sci. 2022, 23, 5709. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Cheng, X.; Yuan, H.; Zhu, S.; Liu, J.; Wen, Q.; Xie, Y.; Liu, J.; Kroemer, G.; et al. PINK1 and PARK2 Suppress Pancreatic Tumorigenesis through Control of Mitochondrial Iron-Mediated Immunometabolism. Dev. Cell 2018, 46, 441–455.E8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, M.; Shen, M.; Kong, D.; Zhang, F.; Shao, J.; Tan, S.; Wang, S.; Chen, A.; Cao, P.; et al. The BRD7-P53-SLC25A28 axis regulates ferroptosis in hepatic stellate cells. Redox Biol. 2020, 36, 101619. [Google Scholar] [CrossRef]
- Hou, W.; Xie, Y.; Song, X.; Sun, X.; Lotze, M.T.; Zeh, H.J., 3rd; Kang, R.; Tang, D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016, 12, 1425–1428. [Google Scholar] [CrossRef]
- Li, W.; Li, W.; Wang, Y.; Leng, Y.; Xia, Z. Inhibition of DNMT-1 alleviates ferroptosis through NCOA4 mediated ferritinophagy during diabetes myocardial ischemia/reperfusion injury. Cell Death Discov. 2021, 7, 267. [Google Scholar] [CrossRef]
- Frey, A.G.; Nandal, A.; Park, J.H.; Smith, P.M.; Yabe, T.; Ryu, M.S.; Ghosh, M.C.; Lee, J.; Rouault, T.A.; Park, M.H.; et al. Iron chaperones PCBP1 and PCBP2 mediate the metallation of the dinuclear iron enzyme deoxyhypusine hydroxylase. Proc. Natl. Acad. Sci. USA 2014, 111, 8031–8036. [Google Scholar] [CrossRef]
- Protchenko, O.; Baratz, E.; Jadhav, S.; Li, F.; Shakoury-Elizeh, M.; Gavrilova, O.; Ghosh, M.C.; Cox, J.E.; Maschek, J.A.; Tyurin, V.A.; et al. Iron Chaperone Poly rC Binding Protein 1 Protects Mouse Liver from Lipid Peroxidation and Steatosis. Hepatology 2021, 73, 1176–1193. [Google Scholar] [CrossRef]
- Yanatori, I.; Richardson, D.R.; Imada, K.; Kishi, F. Iron Export through the Transporter Ferroportin 1 Is Modulated by the Iron Chaperone PCBP2. J. Biol. Chem. 2016, 291, 17303–17318. [Google Scholar] [CrossRef]
- Ward, D.M.; Kaplan, J. Ferroportin-mediated iron transport: Expression and regulation. Biochim. Biophys. Acta 2012, 1823, 1426–1433. [Google Scholar] [CrossRef]
- Nemeth, E.; Tuttle, M.S.; Powelson, J.; Vaughn, M.B.; Donovan, A.; Ward, D.M.; Ganz, T.; Kaplan, J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 2004, 306, 2090–2093. [Google Scholar] [CrossRef]
- Brown, C.W.; Amante, J.J.; Chhoy, P.; Elaimy, A.L.; Liu, H.; Zhu, L.J.; Baer, C.E.; Dixon, S.J.; Mercurio, A.M. Prominin2 Drives Ferroptosis Resistance by Stimulating Iron Export. Dev. Cell 2019, 51, 575–586.E4. [Google Scholar] [CrossRef]
- Brown, C.W.; Chhoy, P.; Mukhopadhyay, D.; Karner, E.R.; Mercurio, A.M. Targeting prominin2 transcription to overcome ferroptosis resistance in cancer. EMBO Mol. Med. 2021, 13, e13792. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Li, X.; Zhang, X.; Kang, R.; Tang, D. CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 2016, 478, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.W.; Sviderskiy, V.O.; Terzi, E.M.; Papagiannakopoulos, T.; Moreira, A.L.; Adams, S.; Sabatini, D.M.; Birsoy, K.; Possemato, R. NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 2017, 551, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Shin, D.; Lee, J.; Jung, A.R.; Roh, J.L. CISD2 inhibition overcomes resistance to sulfasalazine-induced ferroptotic cell death in head and neck cancer. Cancer Lett. 2018, 432, 180–190. [Google Scholar] [CrossRef]
- Rouault, T.A. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006, 2, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.L.; Ghosh, M.C.; Rouault, T.A. The physiological functions of iron regulatory proteins in iron homeostasis—an update. Front. Pharmacol. 2014, 5, 124. [Google Scholar] [CrossRef]
- Anderson, G.J.; Frazer, D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017, 106, 1559S–1566S. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, W.K.; Bae, K.H.; Lee, S.C.; Lee, E.W. Lipid Metabolism and Ferroptosis. Biology 2021, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Minikes, A.M.; Jiang, X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol. Cell 2022, 82, 2215–2227. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Shchepinov, M.S.; Pratt, D.A. Resolving the Role of Lipoxygenases in the Initiation and Execution of Ferroptosis. ACS Cent. Sci. 2018, 4, 387–396. [Google Scholar] [CrossRef]
- Lei, P.; Bai, T.; Sun, Y. Mechanisms of Ferroptosis and Relations with Regulated Cell Death: A Review. Front. Physiol. 2019, 10, 139. [Google Scholar] [CrossRef]
- Xiao, X.; Jiang, Y.; Liang, W.; Wang, Y.; Cao, S.; Yan, H.; Gao, L.; Zhang, L. miR-212-5p attenuates ferroptotic neuronal death after traumatic brain injury by targeting Ptgs2. Mol. Brain 2019, 12, 78. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, Y.; Li, K.; Yuan, D.; Yang, S.; Zhou, L.; Zhao, Y.; Miao, S.; Lv, C.; Zhao, J. COX-2/PGE2 Pathway Inhibits the Ferroptosis Induced by Cerebral Ischemia Reperfusion. Mol. Neurobiol. 2022, 59, 1619–1631. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, S.; Song, X.; Sun, X.; Fan, Y.; Liu, J.; Zhong, M.; Yuan, H.; Zhang, L.; Billiar, T.R.; et al. The Tumor Suppressor p53 Limits Ferroptosis by Blocking DPP4 Activity. Cell Rep. 2017, 20, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Ding, C.C.; Sun, T.; Rupprecht, G.; Lin, C.C.; Hsu, D.; Chi, J.T. The Hippo Pathway Effector TAZ Regulates Ferroptosis in Renal Cell Carcinoma. Cell Rep. 2019, 28, 2501–2508.e4. [Google Scholar] [CrossRef]
- Zou, Y.; Palte, M.J.; Deik, A.A.; Li, H.; Eaton, J.K.; Wang, W.; Tseng, Y.Y.; Deasy, R.; Kost-Alimova, M.; Dančík, V.; et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 2019, 10, 1617. [Google Scholar] [CrossRef]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Magtanong, L.; Ko, P.J.; To, M.; Cao, J.Y.; Forcina, G.C.; Tarangelo, A.; Ward, C.C.; Cho, K.; Patti, G.J.; Nomura, D.K.; et al. Exogenous Monounsaturated Fatty Acids Promote a Ferroptosis-Resistant Cell State. Cell Chem. Biol. 2019, 26, 420–432.e9. [Google Scholar] [CrossRef] [PubMed]
- Tesfay, L.; Paul, B.T.; Konstorum, A.; Deng, Z.; Cox, A.O.; Lee, J.; Furdui, C.M.; Hegde, P.; Torti, F.M.; Torti, S.V. Stearoyl-CoA Desaturase 1 Protects Ovarian Cancer Cells from Ferroptotic Cell Death. Cancer Res. 2019, 79, 5355–5366. [Google Scholar] [CrossRef]
- Liang, D.; Feng, Y.; Zandkarimi, F.; Wang, H.; Zhang, Z.; Kim, J.; Cai, Y.; Gu, W.; Stockwell, B.R.; Jiang, X. Ferroptosis surveillance independent of GPX4 and differentially regulated by sex hormones. Cell 2023, 186, 2748–2764.E22. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Nam, M.; Son, H.Y.; Hyun, K.; Jang, S.Y.; Kim, J.W.; Kim, M.W.; Jung, Y.; Jang, E.; Yoon, S.J.; et al. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proc. Natl. Acad. Sci. USA 2020, 117, 32433–32442. [Google Scholar] [CrossRef]
- Vriens, K.; Christen, S.; Parik, S.; Broekaert, D.; Yoshinaga, K.; Talebi, A.; Dehairs, J.; Escalona-Noguero, C.; Schmieder, R.; Cornfield, T.; et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 2019, 566, 403–406. [Google Scholar] [CrossRef]
- Ates, G.; Goldberg, J.; Currais, A.; Maher, P. CMS121, a fatty acid synthase inhibitor, protects against excess lipid peroxidation and inflammation and alleviates cognitive loss in a transgenic mouse model of Alzheimer’s disease. Redox Biol. 2020, 36, 101648. [Google Scholar] [CrossRef]
- Bartolacci, C.; Andreani, C.; Vale, G.; Berto, S.; Melegari, M.; Crouch, A.C.; Baluya, D.L.; Kemble, G.; Hodges, K.; Starrett, J.; et al. Targeting de novo lipogenesis and the Lands cycle induces ferroptosis in KRAS-mutant lung cancer. Nat. Commun. 2022, 13, 4327. [Google Scholar] [CrossRef]
- Yi, J.; Zhu, J.; Wu, J.; Thompson, C.B.; Jiang, X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 31189–31197. [Google Scholar] [CrossRef]
- Nagarajan, S.R.; Butler, L.M.; Hoy, A.J. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metab. 2021, 9, 2. [Google Scholar] [CrossRef]
- Veglia, F.; Tyurin, V.A.; Blasi, M.; De Leo, A.; Kossenkov, A.V.; Donthireddy, L.; To, T.K.J.; Schug, Z.; Basu, S.; Wang, F.; et al. Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature 2019, 569, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Xiao, L.; Liu, L.; Ye, L.; Su, P.; Bi, E.; Wang, Q.; Yang, M.; Qian, J.; Yi, Q. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab. 2021, 33, 1001–1012.e5. [Google Scholar] [CrossRef]
- Yang, M.; Chen, P.; Liu, J.; Zhu, S.; Kroemer, G.; Klionsky, D.J.; Lotze, M.T.; Zeh, H.J.; Kang, R.; Tang, D. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci. Adv. 2019, 5, eaaw2238. [Google Scholar] [CrossRef]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Pratt, D.A. The chemical basis of ferroptosis. Nat. Chem. Biol. 2019, 15, 1137–1147. [Google Scholar] [CrossRef]
- Shimada, K.; Skouta, R.; Kaplan, A.; Yang, W.S.; Hayano, M.; Dixon, S.J.; Brown, L.M.; Valenzuela, C.A.; Wolpaw, A.J.; Stockwell, B.R. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat. Chem. Biol. 2016, 12, 497–503. [Google Scholar] [CrossRef]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Porto Freitas, F.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422.e21. [Google Scholar] [CrossRef]
- Xuan, Y.; Wang, H.; Yung, M.M.; Chen, F.; Chan, W.S.; Chan, Y.S.; Tsui, S.K.; Ngan, H.Y.; Chan, K.K.; Chan, D.W. SCD1/FADS2 fatty acid desaturases equipoise lipid metabolic activity and redox-driven ferroptosis in ascites-derived ovarian cancer cells. Theranostics 2022, 12, 3534–3552. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S.; Wang, S.F.; Hsu, C.Y.; Yin, P.H.; Yeh, T.S.; Lee, H.C.; Tseng, L.M. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2alpha-ATF4 pathway. Oncotarget 2017, 8, 114588–114602. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, D.; Fu, Q.; Hao, S.; Gu, Y.; Zhao, W.; Chen, S.; Sheng, F.; Xu, Y.; Chen, Z.; et al. CHAC1 as a Novel Contributor of Ferroptosis in Retinal Pigment Epithelial Cells with Oxidative Damage. Int. J. Mol. Sci. 2023, 24, 1582. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef]
- Liu, L.; Liu, R.; Liu, Y.; Li, G.; Chen, Q.; Liu, X.; Ma, S. Cystine-glutamate antiporter xCT as a therapeutic target for cancer. Cell Biochem. Funct. 2021, 39, 174–179. [Google Scholar] [CrossRef]
- Tu, H.; Tang, L.J.; Luo, X.J.; Ai, K.L.; Peng, J. Insights into the novel function of system Xc- in regulated cell death. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1650–1662. [Google Scholar] [CrossRef]
- Liu, M.R.; Zhu, W.T.; Pei, D.S. System Xc(-): A key regulatory target of ferroptosis in cancer. Investig. New Drugs 2021, 39, 1123–1131. [Google Scholar] [CrossRef]
- Hayano, M.; Yang, W.S.; Corn, C.K.; Pagano, N.C.; Stockwell, B.R. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016, 23, 270–278. [Google Scholar] [CrossRef]
- Shimada, K.; Stockwell, B.R. tRNA synthase suppression activates de novo cysteine synthesis to compensate for cystine and glutathione deprivation during ferroptosis. Mol. Cell Oncol. 2016, 3, e1091059. [Google Scholar] [CrossRef][Green Version]
- Liu, N.; Lin, X.; Huang, C. Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br. J. Cancer 2020, 122, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cai, H.; Hu, Y.; Liu, F.; Huang, S.; Zhou, Y.; Yu, J.; Xu, J.; Wu, F. A pharmacological probe identifies cystathionine beta-synthase as a new negative regulator for ferroptosis. Cell Death Dis. 2018, 9, 1005. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Monian, P.; Quadri, N.; Ramasamy, R.; Jiang, X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol. Cell 2015, 59, 298–308. [Google Scholar] [CrossRef] [PubMed]
- McGivan, J.D.; Bungard, C.I. The transport of glutamine into mammalian cells. Front. Biosci. 2007, 12, 874–882. [Google Scholar] [CrossRef]
- Liu, L.; Su, S.; Ye, D.; Yu, Z.; Lu, W.; Li, X. Long non-coding RNA OGFRP1 regulates cell proliferation and ferroptosis by miR-299-3p/SLC38A1 axis in lung cancer. Anticancer Drugs 2022, 33, 826–839. [Google Scholar] [CrossRef]
- Venkateswaran, G.; McDonald, P.C.; Chafe, S.C.; Brown, W.S.; Gerbec, Z.J.; Awrey, S.J.; Parker, S.J.; Dedhar, S. A Carbonic Anhydrase IX/SLC1A5 axis regulates glutamine metabolism dependent ferroptosis in hypoxic tumor cells. Mol. Cancer Ther. 2023, MCT-23-0041. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, C.; Dong, X.; Wang, H. A novel miR-338-3p/SLC1A5 axis reprograms retinal pigment epithelium to increases its resistance to high glucose-induced cell ferroptosis. J. Mol. Histol. 2022, 53, 561–571. [Google Scholar] [CrossRef]
- Kremer, D.M.; Nelson, B.S.; Lin, L.; Yarosz, E.L.; Halbrook, C.J.; Kerk, S.A.; Sajjakulnukit, P.; Myers, A.; Thurston, G.; Hou, S.W.; et al. GOT1 inhibition promotes pancreatic cancer cell death by ferroptosis. Nat. Commun. 2021, 12, 4860. [Google Scholar] [CrossRef]
- Luo, M.; Wu, L.; Zhang, K.; Wang, H.; Zhang, T.; Gutierrez, L.; O’Connell, D.; Zhang, P.; Li, Y.; Gao, T.; et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ. 2018, 25, 1457–1472. [Google Scholar] [CrossRef]
- Xu, F.; Wang, H.; Pei, H.; Zhang, Z.; Liu, L.; Tang, L.; Wang, S.; Ren, B.C. SLC1A5 Prefers to Play as an Accomplice Rather Than an Opponent in Pancreatic Adenocarcinoma. Front. Cell Dev. Biol. 2022, 10, 800925. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, P.; Liu, N.; Yang, J.Y.; Wang, H.M.; Geng, Q. TRIM6 Reduces Ferroptosis and Chemosensitivity by Targeting SLC1A5 in Lung Cancer. Oxid. Med. Cell Longev. 2023, 2023, 9808100. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tai, W.; Lu, N.; Li, T.; Liu, Y.; Wu, W.; Li, Z.; Pu, L.; Zhao, X.; Zhang, T.; et al. lncRNA ZFAS1 promotes lung fibroblast-to-myofibroblast transition and ferroptosis via functioning as a ceRNA through miR-150-5p/SLC38A1 axis. Aging 2020, 12, 9085–9102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhuo, M.; Zhang, Y.; Shi, E.; Ma, X.; Li, H. miR-190a-5p regulates cardiomyocytes response to ferroptosis via directly targeting GLS2. Biochem. Biophys. Res. Commun. 2021, 566, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Bai, R.; Liu, Y.; Bi, H.; Shi, X.; Qu, C. Long non-coding RNA ATXN8OS promotes ferroptosis and inhibits the temozolomide-resistance of gliomas through the ADAR/GLS2 pathway. Brain Res. Bull. 2022, 186, 27–37. [Google Scholar] [CrossRef]
- Wu, F.; Shang, C.; Jin, T.; Shi, L. Hispidin Inhibits Ferroptosis Induced by High Glucose via the miR-15b-5p/GLS2 Axis in Pancreatic Beta Cells. Evid. Based Complement. Altern. Med. 2023, 2023, 9428241. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, C.; Zhang, M.; Liu, Y.; Wang, L.; Xie, Y.; Qi, H.; Ba, L.; Shi, P.; Cao, Y.; et al. Super-enhancer-driven lncRNA Snhg7 aggravates cardiac hypertrophy via Tbx5/GLS2/ferroptosis axis. Eur. J. Pharmacol. 2023, 953, 175822. [Google Scholar] [CrossRef]
- Shin, D.; Lee, J.; You, J.H.; Kim, D.; Roh, J.L. Dihydrolipoamide dehydrogenase regulates cystine deprivation-induced ferroptosis in head and neck cancer. Redox Biol. 2020, 30, 101418. [Google Scholar] [CrossRef]
- Suzuki, S.; Venkatesh, D.; Kanda, H.; Nakayama, A.; Hosokawa, H.; Lee, E.; Miki, T.; Stockwell, B.R.; Yokote, K.; Tanaka, T.; et al. GLS2 Is a Tumor Suppressor and a Regulator of Ferroptosis in Hepatocellular Carcinoma. Cancer Res. 2022, 82, 3209–3222. [Google Scholar] [CrossRef]
- Bersuker, K.; Hendricks, J.M.; Li, Z.; Magtanong, L.; Ford, B.; Tang, P.H.; Roberts, M.A.; Tong, B.; Maimone, T.J.; Zoncu, R.; et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019, 575, 688–692. [Google Scholar] [CrossRef]
- Doll, S.; Freitas, F.P.; Shah, R.; Aldrovandi, M.; da Silva, M.C.; Ingold, I.; Goya Grocin, A.; Xavier da Silva, T.N.; Panzilius, E.; Scheel, C.H.; et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 2019, 575, 693–698. [Google Scholar] [CrossRef]
- Dai, E.; Meng, L.; Kang, R.; Wang, X.; Tang, D. ESCRT-III-dependent membrane repair blocks ferroptosis. Biochem. Biophys. Res. Commun. 2020, 522, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Kraft, V.A.N.; Bezjian, C.T.; Pfeiffer, S.; Ringelstetter, L.; Muller, C.; Zandkarimi, F.; Merl-Pham, J.; Bao, X.; Anastasov, N.; Kossl, J.; et al. GTP Cyclohydrolase 1/Tetrahydrobiopterin Counteract Ferroptosis through Lipid Remodeling. ACS Cent. Sci. 2020, 6, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Soula, M.; Weber, R.A.; Zilka, O.; Alwaseem, H.; La, K.; Yen, F.; Molina, H.; Garcia-Bermudez, J.; Pratt, D.A.; Birsoy, K. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat. Chem. Biol. 2020, 16, 1351–1360. [Google Scholar] [CrossRef]
- Mao, C.; Liu, X.; Zhang, Y.; Lei, G.; Yan, Y.; Lee, H.; Koppula, P.; Wu, S.; Zhuang, L.; Fang, B.; et al. Author Correction: DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature 2021, 596, E13. [Google Scholar] [CrossRef] [PubMed]
- Lovatt, M.; Adnan, K.; Kocaba, V.; Dirisamer, M.; Peh, G.S.L.; Mehta, J.S. Peroxiredoxin-1 regulates lipid peroxidation in corneal endothelial cells. Redox Biol. 2020, 30, 101417. [Google Scholar] [CrossRef]
- Qi, W.; Li, Z.; Xia, L.; Dai, J.; Zhang, Q.; Wu, C.; Xu, S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci. Rep. 2019, 9, 16185. [Google Scholar] [CrossRef]
- Lu, B.; Chen, X.B.; Hong, Y.C.; Zhu, H.; He, Q.J.; Yang, B.; Ying, M.D.; Cao, J. Identification of PRDX6 as a regulator of ferroptosis. Acta Pharmacol. Sin. 2019, 40, 1334–1342. [Google Scholar] [CrossRef]
- Liu, D.; Liang, C.H.; Huang, B.; Zhuang, X.; Cui, W.; Yang, L.; Yang, Y.; Zhang, Y.; Fu, X.; Zhang, X.; et al. Tryptophan Metabolism Acts as a New Anti-Ferroptotic Pathway to Mediate Tumor Growth. Adv. Sci. 2023, 10, e2204006. [Google Scholar] [CrossRef]
- Anandhan, A.; Dodson, M.; Schmidlin, C.J.; Liu, P.; Zhang, D.D. Breakdown of an Ironclad Defense System: The Critical Role of NRF2 in Mediating Ferroptosis. Cell Chem. Biol. 2020, 27, 436–447. [Google Scholar] [CrossRef]
- Shakya, A.; McKee, N.W.; Dodson, M.; Chapman, E.; Zhang, D.D. Anti-Ferroptotic Effects of Nrf2: Beyond the Antioxidant Response. Mol. Cells 2023, 46, 165–175. [Google Scholar] [CrossRef]
- Shimano, H.; Sato, R. SREBP-regulated lipid metabolism: Convergent physiology—Divergent pathophysiology. Nat. Rev. Endocrinol. 2017, 13, 710–730. [Google Scholar] [CrossRef] [PubMed]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef]
- Brown, C.W.; Amante, J.J.; Goel, H.L.; Mercurio, A.M. The alpha6beta4 integrin promotes resistance to ferroptosis. J. Cell Biol. 2017, 216, 4287–4297. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, K. The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin. Cell Biol. Int. 2019, 43, 1245–1256. [Google Scholar] [CrossRef]
- Qiang, Z.; Dong, H.; Xia, Y.; Chai, D.; Hu, R.; Jiang, H. Nrf2 and STAT3 Alleviates Ferroptosis-Mediated IIR-ALI by Regulating SLC7A11. Oxid. Med. Cell Longev. 2020, 2020, 5146982. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, H.; Yang, X.; Wu, Q.; An, P.; Jin, X.; Liu, W.; Huang, X.; Li, Y.; Yan, S.; et al. Auranofin mitigates systemic iron overload and induces ferroptosis via distinct mechanisms. Signal Transduct. Target. Ther. 2020, 5, 138. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wang, J.; Hu, W.; Feng, Z. The Regulation of Ferroptosis by Tumor Suppressor p53 and its Pathway. Int. J. Mol. Sci. 2020, 21, 8387. [Google Scholar] [CrossRef]
- Xu, R.; Wang, W.; Zhang, W. Ferroptosis and the bidirectional regulatory factor p53. Cell Death Discov. 2023, 9, 197. [Google Scholar] [CrossRef]
- Zheng, X.; Liang, Y.; Zhang, C. Ferroptosis Regulated by Hypoxia in Cells. Cells 2023, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yan, J.; Zhao, Q.; Zhang, Y.; Zhang, Y. ATF3 promotes ferroptosis in sorafenib-induced cardiotoxicity by suppressing Slc7a11 expression. Front. Pharmacol. 2022, 13, 904314. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Y.; Du, T.; Yang, H.; Lei, L.; Guo, M.; Ding, H.F.; Zhang, J.; Wang, H.; Chen, X.; et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc. Cell Death Differ. 2020, 27, 662–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Fan, Z.; Rauh, M.; Buchfelder, M.; Eyupoglu, I.Y.; Savaskan, N. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene 2017, 36, 5593–5608. [Google Scholar] [CrossRef]
- Chen, D.; Rauh, M.; Buchfelder, M.; Eyupoglu, I.Y.; Savaskan, N. The oxido-metabolic driver ATF4 enhances temozolamide chemo-resistance in human gliomas. Oncotarget 2017, 8, 51164–51176. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Zhang, P.; Liu, J.; Wang, R.; Kaufman, R.J.; Yaden, B.C.; Karin, M. ATF4 suppresses hepatocarcinogenesis by inducing SLC7A11 (xCT) to block stress-related ferroptosis. J. Hepatol. 2023, 79, 362–377. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, X.Z.; He, C.; Wang, L.; Liang, S.P.; Wang, C.C.; Li, C.; Luo, T.F.; Feng, C.S.; Wang, Z.C.; et al. ATF3 contributes to brucine-triggered glioma cell ferroptosis via promotion of hydrogen peroxide and iron. Acta Pharmacol. Sin. 2021, 42, 1690–1702. [Google Scholar] [CrossRef]
- Liu, H.; Mo, H.; Yang, C.; Mei, X.; Song, X.; Lu, W.; Xiao, H.; Yan, J.; Wang, X.; Yan, J.; et al. A novel function of ATF3 in suppression of ferroptosis in mouse heart suffered ischemia/reperfusion. Free Radic. Biol. Med. 2022, 189, 122–135. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, Q.; Yu, M.; Liu, Y.; Teng, X.; Gu, X. 4-tert-butylphenol triggers common carp hepatocytes ferroptosis via oxidative stress, iron overload, SLC7A11/GSH/GPX4 axis, and ATF4/HSPA5/GPX4 axis. Ecotoxicol. Environ. Saf. 2022, 242, 113944. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, Q.; Sun, X.; Zeh, H.J., 3rd; Lotze, M.T.; Kang, R.; Tang, D. HSPA5 Regulates Ferroptotic Cell Death in Cancer Cells. Cancer Res. 2017, 77, 2064–2077. [Google Scholar] [CrossRef]
- Chen, J.; Zaal, E.A.; Berkers, C.R.; Ruijtenbeek, R.; Garssen, J.; Redegeld, F.A. Omega-3 Fatty Acids DHA and EPA Reduce Bortezomib Resistance in Multiple Myeloma Cells by Promoting Glutathione Degradation. Cells 2021, 10, 2287. [Google Scholar] [CrossRef]
- Wang, N.; Zeng, G.Z.; Yin, J.L.; Bian, Z.X. Artesunate activates the ATF4-CHOP-CHAC1 pathway and affects ferroptosis in Burkitt’s Lymphoma. Biochem. Biophys. Res. Commun. 2019, 519, 533–539. [Google Scholar] [CrossRef]
- Liang, Y.; Chen, S.; Han, S.; Luo, L.; Shen, F.; Huang, Z. Toosendanin induced hepatotoxicity via triggering PERK-eIF2alpha-ATF4 mediated ferroptosis. Toxicol. Lett. 2023, 377, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wu, Z.; Chen, J.; Guo, S.; You, W.; Wang, S.; Ma, J.; Wang, H.; Wang, X.; Wang, H.; et al. Nanoparticle delivery of miR-21-3p sensitizes melanoma to anti-PD-1 immunotherapy by promoting ferroptosis. J. Immunother. Cancer 2022, 10, e004381. [Google Scholar] [CrossRef]
- Cho, I.J.; Kim, D.; Kim, E.O.; Jegal, K.H.; Kim, J.K.; Park, S.M.; Zhao, R.; Ki, S.H.; Kim, S.C.; Ku, S.K. Cystine and Methionine Deficiency Promotes Ferroptosis by Inducing B-Cell Translocation Gene 1. Antioxid. 2021, 10, 1543. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Lam, M.T.; Glass, C.K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011, 90, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Green, D.; Dalmay, T.; Chapman, T. Microguards and micromessengers of the genome. Heredity 2016, 116, 125–134. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Lu, J.; Xu, F.; Lu, H. LncRNA PVT1 regulates ferroptosis through miR-214-mediated TFR1 and p53. Life Sci. 2020, 260, 118305. [Google Scholar] [CrossRef]
- Ma, L.L.; Liang, L.; Zhou, D.; Wang, S.W. Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma 2021, 68, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Ou, R.; Lu, S.; Wang, L.; Wang, Y.; Lv, M.; Li, T.; Xu, Y.; Lu, J.; Ge, R.S. Circular RNA circLMO1 Suppresses Cervical Cancer Growth and Metastasis by Triggering miR-4291/ACSL4-Mediated Ferroptosis. Front. Oncol. 2022, 12, 858598. [Google Scholar] [CrossRef]
- Bao, W.D.; Zhou, X.T.; Zhou, L.T.; Wang, F.; Yin, X.; Lu, Y.; Zhu, L.Q.; Liu, D. Targeting miR-124/Ferroportin signaling ameliorated neuronal cell death through inhibiting apoptosis and ferroptosis in aged intracerebral hemorrhage murine model. Aging Cell 2020, 19, e13235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, Y.; Wang, J.X.; Chen, M.H.; Xu, J.J.; Jiang, M.H.; Feng, Y.L.; Gu, Y.F. miR-30-5p-mediated ferroptosis of trophoblasts is implicated in the pathogenesis of preeclampsia. Redox Biol. 2020, 29, 101402. [Google Scholar] [CrossRef]
- Xu, Q.; Zhou, L.; Yang, G.; Meng, F.; Wan, Y.; Wang, L.; Zhang, L. CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biol. Int. 2020, 44, 2344–2356. [Google Scholar] [CrossRef]
- Hou, Y.; Cai, S.; Yu, S.; Lin, H. Metformin induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast cancer. Acta Biochim. Biophys. Sin 2021, 53, 333–341. [Google Scholar] [CrossRef]
- Yu, R.; Zhou, Y.; Shi, S.; Wang, X.; Huang, S.; Ren, Y. Icariside II induces ferroptosis in renal cell carcinoma cells by regulating the miR-324-3p/GPX4 axis. Phytomedicine 2022, 102, 154182. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, S.; Wang, S.; Li, X.; Hou, D.; Li, H.; Wang, L.; Xu, Y.; Ma, B.; Wang, H.; et al. LncRNA OIP5-AS1 inhibits ferroptosis in prostate cancer with long-term cadmium exposure through miR-128-3p/SLC7A11 signaling. Ecotoxicol. Environ. Saf. 2021, 220, 112376. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, N.; Ling, X.; Pan, M.; Du, W.; Gao, S. MiR-27a-3p Promotes Non-Small Cell Lung Cancer Through SLC7A11-Mediated-Ferroptosis. Front. Oncol. 2021, 11, 759346. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Qin, H.; Sun, C.; Liu, Y.; Ruan, G.; Guo, Q.; Xi, T.; Xing, Y.; Zheng, L. MiR-375 reduces the stemness of gastric cancer cells through triggering ferroptosis. Stem Cell Res. Ther. 2021, 12, 325. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Smillie, C.L.; Sirey, T.; Ponting, C.P. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 231–245. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Perkel, J.M. Visiting “noncodarnia”. Biotechniques 2013, 54, 301, 303–304. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Song, J.; Gao, Y.; Huang, S.; Dou, R.; Zhong, P.; Huang, G.; Han, L.; Zheng, J.; Zhang, X.; et al. Hypoxia-induced HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol. 2022, 52, 102312. [Google Scholar] [CrossRef]
- Zhang, B.; Bao, W.; Zhang, S.; Chen, B.; Zhou, X.; Zhao, J.; Shi, Z.; Zhang, T.; Chen, Z.; Wang, L.; et al. LncRNA HEPFAL accelerates ferroptosis in hepatocellular carcinoma by regulating SLC7A11 ubiquitination. Cell Death Dis. 2022, 13, 734. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Wang, X.; Liu, Y.; Wang, M.; Yan, B.; Jiang, Y.; Shi, Y.; Shen, Y.; Liu, X.; Lai, W.; et al. A G3BP1-Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. 2018, 78, 3484–3496. [Google Scholar] [CrossRef] [PubMed]
- Dawoud, A.; Ihab Zakaria, Z.; Hisham Rashwan, H.; Braoudaki, M.; Youness, R.A. Circular RNAs: New layer of complexity evading breast cancer heterogeneity. Noncoding RNA Res. 2023, 8, 60–74. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Nisar, S.; Bhat, A.A.; Singh, M.; Karedath, T.; Rizwan, A.; Hashem, S.; Bagga, P.; Reddy, R.; Jamal, F.; Uddin, S.; et al. Insights into the Role of CircRNAs: Biogenesis, Characterization, Functional, and Clinical Impact in Human Malignancies. Front. Cell Dev. Biol. 2021, 9, 617281. [Google Scholar] [CrossRef]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef]
- Ren, S.; Lin, P.; Wang, J.; Yu, H.; Lv, T.; Sun, L.; Du, G. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol. Ther. Methods Clin. Dev. 2020, 18, 215–229. [Google Scholar] [CrossRef]
- Wei, G.; Zhu, J.; Hu, H.B.; Liu, J.Q. Circular RNAs: Promising biomarkers for cancer diagnosis and prognosis. Gene 2021, 771, 145365. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.J.; Culver, H.R.; Prieto, T.R.; Martinez, P.J.; Sinha, J.; Bryant, S.J.; Bowman, C.N. Messenger RNA enrichment using synthetic oligo(T) click nucleic acids. Chem. Commun. 2020, 56, 13987–13990. [Google Scholar] [CrossRef] [PubMed]
- Jie, M.; Wu, Y.; Gao, M.; Li, X.; Liu, C.; Ouyang, Q.; Tang, Q.; Shan, C.; Lv, Y.; Zhang, K.; et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. Cancer 2020, 19, 56. [Google Scholar] [CrossRef]
- Prats, A.C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, 8591. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, Q.; Wang, X.; Xu, Z.; Wei, X.; Li, J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020, 6, 72. [Google Scholar] [CrossRef]
- Wang, W.; Xu, R.; Zhao, H.; Xiong, Y.; He, P. CircEXOC5 promotes ferroptosis by enhancing ACSL4 mRNA stability via binding to PTBP1 in sepsis-induced acute lung injury. Immunobiology 2022, 227, 152219. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, S.; Wei, S.; Cheng, S.; Shi, R.; Zhao, R.; Zhang, W.; Zhang, Q.; Hua, T.; Feng, D.; et al. CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox Biol. 2022, 57, 102493. [Google Scholar] [CrossRef]
- Wang, L.; Wu, S.; He, H.; Ai, K.; Xu, R.; Zhang, L.; Zhu, X. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab. Investig. 2022, 102, 1323–1334. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhao, J.; Li, R.; Liu, Y.; Zhou, L.; Wang, C.; Lv, C.; Gao, L.; Cui, D. CircLRFN5 inhibits the progression of glioblastoma via PRRX2/GCH1 mediated ferroptosis. J. Exp. Clin. Cancer Res. 2022, 41, 307. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Ma, L.; Yu, K.; Niu, Y.; Xu, X.; Shi, Y.; Guo, S.; Xue, X.; Wang, Y.; et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun. 2022, 42, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Halbach, R.; Miesen, P.; Joosten, J.; Taskopru, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Ye, Y.; Li, M.; Pan, L.; Su, J.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Meng, X.; Pan, C.; Qu, F.; Gan, W.; Xiang, Z.; Han, X.; Li, D. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cell Signal 2020, 67, 109501. [Google Scholar] [CrossRef]
- Tan, X.; Huang, X.; Niu, B.; Guo, X.; Lei, X.; Qu, B. Targeting GSTP1-dependent ferroptosis in lung cancer radiotherapy: Existing evidence and future directions. Front. Mol. Biosci. 2022, 9, 1102158. [Google Scholar] [CrossRef]
- Sharp, S.J.; Schaack, J.; Cooley, L.; Burke, D.J.; Soll, D. Structure and transcription of eukaryotic tRNA genes. CRC Crit. Rev. Biochem. 1985, 19, 107–144. [Google Scholar] [CrossRef]
- Becker, N.P.; Martitz, J.; Renko, K.; Stoedter, M.; Hybsier, S.; Cramer, T.; Schomburg, L. Hypoxia reduces and redirects selenoprotein biosynthesis. Metallomics 2014, 6, 1079–1086. [Google Scholar] [CrossRef]
- De Spirt, S.; Eckers, A.; Wehrend, C.; Micoogullari, M.; Sies, H.; Stahl, W.; Steinbrenner, H. Interplay between the chalcone cardamonin and selenium in the biosynthesis of Nrf2-regulated antioxidant enzymes in intestinal Caco-2 cells. Free Radic. Biol. Med. 2016, 91, 164–171. [Google Scholar] [CrossRef]
- Kipp, A.P.; Frombach, J.; Deubel, S.; Brigelius-Flohe, R. Selenoprotein W as biomarker for the efficacy of selenium compounds to act as source for selenoprotein biosynthesis. Methods Enzym. 2013, 527, 87–112. [Google Scholar] [CrossRef]
- Pathak, C.; Jaiswal, Y.K.; Vinayak, M. Queuine promotes antioxidant defence system by activating cellular antioxidant enzyme activities in cancer. Biosci. Rep. 2008, 28, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Hiramoto, K.; Satoh, H.; Suzuki, T.; Moriguchi, T.; Pi, J.; Shimosegawa, T.; Yamamoto, M. Myeloid lineage-specific deletion of antioxidant system enhances tumor metastasis. Cancer Prev. Res. 2014, 7, 835–844. [Google Scholar] [CrossRef]
- Aubert, M.; O’Donohue, M.F.; Lebaron, S.; Gleizes, P.E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules 2018, 8, 123. [Google Scholar] [CrossRef]
- Li, W.; Thakor, N.; Xu, E.Y.; Huang, Y.; Chen, C.; Yu, R.; Holcik, M.; Kong, A.N. An internal ribosomal entry site mediates redox-sensitive translation of Nrf2. Nucleic Acids Res. 2010, 38, 778–788. [Google Scholar] [CrossRef]
- Perez, M.J.; Gonzalez-Sanchez, E.; Gonzalez-Loyola, A.; Gonzalez-Buitrago, J.M.; Marin, J.J. Mitochondrial genome depletion dysregulates bile acid- and paracetamol-induced expression of the transporters Mdr1, Mrp1 and Mrp4 in liver cells. Br. J. Pharmacol. 2011, 162, 1686–1699. [Google Scholar] [CrossRef]
- Jayaraman, S.; Chittiboyina, S.; Bai, Y.; Abad, P.C.; Vidi, P.A.; Stauffacher, C.V.; Lelievre, S.A. The nuclear mitotic apparatus protein NuMA controls rDNA transcription and mediates the nucleolar stress response in a p53-independent manner. Nucleic Acids Res. 2017, 45, 11725–11742. [Google Scholar] [CrossRef]
- Yung, B.Y.; Yang, Y.H.; Bor, A.M. Nucleolar protein B23 translocation after deferoxamine treatment in a human leukemia cell line. Int. J. Cancer 1991, 48, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Hu, L.; Song, Q.; Shan, Y.; Yin, G.; Zhu, H.; Kong, W.; Zhou, C. miR-545 promotes colorectal cancer by inhibiting transferring in the non-normal ferroptosis signaling. Aging 2021, 13, 26137–26147. [Google Scholar] [CrossRef]
- Huang, J.; Deng, C.; Guo, T.; Chen, X.; Chen, P.; Du, S.; Lu, M. Cinobufotalin Induces Ferroptosis to Suppress Lung Cancer Cell Growth by lncRNA LINC00597/hsa-miR-367-3p/TFRC Pathway via Resibufogenin. Anticancer. Agents Med. Chem. 2023, 23, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Lei, D.; Li, B.; Isa, Z.; Ma, X.; Zhang, B. Hypoxia-elicited cardiac microvascular endothelial cell-derived exosomal miR-210-3p alleviate hypoxia/reoxygenation-induced myocardial cell injury through inhibiting transferrin receptor 1-mediated ferroptosis. Tissue Cell 2022, 79, 101956. [Google Scholar] [CrossRef]
- Wu, L.; Tian, X.; Zuo, H.; Zheng, W.; Li, X.; Yuan, M.; Tian, X.; Song, H. miR-124-3p delivered by exosomes from heme oxygenase-1 modified bone marrow mesenchymal stem cells inhibits ferroptosis to attenuate ischemia-reperfusion injury in steatotic grafts. J. Nanobiotechnol. 2022, 20, 196. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gu, Q.; Chen, R. Promotive role of IRF7 in ferroptosis of colonic epithelial cells in ulcerative colitis by the miR-375-3p/SLC11A2 axis. Biomol. Biomed. 2023, 23, 437–449. [Google Scholar] [CrossRef]
- Song, Y.; Wang, B.; Zhu, X.; Hu, J.; Sun, J.; Xuan, J.; Ge, Z. Human umbilical cord blood-derived MSCs exosome attenuate myocardial injury by inhibiting ferroptosis in acute myocardial infarction mice. Cell Biol. Toxicol. 2021, 37, 51–64. [Google Scholar] [CrossRef]
- Tomita, K.; Fukumoto, M.; Itoh, K.; Kuwahara, Y.; Igarashi, K.; Nagasawa, T.; Suzuki, M.; Kurimasa, A.; Sato, T. MiR-7-5p is a key factor that controls radioresistance via intracellular Fe(2+) content in clinically relevant radioresistant cells. Biochem. Biophys. Res. Commun. 2019, 518, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Ke, Y.Q.; Duan, P.; Zhou, L.; Wang, C.Y.; Cao, P. MicroRNA-302a-3p induces ferroptosis of non-small cell lung cancer cells via targeting ferroportin. Free Radic. Res. 2021, 55, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Song, Z.; Chen, Z.; Lin, T.; Lin, H.; Xu, Z.; Ai, F.; Zheng, S. MicroRNA-4735-3p Facilitates Ferroptosis in Clear Cell Renal Cell Carcinoma by Targeting SLC40A1. Anal. Cell. Pathol. 2022, 2022, 4213401. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Ge, F.H.; Li, W.X.; Xu, Z.; Wang, X.L.; Shen, J.L.; Xu, A.B.; Hao, R.R. MicroRNA-147a Targets SLC40A1 to Induce Ferroptosis in Human Glioblastoma. Anal. Cell. Pathol. 2022, 2022, 2843990. [Google Scholar] [CrossRef]
- Zheng, R.; Lin, C.; Mao, Y.; Jin, F. miR-761-hepcidin/Gpx4 pathway contribute to unexplained liver dysfunction in polycystic ovary syndrome by regulating liver iron overload and ferroptosis. Gynecol. Endocrinol. 2023, 39, 2166483. [Google Scholar] [CrossRef]
- Xiang, L.; Zeng, Q.; Liu, J.; Xiao, M.; He, D.; Zhang, Q.; Xie, D.; Deng, M.; Zhu, Y.; Liu, Y.; et al. MAFG-AS1/MAFG positive feedback loop contributes to cisplatin resistance in bladder urothelial carcinoma through antagonistic ferroptosis. Sci. Bull. 2021, 66, 1773–1788. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wang, J.; Xu, W.; Ma, C.; Wan, F.; Huang, Y.; Yao, M.; Zhang, H.; Qu, Y.; Ye, D.; et al. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis. 2021, 12, 1043. [Google Scholar] [CrossRef]
- Li, X.; Si, W.; Li, Z.; Tian, Y.; Liu, X.; Ye, S.; Huang, Z.; Ji, Y.; Zhao, C.; Hao, X.; et al. miR-335 promotes ferroptosis by targeting ferritin heavy chain 1 in in vivo and in vitro models of Parkinson’s disease. Int. J. Mol. Med. 2021, 47, 61. [Google Scholar] [CrossRef]
- Zheng, H.; Shi, L.; Tong, C.; Liu, Y.; Hou, M. circSnx12 Is Involved in Ferroptosis During Heart Failure by Targeting miR-224-5p. Front. Cardiovasc. Med. 2021, 8, 656093. [Google Scholar] [CrossRef]
- Zhang, R.; Pan, T.; Xiang, Y.; Zhang, M.; Xie, H.; Liang, Z.; Chen, B.; Xu, C.; Wang, J.; Huang, X.; et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact. Mater. 2022, 13, 23–36. [Google Scholar] [CrossRef]
- Gong, H.; Gao, M.; Lin, Y.; Liu, J.; Hu, Z.; Liu, J. TUG1/MAZ/FTH1 Axis Attenuates the Antiglioma Effect of Dihydroartemisinin by Inhibiting Ferroptosis. Oxid. Med. Cell Longev. 2022, 2022, 7843863. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Gao, H.; Zhou, W.; Cai, H.; Liao, L.; Wang, C. Circular RNA HIPK3 facilitates ferroptosis in gestational diabetes mellitus by regulating glutathione peroxidase 4 DNA methylation. J. Gene Med. 2023, e3526. [Google Scholar] [CrossRef]
- Qiu, X.; Shi, Q.; Zhang, X.; Shi, X.; Jiang, H.; Qin, S. LncRNA A2M-AS1 Promotes Ferroptosis in Pancreatic Cancer via Interacting with PCBP3. Mol. Cancer Res. 2022, 20, 1636–1645. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Ai, R.; Mu, S.; Niu, X.; Guo, Z.; Liu, L. MiR-19a suppresses ferroptosis of colorectal cancer cells by targeting IREB2. Bioengineered 2022, 13, 12021–12029. [Google Scholar] [CrossRef]
- Xia, H.; Wu, Y.; Zhao, J.; Cheng, C.; Lin, J.; Yang, Y.; Lu, L.; Xiang, Q.; Bian, T.; Liu, Q. N6-Methyladenosine-modified circSAV1 triggers ferroptosis in COPD through recruiting YTHDF1 to facilitate the translation of IREB2. Cell Death Differ. 2023, 30, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Shen, Y.; Wu, D.; Zhang, J.; Lin, C.; Wang, L.; Yu, C.; Yu, B.; Shen, W. CircBCAR3 accelerates esophageal cancer tumorigenesis and metastasis via sponging miR-27a-3p. Mol. Cancer 2022, 21, 145. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, L.; Chew, S.H.; Hirayama, T.; Sekido, Y.; Toyokuni, S. Carbonic anhydrase 9 confers resistance to ferroptosis/apoptosis in malignant mesothelioma under hypoxia. Redox Biol. 2019, 26, 101297. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Guo, S.; Zhang, Y.; Zhao, Y.; Li, X.; Jia, Y.; Xu, Y.; Ma, B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal 2020, 65, 109422. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J. Int. Med. Res. 2021, 49, 300060521996183. [Google Scholar] [CrossRef]
- Bao, C.; Zhang, J.; Xian, S.Y.; Chen, F. MicroRNA-670-3p suppresses ferroptosis of human glioblastoma cells through targeting ACSL4. Free Radic. Res. 2021, 55, 853–864. [Google Scholar] [CrossRef]
- Lu, Y.; Chan, Y.T.; Tan, H.Y.; Zhang, C.; Guo, W.; Xu, Y.; Sharma, R.; Chen, Z.S.; Zheng, Y.C.; Wang, N.; et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2022, 41, 3. [Google Scholar] [CrossRef]
- Liu, B.; Ma, H.; Liu, X.; Xing, W. CircSCN8A suppresses malignant progression and induces ferroptosis in non-small cell lung cancer by regulating miR-1290/ACSL4 axis. Cell Cycle 2023, 22, 758–776. [Google Scholar] [CrossRef]
- Mao, R.; Liu, H. Depletion of mmu_circ_0001751 (circular RNA Carm1) protects against acute cerebral infarction injuries by binding with microRNA-3098-3p to regulate acyl-CoA synthetase long-chain family member 4. Bioengineered 2022, 13, 4063–4075. [Google Scholar] [CrossRef]
- Shi, L.; Song, Z.; Li, Y.; Huang, J.; Zhao, F.; Luo, Y.; Wang, J.; Deng, F.; Shadekejiang, H.; Zhang, M.; et al. MiR-20a-5p alleviates kidney ischemia/reperfusion injury by targeting ACSL4-dependent ferroptosis. Am. J. Transpl. 2023, 23, 11–25. [Google Scholar] [CrossRef]
- Chen, H.; Ren, L.; Ma, W. Mechanism of SOX10 in ferroptosis of hippocampal neurons after intracerebral hemorrhage via the miR-29a-3p/ACSL4 axis. J. Neurophysiol. 2023, 129, 862–871. [Google Scholar] [CrossRef]
- Kannan, M.; Sil, S.; Oladapo, A.; Thangaraj, A.; Periyasamy, P.; Buch, S. HIV-1 Tat-mediated microglial ferroptosis involves the miR-204-ACSL4 signaling axis. Redox Biol. 2023, 62, 102689. [Google Scholar] [CrossRef] [PubMed]
- Qi, R.; Bai, Y.; Li, K.; Liu, N.; Xu, Y.; Dal, E.; Wang, Y.; Lin, R.; Wang, H.; Liu, Z.; et al. Cancer-associated fibroblasts suppress ferroptosis and induce gemcitabine resistance in pancreatic cancer cells by secreting exosome-derived ACSL4-targeting miRNAs. Drug Resist. Updat. 2023, 68, 100960. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.J.; Zhang, D.; Wu, Y.; Jia, Q.H.; Zhang, L.; Li, Y.X.; Yang, Y.F.; Wang, H.; Wu, C.T.; Wang, L.S. miRNA-17-92 protects endothelial cells from erastin-induced ferroptosis through targeting the A20-ACSL4 axis. Biochem. Biophys. Res. Commun. 2019, 515, 448–454. [Google Scholar] [CrossRef]
- Sun, Z.; Wu, J.; Bi, Q.; Wang, W. Exosomal lncRNA TUG1 derived from human urine-derived stem cells attenuates renal ischemia/reperfusion injury by interacting with SRSF1 to regulate ASCL4-mediated ferroptosis. Stem Cell Res. Ther. 2022, 13, 297. [Google Scholar] [CrossRef]
- Yang, H.; Hu, Y.; Weng, M.; Liu, X.; Wan, P.; Hu, Y.; Ma, M.; Zhang, Y.; Xia, H.; Lv, K. Hypoxia inducible lncRNA-CBSLR modulates ferroptosis through m6A-YTHDF2-dependent modulation of CBS in gastric cancer. J. Adv. Res. 2022, 37, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Nagasawa, T.; Kuwahara, Y.; Torii, S.; Igarashi, K.; Roudkenar, M.H.; Roushandeh, A.M.; Kurimasa, A.; Sato, T. MiR-7-5p Is Involved in Ferroptosis Signaling and Radioresistance Thru the Generation of ROS in Radioresistant HeLa and SAS Cell Lines. Int. J. Mol. Sci. 2021, 22, 8300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M.; et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef]
- Yang, X.; Liu, J.; Wang, C.; Cheng, K.K.; Xu, H.; Li, Q.; Hua, T.; Jiang, X.; Sheng, L.; Mao, J.; et al. miR-18a promotes glioblastoma development by down-regulating ALOXE3-mediated ferroptotic and anti-migration activities. Oncogenesis 2021, 10, 15. [Google Scholar] [CrossRef]
- Yi, T.T.; Zhang, L.M.; Huang, X.N. Glycyrrhizic acid protects against temporal lobe epilepsy in young rats by regulating neuronal ferroptosis through the miR-194-5p/PTGS2 axis. Kaohsiung J. Med. Sci. 2023, 39, 154–165. [Google Scholar] [CrossRef]
- Liu, C.; Lu, J.; Yuan, T.; Xie, L.; Zhang, L. EPC-exosomal miR-26a-5p improves airway remodeling in COPD by inhibiting ferroptosis of bronchial epithelial cells via PTGS2/PGE2 signaling pathway. Sci. Rep. 2023, 13, 6126. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, S.; Wei, J.; Zhou, H.; Wang, W.; Yang, J.; Deng, Q.; Wang, H.; Fu, Z. Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/beta-catenin-TFE3 feedback loop signalling. Clin. Transl. Med. 2022, 12, e752. [Google Scholar] [CrossRef] [PubMed]
- Long, F.; Lin, Z.; Long, Q.; Lu, Z.; Zhu, K.; Zhao, M.; Yang, M. CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD. Cancers 2023, 15, 459. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.; He, Y.; Deng, T.; Liu, R.; Wang, W.; Zhu, K.; Bai, M.; Ning, T.; Yang, H.; et al. Chemotoxicity-induced exosomal lncFERO regulates ferroptosis and stemness in gastric cancer stem cells. Cell Death Dis. 2021, 12, 1116. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ding, J.; Li, B.; Sun, T.; You, X.; He, Q.; Sheng, W. Effects of icariin and curcumol on autophagy, ferroptosis, and lipid metabolism based on miR-7/m-TOR/SREBP1 pathway on prostate cancer. BioFactors 2023, 49, 438–456. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Li, C.; Ye, D.M.; Yu, K.; Li, Y.; Tang, H.; Xu, G.; Yi, S.; Zhang, Z. Circular RNA circEPSTI1 accelerates cervical cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging 2021, 13, 4663–4673. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, H.; Deng, J.; Jiang, R.; Meng, E.; Wu, H. CircRPPH1 promotes the stemness of gastric cancer cells by targeting miR-375/SLC7A11 axis. Environ. Toxicol. 2023, 38, 115–125. [Google Scholar] [CrossRef]
- Mao, S.H.; Zhu, C.H.; Nie, Y.; Yu, J.; Wang, L. Levobupivacaine Induces Ferroptosis by miR-489-3p/SLC7A11 Signaling in Gastric Cancer. Front. Pharmacol. 2021, 12, 681338. [Google Scholar] [CrossRef]
- Lyu, N.; Zeng, Y.; Kong, Y.; Chen, Q.; Deng, H.; Ou, S.; Bai, Y.; Tang, H.; Wang, X.; Zhao, M. Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann. Transl. Med. 2021, 9, 675. [Google Scholar] [CrossRef]
- Wang, H.H.; Ma, J.N.; Zhan, X.R. Circular RNA Circ_0067934 Attenuates Ferroptosis of Thyroid Cancer Cells by miR-545-3p/SLC7A11 Signaling. Front Endocrinol. 2021, 12, 670031. [Google Scholar] [CrossRef]
- Yadav, P.; Sharma, P.; Sundaram, S.; Venkatraman, G.; Bera, A.K.; Karunagaran, D. SLC7A11/ xCT is a target of miR-5096 and its restoration partially rescues miR-5096-mediated ferroptosis and anti-tumor effects in human breast cancer cells. Cancer Lett. 2021, 522, 211–224. [Google Scholar] [CrossRef]
- Yang, J.; Cao, X.H.; Luan, K.F.; Huang, Y.D. Circular RNA FNDC3B Protects Oral Squamous Cell Carcinoma Cells from Ferroptosis and Contributes to the Malignant Progression by Regulating miR-520d-5p/SLC7A11 Axis. Front. Oncol. 2021, 11, 672724. [Google Scholar] [CrossRef]
- Sun, K.; Ren, W.; Li, S.; Zheng, J.; Huang, Y.; Zhi, K.; Gao, L. MiR-34c-3p upregulates erastin-induced ferroptosis to inhibit proliferation in oral squamous cell carcinomas by targeting SLC7A11. Pathol. Res. Pr. 2022, 231, 153778. [Google Scholar] [CrossRef]
- Yu, Y.; MohamedAl-Sharani, H.; Zhang, B. EZH2-mediated SLC7A11 upregulation via miR-125b-5p represses ferroptosis of TSCC. Oral. Dis. 2023, 29, 880–891. [Google Scholar] [CrossRef]
- Xu, F.; Ji, S.; Yang, L.; Li, Y.; Shen, P. Potential upstream lncRNA-miRNA-mRNA regulatory network of the ferroptosis-related gene SLC7A11 in renal cell carcinoma. Transl. Androl. Urol. 2023, 12, 33–57. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Z.; Zhu, H.C.; Du, Y.; Zhao, H.C.; Wang, L. Silencing lncRNA SLC16A1-AS1 Induced Ferroptosis in Renal Cell Carcinoma Through miR-143-3p/SLC7A11 Signaling. Technol. Cancer Res. Treat. 2022, 21, 15330338221077803. [Google Scholar] [CrossRef]
- Qin, K.; Zhang, F.; Wang, H.; Wang, N.; Qiu, H.; Jia, X.; Gong, S.; Zhang, Z. circRNA circSnx12 confers Cisplatin chemoresistance to ovarian cancer by inhibiting ferroptosis through a miR-194-5p/SLC7A11 axis. BMB Rep. 2023, 56, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Hu, X.; Ye, L.; Bai, P.; Jie, Y.; Shu, K. Long non-coding RNA ADAMTS9-AS1 attenuates ferroptosis by Targeting microRNA-587/solute carrier family 7 member 11 axis in epithelial ovarian cancer. Bioengineered 2022, 13, 8226–8239. [Google Scholar] [CrossRef]
- Wang, W.; Xie, Y.; Malhotra, A. Potential of Curcumin and Quercetin in Modulation of Premature Mitochondrial Senescence and Related Changes during Lung Carcinogenesis. J. Environ. Pathol. Toxicol. Oncol. 2021, 40, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Bi, G.; Liang, J.; Zhao, M.; Zhang, H.; Jin, X.; Lu, T.; Zheng, Y.; Bian, Y.; Chen, Z.; Huang, Y.; et al. miR-6077 promotes cisplatin/pemetrexed resistance in lung adenocarcinoma via CDKN1A/cell cycle arrest and KEAP1/ferroptosis pathways. Mol. Ther. Nucleic Acids 2022, 28, 366–386. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.F.; Wei, K.; Ma, Z.J.; He, Y.Z.; Huang, J.J.; Guo, Z.Z.; Chen, Z.P.; Barr, M.P.; Shackelford, R.E.; Xia, Y.; et al. CircP4HB regulates ferroptosis via SLC7A11-mediated glutathione synthesis in lung adenocarcinoma. Transl. Lung Cancer Res. 2022, 11, 366–380. [Google Scholar] [CrossRef]
- Zhao, Y.; Cui, Q.; Shen, J.; Shen, W.; Weng, Y. Hsa_circ_0070440 promotes lung adenocarcinoma progression by SLC7A11-mediated-ferroptosis. Histol. Histopathol. 2023, 18597. [Google Scholar] [CrossRef]
- Jiang, X.; Guo, S.; Xu, M.; Ma, B.; Liu, R.; Xu, Y.; Zhang, Y. TFAP2C-Mediated lncRNA PCAT1 Inhibits Ferroptosis in Docetaxel-Resistant Prostate Cancer Through c-Myc/miR-25-3p/SLC7A11 Signaling. Front. Oncol. 2022, 12, 862015. [Google Scholar] [CrossRef]
- Pan, C.; Chen, G.; Zhao, X.; Xu, X.; Liu, J. lncRNA BBOX1-AS1 silencing inhibits esophageal squamous cell cancer progression by promoting ferroptosis via miR-513a-3p/SLC7A11 axis. Eur. J. Pharmacol. 2022, 934, 175317. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.; Gao, Y.; Hu, Y.; Wang, R.; Wang, S.; Li, Y.; He, Y.; Yuan, C. LncSNHG14 promotes nutlin3a resistance by inhibiting ferroptosis via the miR-206 /SLC7A11 axis in osteosarcoma cells. Cancer Gene Ther. 2023, 30, 704–715. [Google Scholar] [CrossRef]
- Li, Q.; Li, K.; Guo, Q.; Yang, T. CircRNA circSTIL inhibits ferroptosis in colorectal cancer via miR-431/SLC7A11 axis. Environ. Toxicol. 2023, 38, 981–989. [Google Scholar] [CrossRef]
- Ding, C.; Ding, X.; Zheng, J.; Wang, B.; Li, Y.; Xiang, H.; Dou, M.; Qiao, Y.; Tian, P.; Xue, W. miR-182-5p and miR-378a-3p regulate ferroptosis in I/R-induced renal injury. Cell Death Dis. 2020, 11, 929. [Google Scholar] [CrossRef]
- Zhu, L.; Feng, Z.; Zhang, J.; Du, L.; Meng, A. MicroRNA-27a Regulates Ferroptosis Through SLC7A11 to Aggravate Cerebral ischemia-reperfusion Injury. Neurochem. Res. 2023, 48, 1370–1381. [Google Scholar] [CrossRef]
- Ye, J.; Lyu, T.J.; Li, L.Y.; Liu, Y.; Zhang, H.; Wang, X.; Xi, X.; Liu, Z.J.; Gao, J.Q. Ginsenoside Re attenuates myocardial ischemia/reperfusion induced ferroptosis via miR-144-3p/SLC7A11. Phytomedicine 2023, 113, 154681. [Google Scholar] [CrossRef]
- Li, H.; Ding, J.; Liu, W.; Wang, X.; Feng, Y.; Guan, H.; Chen, Z. Plasma exosomes from patients with acute myocardial infarction alleviate myocardial injury by inhibiting ferroptosis through miR-26b-5p/SLC7A11 axis. Life Sci. 2023, 322, 121649. [Google Scholar] [CrossRef]
- Zhang, K.; Gu, X.; Xia, Y.; Zhao, X.; Khoso Pervez, A.; Li, S. MiR-129-3p regulates ferroptosis in the liver of Selenium-deficient broilers by targeting SLC7A11. Poult. Sci. 2023, 102, 102271. [Google Scholar] [CrossRef]
- Chen, Y.; Deng, Y.; Chen, L.; Huang, Z.; Yan, Y.; Huang, Z. miR-16-5p Regulates Ferroptosis by Targeting SLC7A11 in Adriamycin-Induced Ferroptosis in Cardiomyocytes. J. Inflamm. Res. 2023, 16, 1077–1089. [Google Scholar] [CrossRef]
- Liu, D.; Yang, M.; Yao, Y.; He, S.; Wang, Y.; Cao, Z.; Chen, H.; Fu, Y.; Liu, H.; Zhao, Q. Cardiac Fibroblasts Promote Ferroptosis in Atrial Fibrillation by Secreting Exo-miR-23a-3p Targeting SLC7A11. Oxid. Med. Cell Longev. 2022, 2022, 3961495. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Han, Y.; Sun, H.; Sun, S.; Wang, Y.; Guo, R.; Guo, J.; Tian, X.; Wang, J.; Wang, J. Mesenchymal stem cell-derived exosomal microRNA-367-3p alleviates experimental autoimmune encephalomyelitis via inhibition of microglial ferroptosis by targeting EZH2. Biomed. Pharmacother. 2023, 162, 114593. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wei, Y.; Wang, J.; Yao, J.; Zhang, C.; Yu, C.; Tang, Y.; Zhu, D.; Yang, J.; Zhou, J. Long Noncoding RNA LINC00578 Inhibits Ferroptosis in Pancreatic Cancer via Regulating SLC7A11 Ubiquitination. Oxid. Med. Cell Longev. 2023, 2023, 1744102. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, Y.; Li, Q.; Li, X.; Feng, X. A novel circular RNA confers trastuzumab resistance in human epidermal growth factor receptor 2-positive breast cancer through regulating ferroptosis. Environ. Toxicol. 2022, 37, 1597–1607. [Google Scholar] [CrossRef]
- An, L.; Huang, J.; Ge, S.; Zhang, X.; Wang, J. lncRNA AGAP2-AS1 Facilitates Tumorigenesis and Ferroptosis Resistance through SLC7A11 by IGF2BP2 Pathway in Melanoma. Comput. Math. Methods Med. 2022, 2022, 1972516. [Google Scholar] [CrossRef]
- Yuan, J.; Liu, Z.; Song, R. Antisense lncRNA As-SLC7A11 suppresses epithelial ovarian cancer progression mainly by targeting SLC7A11. Pharmazie 2017, 72, 402–407. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, X.; Liu, N.; Shi, Y.; Liu, Y.; Ouyang, L.; Tam, S.; Xiao, D.; Liu, S.; Wen, F.; et al. A Nuclear Long Non-Coding RNA LINC00618 Accelerates Ferroptosis in a Manner Dependent upon Apoptosis. Mol. Ther. 2021, 29, 263–274. [Google Scholar] [CrossRef]
- He, G.N.; Bao, N.R.; Wang, S.; Xi, M.; Zhang, T.H.; Chen, F.S. Ketamine Induces Ferroptosis of Liver Cancer Cells by Targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des. Devel Ther. 2021, 15, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Fu, J.; Chen, Y.; Li, Y.; Ning, L.; Huang, D.; Yan, S.; Zhang, Q. Circular RNA circKIF4A facilitates the malignant progression and suppresses ferroptosis by sponging miR-1231 and upregulating GPX4 in papillary thyroid cancer. Aging 2021, 13, 16500–16512. [Google Scholar] [CrossRef]
- Han, B.; Liu, Y.; Zhang, Q.; Liang, L. Propofol decreases cisplatin resistance of non-small cell lung cancer by inducing GPX4-mediated ferroptosis through the miR-744-5p/miR-615-3p axis. J. Proteom. 2023, 274, 104777. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ma, H.; Fang, J.; Yu, Y.; Ren, Y.; Yu, R. CircDTL Functions as an Oncogene and Regulates Both Apoptosis and Ferroptosis in Non-small Cell Lung Cancer Cells. Front. Genet. 2021, 12, 743505. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, L.; Wang, C.; Zhang, L.; Xu, W. MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4. Free Radic. Res. 2021, 55, 1119–1129. [Google Scholar] [CrossRef]
- Xu, P.; Wang, Y.; Deng, Z.; Tan, Z.; Pei, X. MicroRNA-15a promotes prostate cancer cell ferroptosis by inhibiting GPX4 expression. Oncol. Lett. 2022, 23, 67. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Yang, Z.; Wen, D.; Hu, Z. Circular RNA circACAP2 Suppresses Ferroptosis of Cervical Cancer during Malignant Progression by miR-193a-5p/GPX4. J. Oncol. 2022, 2022, 5228874. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.; Cao, W.; Zeng, Z.; Zhang, Z.; Jin, B.; Tian, Q.; Wu, Y.; Zhang, T.; Li, D.; Hu, C.; et al. JUND/linc00976 promotes cholangiocarcinoma progression and metastasis, inhibits ferroptosis by regulating the miR-3202/GPX4 axis. Cell Death Dis. 2022, 13, 967. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shi, R.; Guo, J.; Wang, H.; Shen, L.; Shi, H.; Yu, P.; Chen, X. miR-135b-3p Promotes Cardiomyocyte Ferroptosis by Targeting GPX4 and Aggravates Myocardial Ischemia/Reperfusion Injury. Front. Cardiovasc. Med. 2021, 8, 663832. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Jin, J.; Liu, S.; Ding, K.; Qian, C. Inhibition of miR-1224 suppresses hypoxia/reoxygenation-induced oxidative stress and apoptosis in cardiomyocytes through targeting GPX4. Exp. Mol. Pathol. 2021, 121, 104645. [Google Scholar] [CrossRef]
- Fan, K.; Huang, W.; Qi, H.; Song, C.; He, C.; Liu, Y.; Zhang, Q.; Wang, L.; Sun, H. The Egr-1/miR-15a-5p/GPX4 axis regulates ferroptosis in acute myocardial infarction. Eur. J. Pharmacol. 2021, 909, 174403. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Li, S.; Zhang, M.; Xu, W.; Chen, Q.; Zheng, L.; Liu, P.; Zou, W. Acupuncture Ameliorates Neuronal Cell Death, Inflammation, and Ferroptosis and Downregulated miR-23a-3p After Intracerebral Hemorrhage in Rats. J. Mol. Neurosci. 2021, 71, 1863–1875. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Y.; Zheng, D.; Liang, M.; He, Q. A Novel Identified Circular RNA, mmu_mmu_circRNA_0000309, Involves in Germacrone-Mediated Improvement of Diabetic Nephropathy Through Regulating Ferroptosis by Targeting miR-188-3p/GPX4 Signaling Axis. Antioxid. Redox Signal 2022, 36, 740–759. [Google Scholar] [CrossRef]
- Guo, Y.; Bie, Z.D.; Li, X. Hypoxic cardiomyocyte-derived exosomes regulate cardiac fibroblast activation, apoptosis, migration and ferroptosis through miR-208a/b. Gen. Physiol. Biophys. 2023, 42, 149–158. [Google Scholar] [CrossRef]
- Zha, J.; Pan, Y.; Liu, X.; Zhu, H.; Liu, Y.; Zeng, W. Exosomes from hypoxia-pretreated adipose-derived stem cells attenuate ultraviolet light-induced skin injury via delivery of circ-Ash1l. Photodermatol. Photoimmunol. Photomed. 2023, 39, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Zhong, S.; Wu, R.; Mo, Z.; Zheng, S.; Xue, J.; Meng, H.; Liu, M.; Chen, X.; Zhang, G.; et al. Suppressing circIDE/miR-19b-3p/RBMS1 axis exhibits promoting-tumour activity through upregulating GPX4 to diminish ferroptosis in hepatocellular carcinoma. Epigenetics 2023, 18, 2192438. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Huo, Y.; Jia, S.; He, F.; Li, H.; Zhou, Q.; Chang, N.; Liu, D.; Li, R.; Hu, Y.; et al. Silenced LINC01134 Enhances Oxaliplatin Sensitivity by Facilitating Ferroptosis Through GPX4 in Hepatocarcinoma. Front. Oncol. 2022, 12, 939605. [Google Scholar] [CrossRef]
- Gomaa, A.; Peng, D.; Chen, Z.; Soutto, M.; Abouelezz, K.; Corvalan, A.; El-Rifai, W. Epigenetic regulation of AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci. Rep. 2019, 9, 16970. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Mao, C.; Ouyang, L.; Liu, Y.; Lai, W.; Liu, N.; Shi, Y.; Chen, L.; Xiao, D.; Yu, F.; et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019, 26, 2329–2343. [Google Scholar] [CrossRef]
- Wu, N.; Zhu, D.; Li, J.; Li, X.; Zhu, Z.; Rao, Q.; Hu, B.; Wang, H.; Zhu, Y. CircOMA1 modulates cabergoline resistance by downregulating ferroptosis in prolactinoma. J. Endocrinol. Investig. 2023, 46, 1573–1587. [Google Scholar] [CrossRef]
- Wang, Y.; Niu, H.; Li, L.; Han, J.; Liu, Z.; Chu, M.; Sha, X.; Zhao, J. Anti-CHAC1 exosomes for nose-to-brain delivery of miR-760-3p in cerebral ischemia/reperfusion injury mice inhibiting neuron ferroptosis. J. Nanobiotechnology 2023, 21, 109. [Google Scholar] [CrossRef]
- Deng, Y.; Lai, W.; Yu, L.; Zhang, W.; Ding, Y. miR-2115-3p inhibits ferroptosis by downregulating the expression of glutamic-oxaloacetic transaminase in preeclampsia. Placenta 2022, 129, 94–103. [Google Scholar] [CrossRef]
- Zhang, K.; Wu, L.; Zhang, P.; Luo, M.; Du, J.; Gao, T.; O’Connell, D.; Wang, G.; Wang, H.; Yang, Y. miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol. Carcinog. 2018, 57, 1566–1576. [Google Scholar] [CrossRef]
- Bazhabayi, M.; Qiu, X.; Li, X.; Yang, A.; Wen, W.; Zhang, X.; Xiao, X.; He, R.; Liu, P. CircGFRA1 facilitates the malignant progression of HER-2-positive breast cancer via acting as a sponge of miR-1228 and enhancing AIFM2 expression. J. Cell Mol. Med. 2021, 25, 10248–10256. [Google Scholar] [CrossRef]
- Shao, C.; Chen, Y.; Yang, T.; Zhao, H.; Li, D. Mesenchymal Stem Cell Derived Exosomes Suppress Neuronal Cell Ferroptosis Via lncGm36569/miR-5627-5p/FSP1 Axis in Acute Spinal Cord Injury. Stem Cell Rev. Rep. 2022, 18, 1127–1142. [Google Scholar] [CrossRef]
- Wang, F.; Li, J.; Zhao, Y.; Guo, D.; Liu, D.; Chang, S.; Qiao, H.; Li, J.; Yang, Y.; Zhang, C.; et al. miR-672-3p Promotes Functional Recovery in Rats with Contusive Spinal Cord Injury by Inhibiting Ferroptosis Suppressor Protein 1. Oxid. Med. Cell Longev. 2022, 2022, 6041612. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Jia, G.; Ma, P.; Cang, S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci. 2021, 276, 119399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, M.; Cui, X.; O’Connell, D.; Yang, Y. Long noncoding RNA NEAT1 promotes ferroptosis by modulating the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ. 2022, 29, 1850–1863. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Jiang, L.J.; Zhang, Y.X.; Xu, S.T.; Liu, S.L.; Ye, J.T.; Liu, P.Q. Inhibition of miR-214-3p attenuates ferroptosis in myocardial infarction via regulating ME2. Biochem. Biophys. Res. Commun. 2023, 661, 64–74. [Google Scholar] [CrossRef]
- Qu, X.; Liu, B.; Wang, L.; Liu, L.; Zhao, W.; Liu, C.; Ding, J.; Zhao, S.; Xu, B.; Yu, H.; et al. Loss of cancer-associated fibroblast-derived exosomal DACT3-AS1 promotes malignant transformation and ferroptosis-mediated oxaliplatin resistance in gastric cancer. Drug Resist. Updat. 2023, 68, 100936. [Google Scholar] [CrossRef]
- Gai, C.; Liu, C.; Wu, X.; Yu, M.; Zheng, J.; Zhang, W.; Lv, S.; Li, W. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis. 2020, 11, 751. [Google Scholar] [CrossRef]
- Shen, K.; Wang, X.; Wang, Y.; Jia, Y.; Zhang, Y.; Wang, K.; Luo, L.; Cai, W.; Li, J.; Li, S.; et al. miR-125b-5p in adipose derived stem cells exosome alleviates pulmonary microvascular endothelial cells ferroptosis via Keap1/Nrf2/GPX4 in sepsis lung injury. Redox Biol. 2023, 62, 102655. [Google Scholar] [CrossRef]
- Han, Y.; Gao, X.; Wu, N.; Jin, Y.; Zhou, H.; Wang, W.; Liu, H.; Chu, Y.; Cao, J.; Jiang, M.; et al. Long noncoding RNA LINC00239 inhibits ferroptosis in colorectal cancer by binding to Keap1 to stabilize Nrf2. Cell Death Dis. 2022, 13, 742. [Google Scholar] [CrossRef]
- Liao, Y.; Jia, X.; Ren, Y.; Deji, Z.; Gesang, Y.; Ning, N.; Feng, H.; Yu, H.; Wei, A. Suppressive role of microRNA-130b-3p in ferroptosis in melanoma cells correlates with DKK1 inhibition and Nrf2-HO-1 pathway activation. Hum. Cell 2021, 34, 1532–1544. [Google Scholar] [CrossRef]
- Dong, L.H.; Huang, J.J.; Zu, P.; Liu, J.; Gao, X.; Du, J.W.; Li, Y.F. CircKDM4C upregulates P53 by sponging hsa-let-7b-5p to induce ferroptosis in acute myeloid leukemia. Environ. Toxicol. 2021, 36, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Liu, S.; Ye, F.; Shen, Y.; Tie, Y.; Zhu, J.; Wei, L.; Jin, Y.; Fu, H.; Wu, Y.; et al. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS ONE 2015, 10, e0139790. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Chen, C.; Huang, Y.; Xia, P.; Zhang, F.; Li, L.; Wang, E.; Guo, Q.; Ye, Z. Long noncoding RNA Meg3 mediates ferroptosis induced by oxygen and glucose deprivation combined with hyperglycemia in rat brain microvascular endothelial cells, through modulating the p53/GPX4 axis. Eur. J. Histochem. 2021, 65, 3224. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, Z.; Li, D.; Lv, Q.; Chen, S.; Zhang, Z.; Wu, M. LncRNA PELATON, a Ferroptosis Suppressor and Prognositic Signature for GBM. Front. Oncol. 2022, 12, 817737. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.J.; Zhou, H.L.; Gao, X.Z.; Xu, C.F. Downregulation of miR-221-3p promotes the ferroptosis in gastric cancer cells via upregulation of ATF3 to mediate the transcription inhibition of GPX4 and HRD1. Transl. Oncol. 2023, 32, 101649. [Google Scholar] [CrossRef]
- Zhao, J.; Yang, S.; Lv, C.; Liu, Y. Cancer-associated fibroblasts suppressed ferroptosis in glioblastoma via upregulating lncRNA DLEU1. Am. J. Physiol. Cell Physiol. 2023, 324, C1039–C1052. [Google Scholar] [CrossRef]
- Guan, L.; Wang, F.; Wang, M.; Han, S.; Cui, Z.; Xi, S.; Xu, H.; Li, S. Downregulation of HULC Induces Ferroptosis in Hepatocellular Carcinoma via Targeting of the miR-3200-5p/ATF4 Axis. Oxid. Med. Cell Longev. 2022, 2022, 9613095. [Google Scholar] [CrossRef]
- Bai, T.; Liang, R.; Zhu, R.; Wang, W.; Zhou, L.; Sun, Y. MicroRNA-214-3p enhances erastin-induced ferroptosis by targeting ATF4 in hepatoma cells. J. Cell Physiol. 2020, 235, 5637–5648. [Google Scholar] [CrossRef]
- Zhang, H.; Ge, Z.; Wang, Z.; Gao, Y.; Wang, Y.; Qu, X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via mir-106a-5p/STAT3 axis in breast cancer. Aging 2021, 13, 8115–8126. [Google Scholar] [CrossRef]
- Liu, Y.P.; Qiu, Z.Z.; Li, X.H.; Li, E.Y. Propofol induces ferroptosis and inhibits malignant phenotypes of gastric cancer cells by regulating miR-125b-5p/STAT3 axis. World J. Gastrointest. Oncol. 2021, 13, 2114–2128. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, X.L. Dexmedetomidine promotes ferroptotic cell death in gastric cancer via hsa_circ_0008035/miR-302a/E2F7 axis. Kaohsiung J. Med. Sci. 2023, 39, 390–403. [Google Scholar] [CrossRef]
- Li, Q.; Meng, X.; Hua, Q. Circ ASAP2 decreased inflammation and ferroptosis in diabetic nephropathy through SOX2/SLC7A11 by miR-770-5p. Acta Diabetol. 2023, 60, 29–42. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Zhang, B.W.; Zhang, Z.B.; Deng, Q.J. Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2585–2600. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, H.; Wei, X. Circ_0007142 downregulates miR-874-3p-mediated GDPD5 on colorectal cancer cells. Eur. J. Clin. Investig. 2021, 51, e13541. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Lu, W.; Qu, X.; Liu, N.; Zhu, C. Silencing long non-coding RNA MEG8 inhibits the proliferation and induces the ferroptosis of hemangioma endothelial cells by regulating miR-497-5p/NOTCH2 axis. Biochem. Biophys. Res. Commun. 2021, 556, 72–78. [Google Scholar] [CrossRef]
- Yang, X.; Li, Y.; Zhang, Y.; Liu, J. Circ_0000745 promotes acute lymphoblastic leukemia progression through mediating miR-494-3p/NET1 axis. Hematology 2022, 27, 11–22. [Google Scholar] [CrossRef]
- Yao, W.; Wang, J.; Meng, F.; Zhu, Z.; Jia, X.; Xu, L.; Zhang, Q.; Wei, L. Circular RNA CircPVT1 Inhibits 5-Fluorouracil Chemosensitivity by Regulating Ferroptosis Through MiR-30a-5p/FZD3 Axis in Esophageal Cancer Cells. Front. Oncol. 2021, 11, 780938. [Google Scholar] [CrossRef] [PubMed]
- Xian, Z.Y.; Hu, B.; Wang, T.; Cai, J.L.; Zeng, J.Y.; Zou, Q.; Zhu, P.X. CircABCB10 silencing inhibits the cell ferroptosis and apoptosis by regulating the miR-326/CCL5 axis in rectal cancer. Neoplasma 2020, 67, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, L.; Li, X.; Chen, Y.; Shi, T.; Wang, Q. LINC00460 Facilitates Cell Proliferation and Inhibits Ferroptosis in Breast Cancer Through the miR-320a/MAL2 Axis. Technol. Cancer Res. Treat. 2023, 22, 15330338231164359. [Google Scholar] [CrossRef]
- Conrad, M.; Kagan, V.E.; Bayir, H.; Pagnussat, G.C.; Head, B.; Traber, M.G.; Stockwell, B.R. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes. Dev. 2018, 32, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Distefano, A.M.; Marchetti, F.; Zabaleta, E.; Pagnussat, G.C. Measuring and Perturbing Ferroptosis in Plants. Methods Mol. Biol. 2022, 2447, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Costa, I.; Barbosa, D.J.; Silva, V.; Benfeito, S.; Borges, F.; Remiao, F.; Silva, R. Research Models to Study Ferroptosis’s Impact in Neurodegenerative Diseases. Pharmaceutics 2023, 15, 1369. [Google Scholar] [CrossRef]
- Guo, Z.; Sun, J.; Lv, X.; Zhang, T.; Yao, H.; Wu, W.; Xing, Z.; Kong, N.; Wang, L.; Song, L. The ferroptosis in haemocytes of Pacific oyster Crassostrea gigas upon erastin treatment. Fish. Shellfish. Immunol. 2023, 133, 108556. [Google Scholar] [CrossRef]
- Wang, W.T.; Han, C.; Sun, Y.M.; Chen, T.Q.; Chen, Y.Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef] [PubMed]
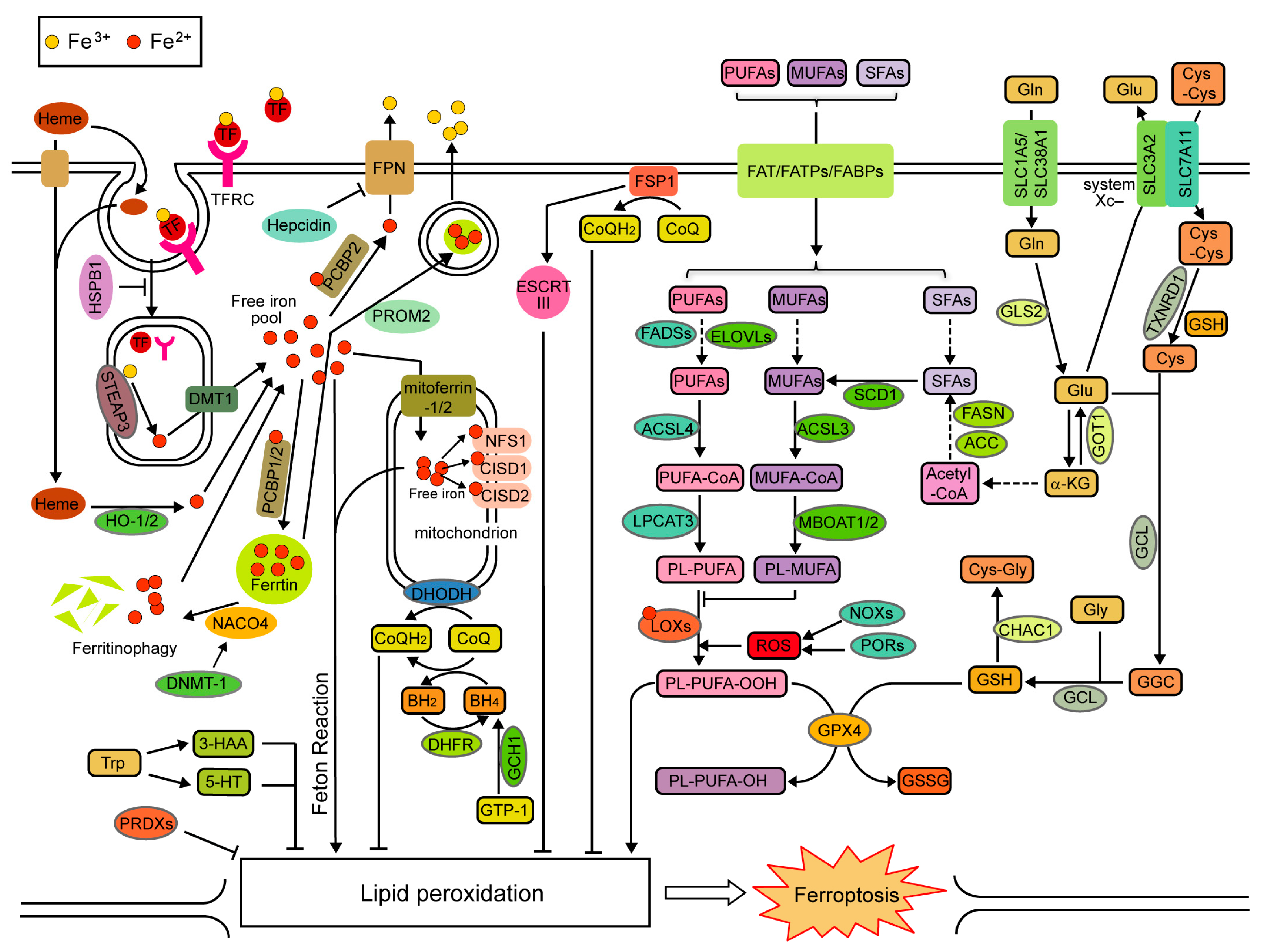
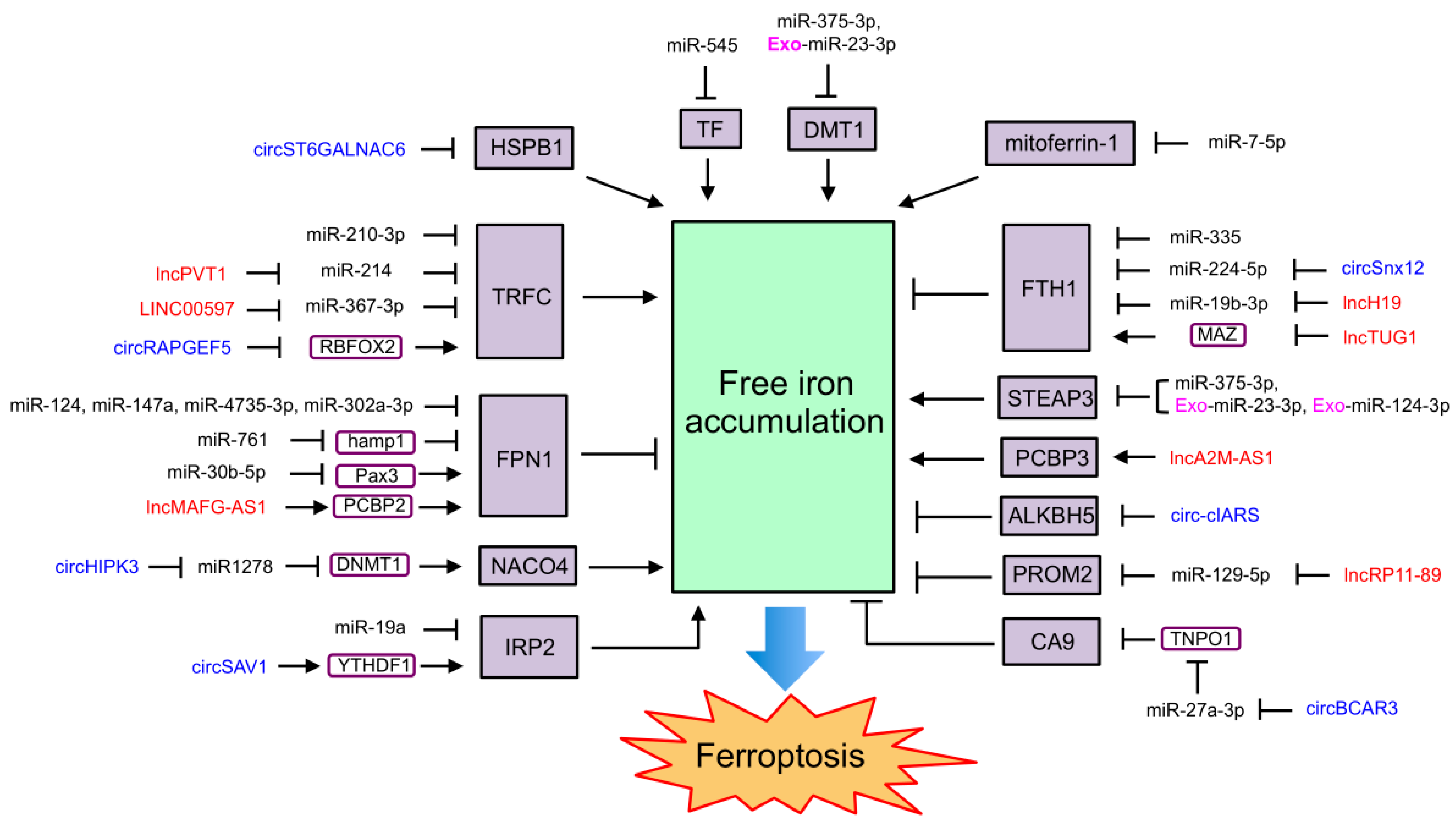
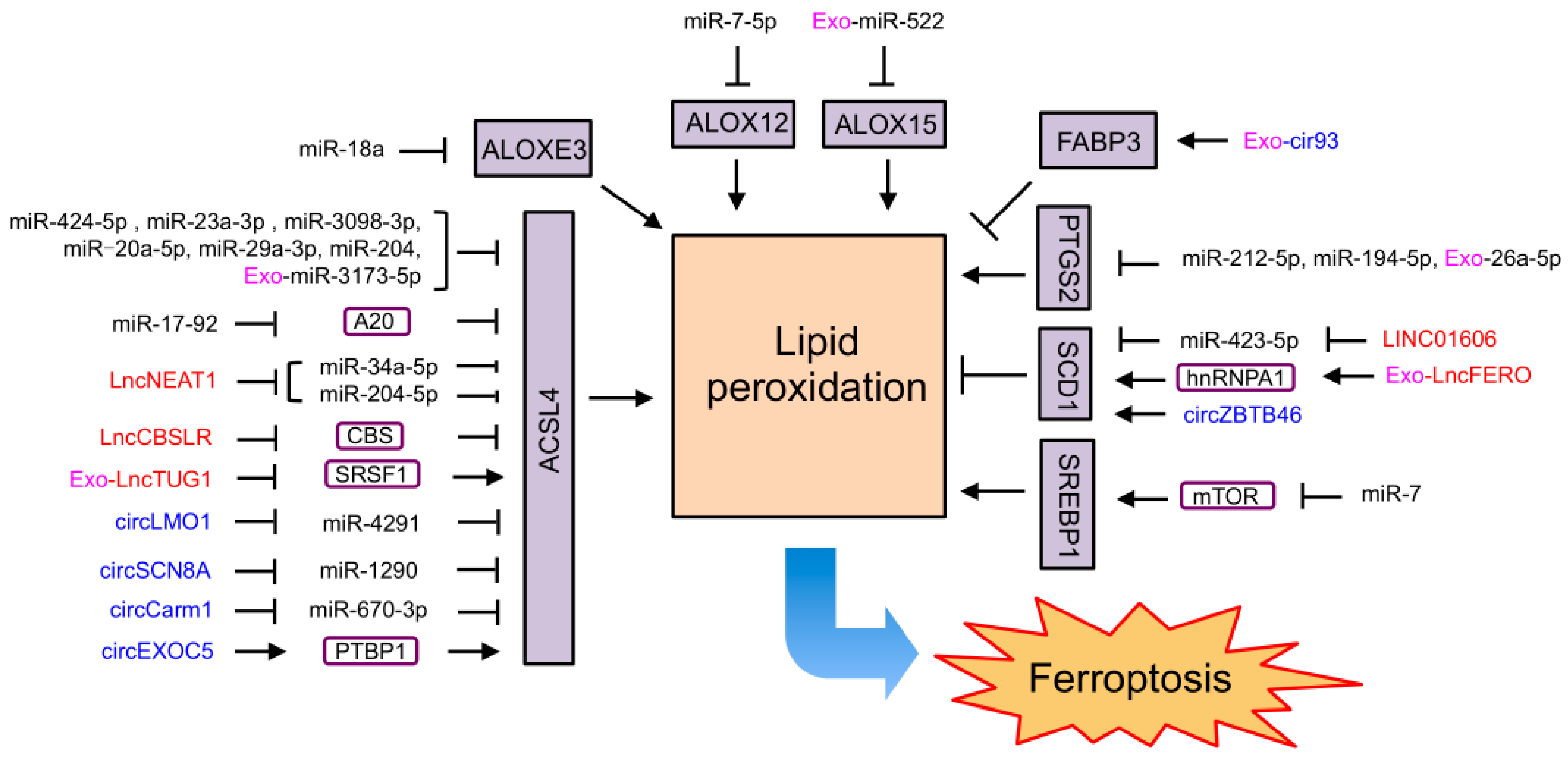
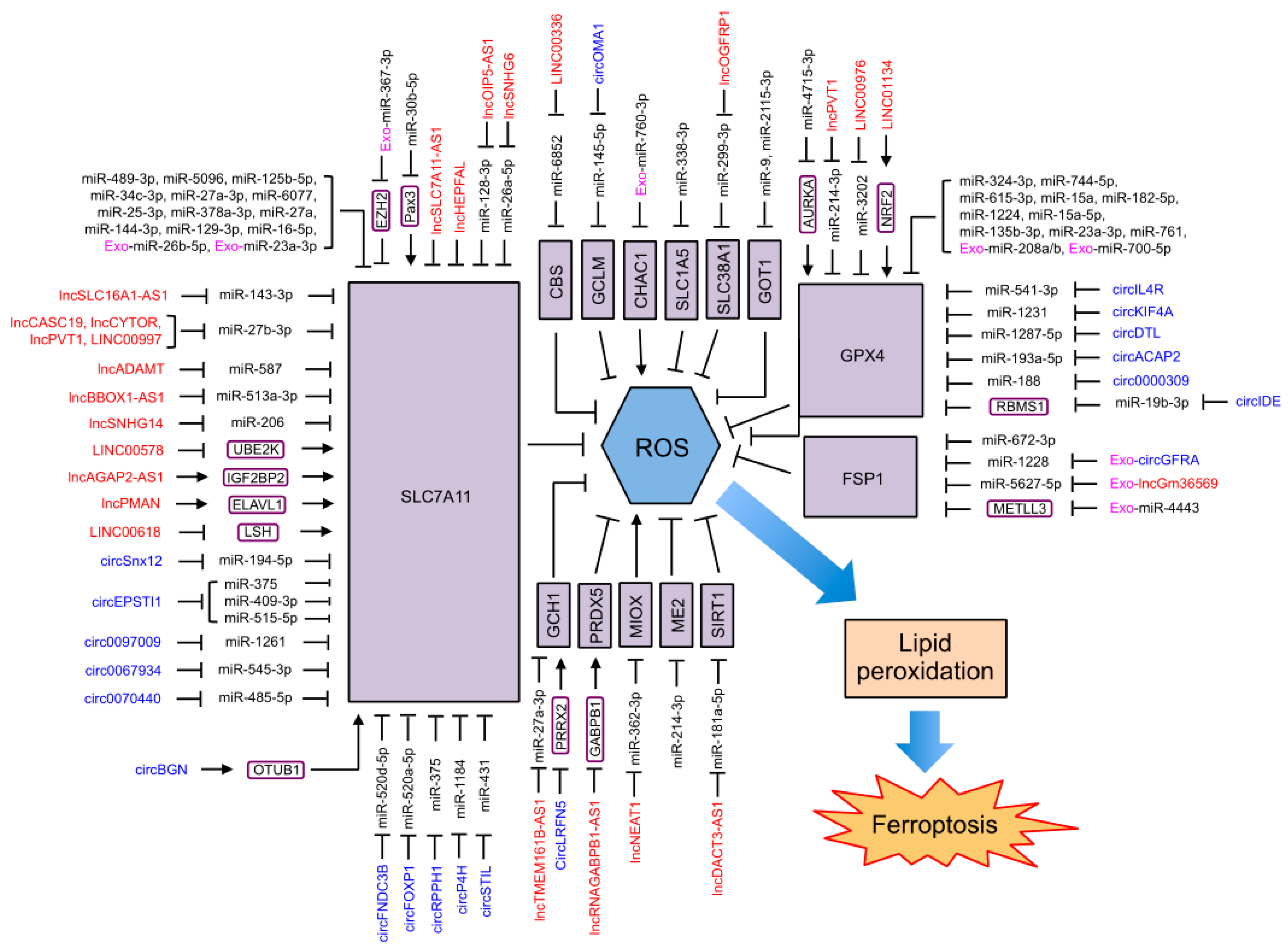
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Zhang, C. The Regulation of Ferroptosis by Noncoding RNAs. Int. J. Mol. Sci. 2023, 24, 13336. https://doi.org/10.3390/ijms241713336
Zheng X, Zhang C. The Regulation of Ferroptosis by Noncoding RNAs. International Journal of Molecular Sciences. 2023; 24(17):13336. https://doi.org/10.3390/ijms241713336
Chicago/Turabian StyleZheng, Xiangnan, and Cen Zhang. 2023. "The Regulation of Ferroptosis by Noncoding RNAs" International Journal of Molecular Sciences 24, no. 17: 13336. https://doi.org/10.3390/ijms241713336
APA StyleZheng, X., & Zhang, C. (2023). The Regulation of Ferroptosis by Noncoding RNAs. International Journal of Molecular Sciences, 24(17), 13336. https://doi.org/10.3390/ijms241713336




