Abstract
Aberrant mucus secretion is a hallmark of chronic obstructive pulmonary disease (COPD). Expression of the membrane-tethered mucins 3A and 3B (MUC3A, MUC3B) in human lung is largely unknown. In this observational cross-sectional study, we recruited subjects 45–65 years old from the general population of Stockholm, Sweden, during the years 2007–2011. Bronchial mucosal biopsies, bronchial brushings, and bronchoalveolar lavage fluid (BALF) were retrieved from COPD patients (n = 38), healthy never-smokers (n = 40), and smokers with normal lung function (n = 40). Protein expression of MUC3A and MUC3B in bronchial mucosal biopsies was assessed by immunohistochemical staining. In a subgroup of subjects (n = 28), MUC3A and MUC3B mRNAs were quantified in bronchial brushings using microarray. Non-parametric tests were used to perform correlation and group comparison analyses. A value of p < 0.05 was considered statistically significant. MUC3A and MUC3B immunohistochemical expression was localized to ciliated cells. MUC3B was also expressed in basal cells. MUC3A and MUC3B immunohistochemical expression was equal in all study groups but subjects with emphysema had higher MUC3A expression, compared to those without emphysema. Smokers had higher mRNA levels of MUC3A and MUC3B than non-smokers. MUC3A and MUC3B mRNA were higher in male subjects and correlated negatively with expiratory air flows. MUC3B mRNA correlated positively with total cell concentration and macrophage percentage, and negatively with CD4/CD8 T cell ratio in BALF. We concluded that MUC3A and MUC3B in large airways may be a marker of disease or may play a role in the pathophysiology of airway obstruction.
Keywords:
bronchoalveolar lavage; COPD; epithelium; immunohistochemistry; large airways; lung function; microarray; mRNA; mucin; smoking 1. Introduction
Increased mucus secretion and accumulation in the airway lumen are clinical hallmarks of various lung diseases such as asthma, cystic fibrosis, and chronic obstructive pulmonary disease (COPD). Many patients with these diseases exhibit increased mucin secretion and mucus hyper-concentration, which leads subsequently to an impaired mucus clearance, persistent neutrophil airway inflammation and bacterial colonization [1], and symptoms of chronic bronchitis. These subjects experience also more exacerbations [2], accelerated lung function decline, and impaired survival [3].
Chronic obstructive pulmonary disease (COPD) remains a significant burden and cost to health-care systems, as well as a substantial source of amenable mortality [4], as it is the third leading cause of death worldwide [5]. It is a heterogeneous lung condition characterized by chronic respiratory symptoms due to abnormalities of the airways (bronchitis, bronchiolitis) and/or alveoli (emphysema) that cause persistent, often progressive, airflow obstruction [6]. Increasing importance is placed on attempts to slow down COPD lung disease progression by improving risk factors. Globally, male sex, smoking, age, body-mass index of less than 18.5 kg/m2, biomass exposure, and occupational exposure to dust or smoke are all substantial risk factors for COPD [7].
The leading etiological factor causing COPD is cigarette smoking [8]. It is estimated that the global prevalence of COPD is 10.3%, and it is appreciably higher in smokers and ex-smokers compared to non-smokers [6]. Smoking alters bronchial-mucus-barrier cell composition and transcriptome and increases mucus production [9]. Thus, in smokers, goblet cell hyperplasia and hypertrophy as well as abnormal patterns of mucin gene expression have been described [10]. In asthmatic patients, exposure to tobacco smoke results in oxidant/antioxidant imbalance, which leads to increases in pro-inflammatory cytokines such as TNFα, IL-6, and IL-8 [11]. Active and second-hand smoke exposure can be quantified by measuring serum and urinary cotinine and exhaled carbon monoxide [12].
Despite progress in the treatment of symptoms and the prevention of acute exacerbations, few advances have been made to ameliorate lung disease progression in COPD. To be able to achieve that, a better understanding of the complex disease mechanisms resulting in COPD, as well as a search for biomarkers, are needed [13]. The 6 min walk distance, heart rate, C-reactive protein, fibrinogen, and white cell count were found to be associated with clinical outcomes in patients with stable COPD [14], with a spillover effect of inflammatory response proposed as the underlying mechanism [15]. COPD patients with emphysema presented increased systemic oxidative stress markers and fibrinogen, compared to COPD patients without emphysema [16]. Moreover, elevated plasma level of Pentraxin 3 was associated with emphysema and mortality in smokers [17]. However, data on airway-derived disease biomarkers in COPD are largely lacking.
The major macromolecular components of mucus are mucin glycoproteins (MUCs). They play important roles in the protection of epithelium, signal transduction, and modulation of the immune system. Airway protein expression is predominated by four membrane-associated mucins (MUC1, MUC4, MUC16, MUC20), one secreted non-gel-forming mucin (MUC7), and two gel-forming mucins (MUC5AC and MUC5B) [18]. Long-term smoking induces enhanced MUC5AC expression [19]. Smoking history and the presence of chronic bronchitis, regardless of airway obstruction, affect both cellular and soluble MUC1 in human airways [20]. In line with that, Kato et al. showed that MUC1 contributed to goblet cell metaplasia and enhanced MUC5AC expression in response to cigarette smoke in vivo [21]. In asthma, the expression of MUC5AC increased remarkably by asthma severity; also, it was associated with airway wall thickness [22]. In ovalbumin-induced asthmatic mice, the key circadian rhythm gene, BMAL1, caused periodic changes in airway MUC1 expression [23]. In addition, protein levels of MUC5AC and MUC8 were significantly elevated by lipopolysaccharide or IL-17A stimulation in human middle ear epithelial cells [24]. COPD exacerbations are often associated with increased sputum production and infection with Haemophilus influenzae. Interestingly, Haemophilus influenzae has been found to upregulate the MUC5AC gene via TLR2-MyD88-dependent p38 mitogen-activated protein kinase pathway [25]. Moreover, neutrophil elastase induces MUC5AC gene expression by an oxidant-dependent mechanism [26], and cytokines, oxidative stress, and endoplasmic reticulum stress stimulate mucus production [27,28,29]. A recent study indicated that the glycosylated extracellular domains of different transmembrane mucins might have protective functions in respiratory cells by restricting SARS-CoV-2 binding and entry [30], and MUC4 was found to be a cellular biomarker of necrotizing bronchiolitis in influenza A virus infection [31]. Taken together, these studies indicate that mucins could be potential biomarkers of airway diseases.
MUC3A and MUC3B are membrane-tethered mucins that were originally described as being expressed in the intestinal epithelium [32]. Non-synonymous single-nucleotide polymorphisms of MUC3A, involving a tyrosine residue with a proposed role in cell signaling, were reported to confer genetic predisposition to Crohn’s disease [33]. Dohrman et al. studied specifically MUC3 mRNA distribution in human bronchus, using an mRNA in situ hybridization technique, and found that it occurred in a diffuse pattern throughout the bronchial epithelium. However, they could not determine whether the labelling was associated with goblet, ciliated, or basal cells [34]. To our knowledge, MUC3A and MUC3B immunohistochemical expression in human airway epithelium in states of health and disease is largely unknown.
Therefore, the aim of this study was to determine the immunohistochemical MUC3A and MUC3B expression and mRNA levels by investigating bronchial mucosal biopsies and bronchial brushing samples of large airway epithelium in never-smokers, current smokers with normal lung function, and in COPD patients. Moreover, we aimed to assess the biomarker potential of MUC3A and MUC3B expression and of their mRNA levels in large airway epithelium.
2. Results
2.1. Demographics of the Study Groups
The four study groups were matched in terms of age, sex, smoking history (>10 pack-years), and current smoking habits (smoking > 10 cigarettes/day past 6 months) (Table 1).

Table 1.
Demographics of the study groups.
2.2. Immunohistochemical MUC3A and MUC3B Expression in Airway Epithelium
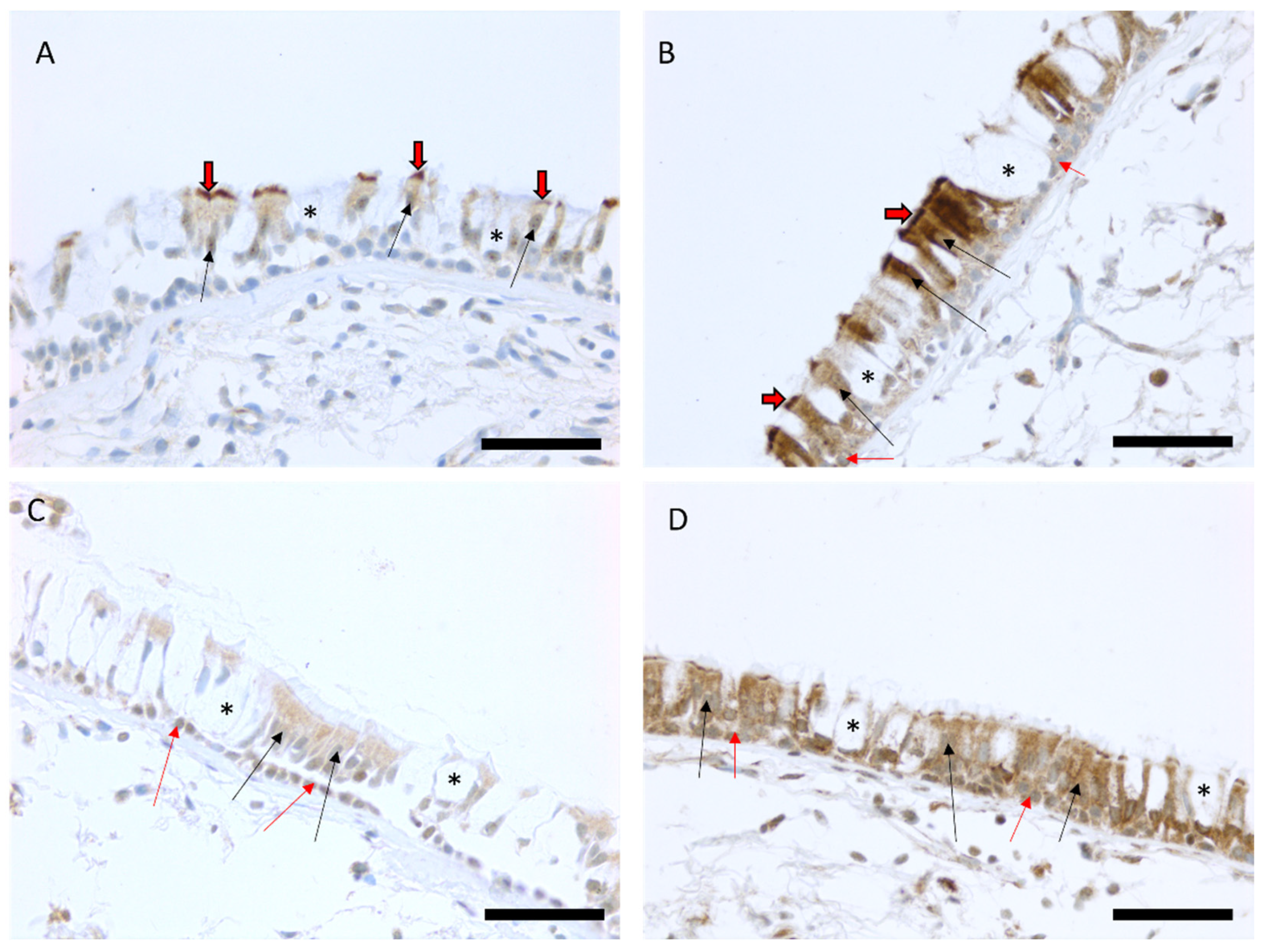
Immunohistochemical MUC3A expression was localized to ciliated cells in 86 out of 96 biopsy samples (90%) and it was found in basal cells in four out of 96 biopsy samples (4%). The intensity of MUC3A expression in ciliated cells varied from weak (100) to strong (300). MUC3B expression was localized to both basal cells and ciliated cells in the majority of the biopsy samples (99% and 99% of biopsies, respectively), and its intensity varied from weak (100) to very strong (400) (Figure 1).
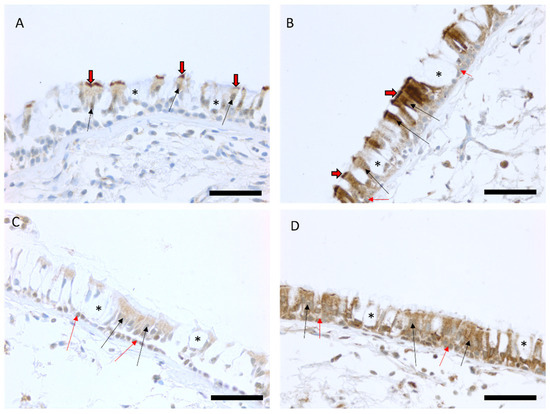
Figure 1.
Representative images of the immunohistochemical stainings for MUC3A and MUC3B proteins in bronchial biopsy samples. Scores of the expression were assessed as negative, faint, moderate, strong, or very strong. Faint expression of MUC3A in the large airways of a smoker with COPD (A) and a strong MUC3A expression in the epithelium of a smoker with normal lung function (B). Faint MUC3B expression in ciliated and basal cells in a never-smoker (C) and a strong expression of MUC3B in epithelial cells in a never-smoker (D). Black arrows show the positive ciliated cells in the epithelium, black/red arrows point to the positive cilia of the cells, red arrows show the positive basal cells, and an asterisk is showing negative goblet cells. Scale bar is 50 µm.
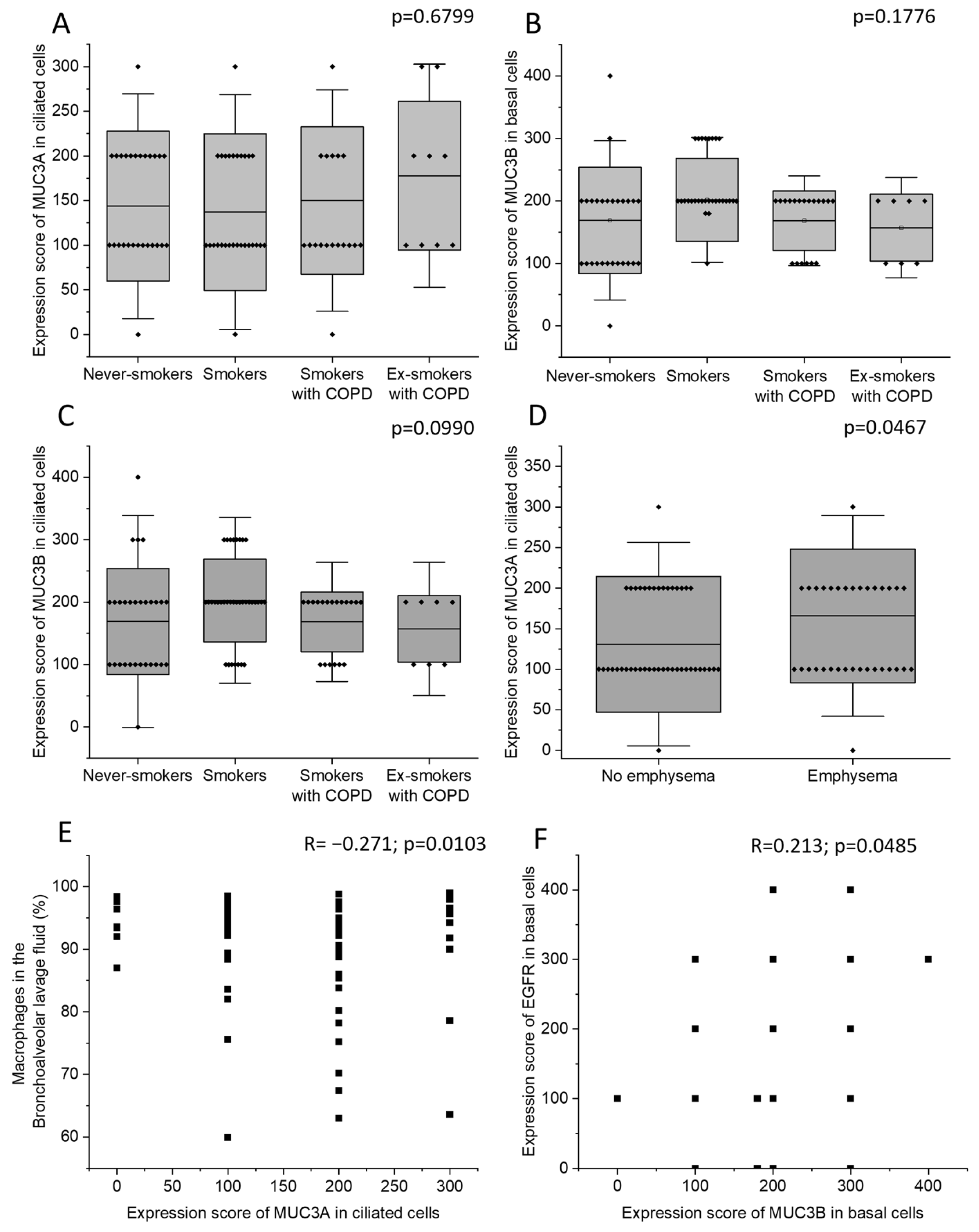
Never-smokers, current smokers with normal lung function, currently smoking COPD patients, and COPD patients who were ex-smokers did not differ in MUC3A expression in ciliated cells. Neither were there any differences in MUC3B expression in basal cells or in ciliated cells (Figure 2A–C). Patients with emphysema had higher MUC3A expression in ciliated cells (Figure 2D), compared to patients without emphysema, but the two groups did not differ in MUC3B expression. COPD diagnosis, smoking history, sex, and chronic bronchitis were neither associated with MUC3A nor with MUC3B expression. However, the four biopsy samples where MUC3A expression was detected in basal cells were all retrieved from female subjects.
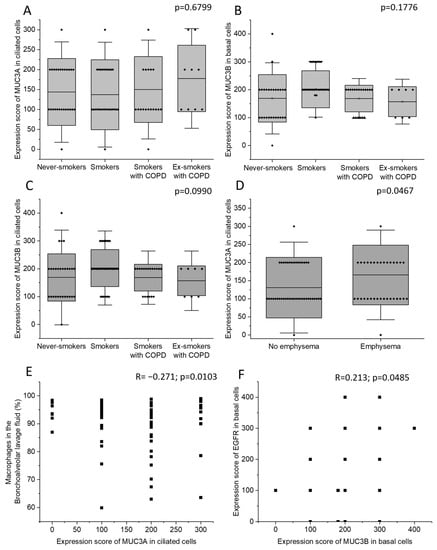
Figure 2.
The immunohistochemical expression of MUC3A in ciliated cells (A), MUC3B in basal cells (B), and MUC3B in ciliated cells (C) in the four study groups: never-smokers, current smokers with normal lung function, currently smoking COPD patients, and COPD patients who were ex-smokers. The immunohistochemical expression of MUC3A in ciliated cells in subjects with and without emphysema (D). Correlation between the immunohistochemical expression of MUC3A in ciliated cells and macrophage percentage in BALF (E). Correlation between the immunohistochemical expression of MUC3B in basal cells and the immunohistochemical EGFR expression in basal cells (F). Bars represent mean and standard deviation. Kruskal–Wallis test in (A–C). Mann–Whitney U Test in (D). Spearman correlation analysis in (E,F). R: Spearman’s rank correlation coefficient.
MUC3A expression in ciliated cells was negatively correlated with macrophage percentage in BALF (Figure 2E). MUC3B immunohistochemical expression in basal cells was positively correlated with epidermal growth factor receptor (EGFR) expression in basal cells (Figure 2F). MUC3B expression in ciliated cells was strongly intercorrelated with MUC3B expression in basal cells (Supplementary Figure S1A). In ciliated cells, MUC3A and MUC3B expression were not correlated (Supplementary Figure S1B). MUC3A and MUC3B immunohistochemical expression did not correlate with expiratory flows.
2.3. MUC3A and MUC3B mRNA Levels across the Study Groups
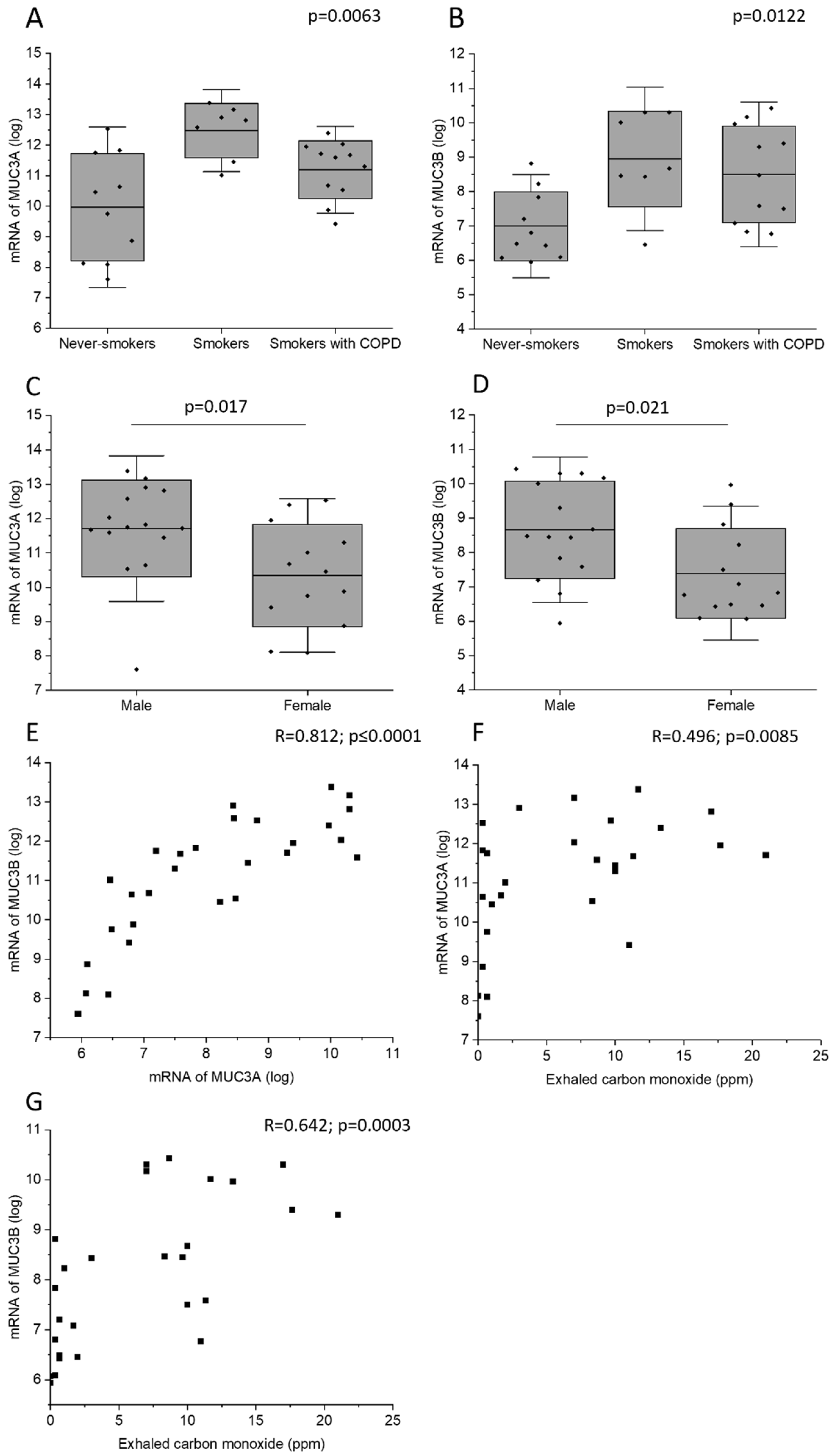
Current smokers with normal lung function had higher MUC3A mRNA level in bronchial brushings than never-smokers (Figure 3A). In addition, current smokers with normal lung function and current smoking COPD patients had higher MUC3B mRNA levels than never-smokers (Figure 3B). Male subjects had higher MUC3A and MUC3B mRNA concentration in bronchial brush samples, as compared with female subjects (Figure 3C,D). Importantly, this difference could have been due to a difference in smoking habits, as 73% of the male subjects who provided mRNA were current smokers, while only 54% of the female subjects who provided mRNA were current smokers (Table 2). However, even when including only current smokers in the analysis, the observed gender difference persisted. MUC3A mRNA level was positively correlated with MUC3B mRNA level (Figure 3E).
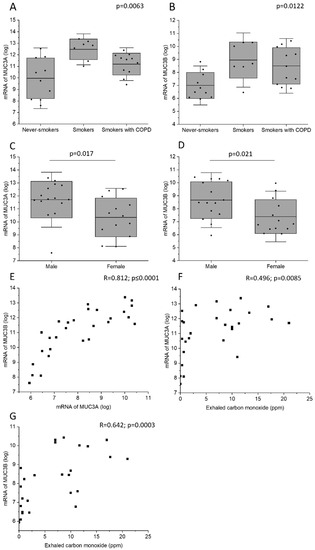
Figure 3.
MUC3A mRNA (A) and MUC3B mRNA (B) in samples acquired by bronchial brushing across the study groups. Due to lack of biospecimens, mRNA samples from study subjects belonging to the study group of ex-smokers with COPD were missing. MUC3A mRNA (C) and MUC3B mRNA (D) in samples acquired by bronchial brushing in male and female study subjects. Correlation between MUC3A mRNA and MUC3B mRNA (E). Correlation between exhaled carbon monoxide and MUC3A mRNA (F) and MUC3B mRNA (G). Kruskal–Wallis test in (A,B). Bars represent mean and standard deviation. Mann–Whitney U Test in (C,D). Spearman correlation analysis in (E–G). R: Spearman’s rank correlation coefficient.

Table 2.
Comparison of the subgroups according to mRNA samples.
In all subjects, MUC3A and MUC3B mRNA levels correlated positively with exhaled carbon monoxide measured before bronchoscopy (Figure 3F,G). However, in the subgroup of current smokers only, MUC3A and MUC3B mRNA did not correlate with exhaled carbon monoxide measured, and COPD diagnosis was associated neither with MUC3A mRNA level nor with MUC3B mRNA level.
2.4. Correlations of MUC3A and MUC3B mRNA Levels with Clinical Parameters and BALF
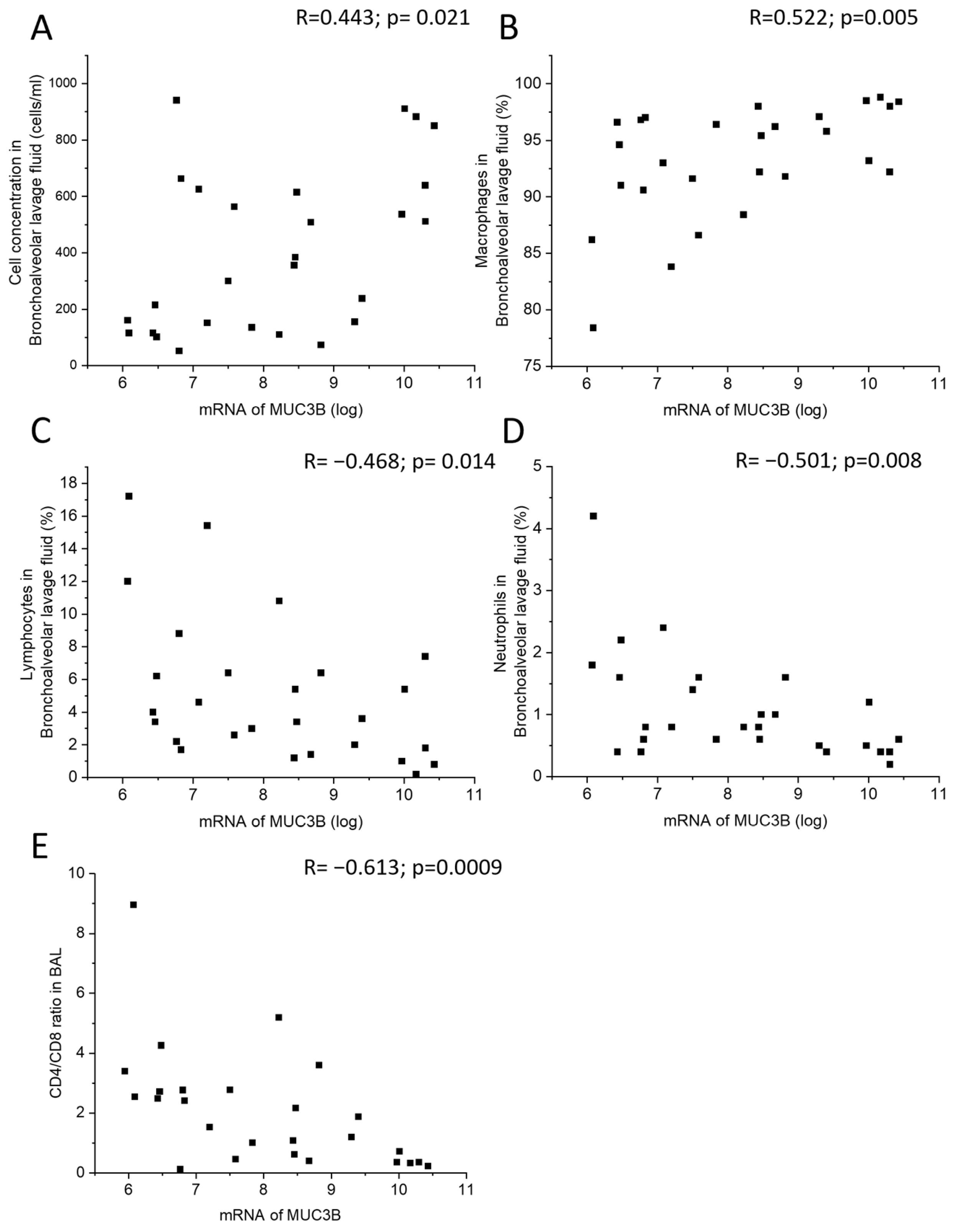
MUC3B mRNA correlated positively with total cell concentration and macrophage percentage in BALF (Figure 4A,B), and negatively with lymphocyte and neutrophil percentage in BALF, as well as with CD4/CD8 T cell ratio in BALF (Figure 4C–E). In the subgroup of current smokers only, MUC3B mRNA correlated negatively with neutrophil percentage in BALF.
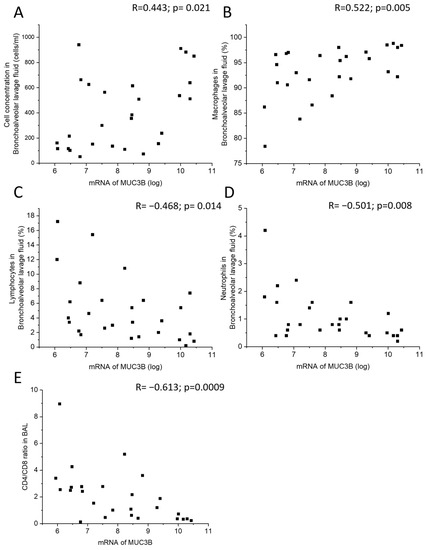
Figure 4.
Correlation between MUC3B mRNA in samples acquired by bronchial brushing and total cell concentration in BALF (A), macrophage percentage (B), lymphocyte percentage (C), neutrophil percentage (D), and CD4/CD8 ratio in BALF (E). Spearman correlation analysis in all. R: Spearman’s rank correlation coefficient.
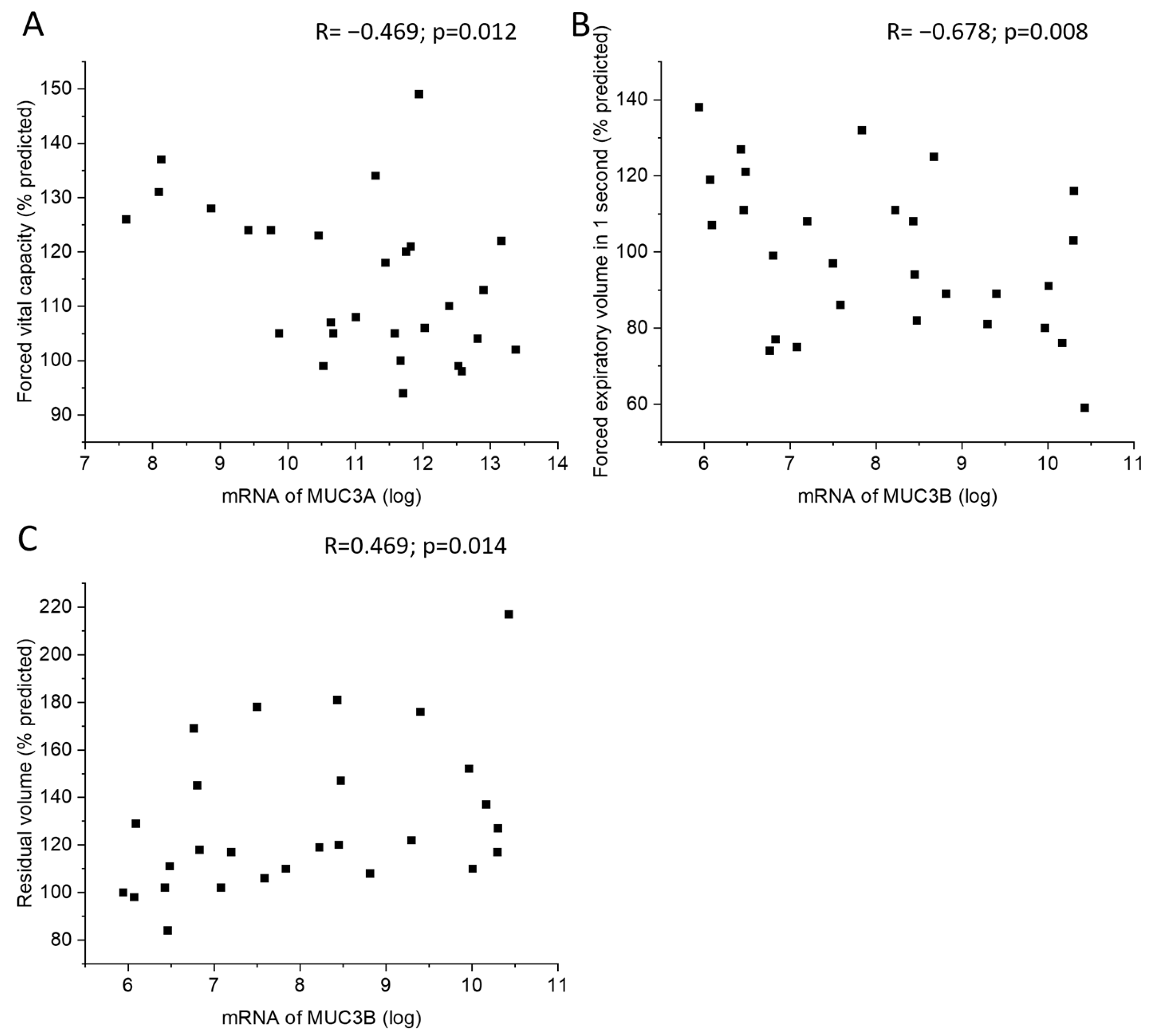
MUC3A mRNA levels in samples acquired by bronchial brushing were negatively correlated with postbronchodilator forced vital capacity (FVC) (Figure 5A). In addition, MUC3B mRNA correlated negatively with postbronchodilator forced expiratory volume in one second (FEV1) and positively with residual volume (RV) (Figure 5B,C). Subjects with emphysema or chronic bronchitis did not differ in MUC3A or MUC3B mRNA levels from subjects without emphysema or chronic bronchitis, respectively.
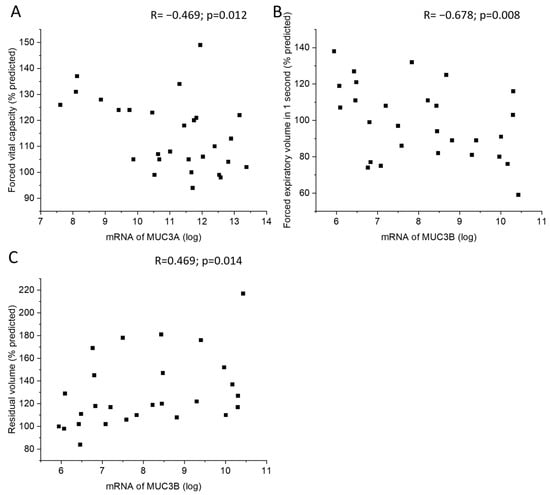
Figure 5.
Correlation between MUC3A mRNA in samples acquired by bronchial brushing and postbronchodilator forced vital capacity in percent predicted (A). Correlation between MUC3B mRNA in samples acquired by bronchial brushing and postbronchodilator forced expiratory volume in 1 s in percent predicted (B) and residual volume in percent of predicted (C), counted according to ECCS reference equations. Spearman correlation analysis in all. R: Spearman’s rank correlation coefficient.
3. Discussion
In the present study, we investigated the immunohistochemical expression and mRNA levels of MUC3A and MUC3B in human large airway epithelium. Here, we showed for the first time that the immunohistochemical expression of both MUC3A and MUC3B in the large airway epithelium was specifically localized to ciliated cells, and MUC3B was also expressed in basal cells. MUC3A and MUC3B mRNA levels were higher in current smokers, regardless of airway obstruction, than in never-smokers. In line with this, they correlated positively with exhaled carbon monoxide and negatively with expiratory air flows. Moreover, patients with emphysema had higher MUC3A protein expression in ciliated cells, compared to patients without emphysema.
Several studies conducted previously failed to detect MUC3 expression in normal adult and fetal respiratory tract by mRNA in situ hybridization and immunohistochemistry [35,36,37]. One unknown type of MUC3 was studied in the bronchial mucosa of COPD patients during exacerbation, but information on the antibodies and concentration was not included in the publication [38]. Here, we showed that MUC3A and MUC3B proteins, detected by immunohistochemistry, were expressed in the large airway epithelium of healthy subjects who had never smoked, as well as in healthy current smokers and COPD patients. The reason for the discrepant results between our study and others is likely due to the fact that our staining protocol and reagents, including antibodies, differed from those used in the previously published studies. We speculate that our method and using the current reagents may be a more sensitive way to detect MUC3 protein expression in human tissues than the previously used methods.
MUC3 was first described as a unique gene but was later separated into two, MUC3A and MUC3B [32,39]. MUC3B has a glycosylated subunit that is longer than that of MUC3A. In the present study, even though MUC3A and MUC3B mRNA levels were both associated with lower expiratory air flows, they behaved in slightly different ways, since they did not show the same pattern in the correlation and group comparison analyses. Moreover, MUC3A and MUC3B immunohistochemical expression in ciliated cells were not correlated. This indicates that MUC3A and MUC3B in large airway epithelium may have two different regulatory pathways, and thus may play different roles in human lung. Our results support the notion that MUC3A and MUC3B indeed are two different mucins, as was appreciated in previous studies [32,39].
In the present study, MUC3B mRNA levels measured by microarray in samples acquired by bronchial brushing were higher in smokers and showed correlations with total cell concentration in BALF. In line with this, MUC3B mRNA levels correlated negatively with the airway obstruction measure FEV1. Taken together, this indicated that MUC3B mRNA in this cohort may be a biomarker associated with variables indicative of worse prognosis. Similarly, the MUC3A mRNA level in samples acquired by bronchial brushing was negatively correlated with postbronchodilator FVC assessed by dynamic spirometry. In addition, patients with emphysema had higher MUC3A expression in ciliated cells, compared with patients without emphysema. Simultaneously, both MUC3A mRNA and MUC3B mRNA were positively correlated with exhaled carbon monoxide being higher in smokers than in never-smokers. It remains unclear whether MUC3A and MUC3B mRNA acted here solely as surrogate markers of smoking exposure, which in its turn contributes to the unfavorable inflammatory profile and worse lung function, or whether MUC3A and MUC3B mRNA may play causal roles. Future studies are warranted to elucidate this.
MUC3A and MUC3B mRNA levels were associated with exhaled carbon monoxide, inflammatory cell pattern in BALF, and lung function. The immunohistochemical MUC3A and MUC3B expressions did not show the same associations, which result is linked to the strong expression profiles of both proteins, since expressions of both MUC3A and MUC3B were quite extensive, and the intensity was scored as moderate or strong in most cases (Figure 1). Nevertheless, exhaled carbon monoxide, inflammatory markers in BALF, and lung function were associated with MUC3A and MUC3B at the mRNA, but not at the protein, level. The results of different kinds of expressions of protein and mRNA levels have been observed also in other studies since a study on lysosomal sulfatases in the airways of COPD patients revealed increased expression of mRNA and decreased expression of proteins [40].
A limitation of this study was the relatively small number of subjects and that we were lacking mRNA data for COPD ex-smokers, as it would have been interesting to see whether MUC3A and MUC3B mRNA levels differed between COPD patients who were ex-smokers and those who were current smokers. In addition, the study design was observational cross-sectional, which enabled us to only create hypotheses based on associations. Future studies with prospective design are needed to evaluate the potential value of mucins as biomarkers of long-term prognosis.
The results of a recent study suggested that female patients with COPD may have a different disease phenotype than male COPD patients [41]. In the present study, female subjects had lower MUC3A and MUC3B mRNA levels in bronchial brush samples than male subjects. However, this difference could have been due to differences in smoking status between the male and female subjects who provided mRNA. Interestingly, all of the four biopsy samples where we are able to detect MUC3A protein expression in basal cells were retrieved from female study participants, and three of those were current smokers. Since the number of subjects in the present study was relatively small and the sex differences might be confounded by smoking, larger studies are needed to evaluate whether female subjects have sputum of different properties than male subjects.
Mucins have recently received significant attention as potential biomarkers. Primarily, mucins have become increasingly recognized as valuable biomarkers in a wide spectrum of cancers, including lung cancer [42,43,44,45,46,47,48,49,50,51,52]. Therefore, mucins present potential molecular targets for cancer therapy [53,54,55,56]. In addition, mucins have been implicated in inflammatory processes. For example, a recent study has found that MUC8 is highly secreted in saliva. This study also found support for the role of MUC8 in the context of inflammatory events and salivary stone formation [57]. In hypersensitivity pneumonitis patients, the T allele of the MUC5B gene predicts lower baseline FVC and its subsequent decrease [58]. Recently, six novel genome-wide significant signals for chronic sputum production have been identified, and these include a chromosome 11 mucin locus containing MUC2, MUC5AC, and MUC5B [59]. In line with these findings, sputum MUC5AC was increased in patients with steroid-untreated mild asthma, and it correlated with markers of airway type 2/eosinophilic inflammation [60]. It is currently being debated whether mucus hypersecretion might aggravate the pathophysiological vicious circle in airway diseases such as COPD and COVID-19, and the literature recommends that future research should focus on targeting mucus hypersecretion as a potential novel therapeutic approach [61,62]. In COPD, total mucin concentration in induced sputum was associated with smoking history, chronic bronchitis, disease severity, and risk of acute exacerbations [63]. A subsequent study showed that increased MUC5AC concentration in the airways was associated with COPD onset and progression [64]. In the studies [60,64], MUC5AC concentration was measured in induced sputum samples, whereas in the present study we assessed the immunohistochemical expression of MUC3A and MUC3B in biopsies of the large airway epithelium. Therefore, the studies are not directly comparable, but interestingly, never-smokers, current smokers with normal lung function, and COPD patients did not differ in MUC3A or MUC3B expression in the collected tissue biopsies. In a state of mucus hypersecretion associated with smoking and COPD, and in light of the results of the studies [60,64], differences in MUC3A and MUC3B expression between the study groups would be expected; however, this was not the case. MUC5AC is a secretory mucin with a relatively well-described role in the respiratory tract, while MUC3A and MUC3B are membrane-tethered mucins that have not yet been studied in the context of pulmonary diseases. This underlines the novelty of the present study and highlights its significance. In the present study, the MUC3A and MUC3B mRNA levels were higher in current smokers compared to never-smokers, and they correlated positively with exhaled carbon monoxide and negatively with expiratory air flows. Additionally, subjects with emphysema had higher MUC3A protein expression in ciliated cells, compared to subjects without emphysema. This indicates that MUC3A and MUC3B may be biomarkers of pulmonary disease and that more research is needed to elucidate the role of these mucins in the respiratory epithelium.
4. Materials and Methods
4.1. Study Subjects
The study design was observational cross-sectional. The subjects included in this study were part of the Karolinska COSMIC (Clinical & Systems Medicine Investigations of Smoking-related Chronic Obstructive Pulmonary Disease) cohort (www.ClinicalTrials.gov/ct2/show/study/NCT02627872 (accessed on 21 August 2023)), as described previously [65,66,67,68,69]. From the general Stockholm population, aged 45–65 years, through advertisements, we recruited healthy never-smokers (n = 40), current smokers with normal lung function (n = 40), and current smokers or ex-smokers with COPD with FEV1/FVC < 0.7 and FEV1 50–100% predicted (n = 38). Exclusion criteria included atopy (defined as positive specific IgE test), asthma, treatment with oral or inhaled glucocorticoids or with antibiotics for a COPD exacerbation within the 3 months prior to study entry, and significant ischemic heart disease or arrhythmia. All the study subjects were recruited during the years 2007–2011.
Current smoking exposure was assessed by exhaled carbon monoxide measurement [70]. All subjects underwent clinical examination, spirometry, computed tomography (CT) scan, and bronchoscopy. Emphysema was evaluated on inspiratory CT scans, by two experienced reviewers (SN and ReKa), and was considered to be present when more than 5% of the whole lung parenchyma was occupied by emphysematous changes. Chronic bronchitis was patient-reported and defined as mucus production for a period of at least three months during at least two years in a row. The study was approved by the Stockholm Regional Ethical Board (Case no 2006-959-31/1; 27 October 2006), and all participants gave informed, written consent.
4.2. Bronchoscopy and Inflammatory Markers
Bronchoscopy was performed as previously described [71,72]. Bronchial mucosal biopsy specimens were taken by use of pulmonary biopsy forceps with smooth-edge jaws (Radial Edge® Biopsy Forceps, Boston Scientific, Boston, MA, USA). Four to six biopsies were taken from each subject, and they were collected from lobar or segmental carinae of the upper lobes or the apical segment of the lower lobes. All biopsies were immediately formalin-fixed and embedded in paraffin. The tissue samples were stained with hematoxylin-eosin and the representativeness of all biopsies was evaluated. Two representative tissue blocks from each subject were selected for immunohistochemical analyses of MUC3A and MUC3B. EGFR immunohistochemical staining results published previously were used also in the present study [20]. The methods of biopsy preparation were performed as previously published [20].
Bronchial brush samples were taken from lobar or segmental bronchi in the lingula segment or the basal lower lobe segment of the left lung. Bronchial brushing was performed by cytology brush (Olympus, BC-202D-2010, Tokyo, Japan) by brushing approximately 10 times back and forth around the circumference. The procedure was repeated five times in various segments, using the same brush, but the brush was gently shaken off between the brushings. The brush samples were taken carefully and a traumatically, for the purpose of only harvesting epithelial cells and not causing bleeding. The brush samples were washed in PBS, and either aliquoted as pellets and frozen at −150 °C until their use in molecular analyses, or prepared as cytospin glasses and frozen at −80 °C until their use in immunocytochemistry analysis.
Bronchoalveolar lavage was performed (5 × 50 mL in the middle lobe) and cells were collected as previously described [71,73]. Total cell concentration, macrophage, lymphocyte, and neutrophil percentage and CD4/CD8 T cell ratio were determined in BALF, as previously described [65,72,74].
4.3. Immunohistochemical Staining and Analysis of Immunohistochemical Protein Expression
Four μm thick sections were cut, deparaffinized with xylene, and rehydrated in a descending ethanol series. The primary antibodies used in the immunostaining were designed for formalin-fixed paraffin-embedded tissues. The antibodies used are summarized in Table 3. MUC3A and EGFR were stained with DAKO REAL EnVision-kit (Dako, Glostrup, Denmark) and MUC3B with FLEX-kit from Dako. All stainings were made according to the manufacturer’s instructions. Before application of the primary antibodies, the sections were heated in a microwave oven in 10 mM citrate buffer, pH 6.0, for 10 min. After overnight incubation at +4 °C with the primary antibody, samples were incubated for 30 min at room temperature with a biotinylated secondary HRP rabbit/mouse antibody (Dako, Envision, or Flex). In all the immunostainings, the color was developed with diaminobenzidine, after which the sections were lightly counterstained with hematoxylin. Representative stainings are shown in Figure 1.

Table 3.
Antibodies used for the immunohistochemical staining.
In the evaluation of immunohistochemical samples, cytosolic expression was considered significant. The intensity of immunostaining was assessed as zero (negative) to four (strong positive) and the extent of the positive staining was estimated from 0% to 100% in each cell type. The score for each antibody correlated total intensity with extent, resulting in a total score with a range between 0 and 400 [20,75,76]. The evaluation was performed blinded to the clinical information of the study subjects and by an experienced researcher (HM), as described previously [20].
4.4. MUC3A and MUC3B mRNA Quantification by Microarray
MUC3A and MUC3B mRNA levels in samples acquired by bronchial brushing were measured by microarray for a subset of individuals, as previously described [77]. RNA was isolated using the NucleoSpin® miRNA kit according to the manufacturer’s instructions (Macherey-Nagel, Düren, Germany). RNA quality and quantity were assessed for concentration and purity by determining UV 260/280 and 230/260 absorbance ratios obtained by the Nanodrop ND-1000 spectrophotometer (Nanodrop, Wilmington, DE, USA). RNA integrity and size distribution were examined by gel electrophoresis on RNA Pico LabChips (Agilent Technologies, Palo Alto, CA, USA) processed on the Agilent 2100 Bioanalyzer. RNA was isolated and amplified using the Low Input Quick Amplification Kit (Agilent Technologies), according to the manufacturer’s protocol, and subsequent Cy3-CTP labeling was performed by using one-color labeling kits (Agilent Technologies). Clean-up of the labeled and amplified probes was performed (Zymo Research Corporation, Irvine, CA, USA). The size distribution and quantity of the amplified product was assessed by Nanodrop. Equal amounts of Cy3-labeled target were hybridized to Agilent human whole-genome 4x44K Ink-jet arrays containing a total of 41,000 probes corresponding to 19,596 entrez genes. Hybridizations were performed at 65 °C for 17 h at a rotation of 10 rpm. Arrays were scanned by using the Agilent microarray G2565BA scanner (Agilent Technologies) with Scan region: Agilent HD (61 × 21.6) and a resolution of 5 µm, TIFF: 16 bit, XDR: 0.10. Raw signal intensities were extracted with Feature Extraction v10.1 software (Agilent Technologies). Flagged outliers were not included in any subsequent analyses. Microarray datasets were normalized using the quantile normalization method according to Bolstad et al. [78]. No background subtraction was performed, and the median feature pixel intensity was used as the raw signal before normalization. All procedures were carried out using functions in the R package limma in Bioconductor [79,80].
Due to the lack of biospecimens (due to a freezer breakdown accident), the total number of mRNA samples was relatively small (Table 1), and we were completely lacking mRNA data on COPD patients who were ex-smokers. Study subjects who provided mRNA samples had lower FEV1 and higher residual volume, as compared with subjects who did not provide mRNA samples (Table 2).
4.5. Statistical Methods
All values are expressed as the mean ± SD. All statistical analyses were done using Tibco Statistica (Version 13) and Graph Pad Prism 9 software (GraphPad, La Jolla, CA, USA). The Kruskal–Wallis-test was used to compare three or more independent groups. For comparison of two independent groups, the unpaired two-tailed Mann–Whitney U Test was used. Correlations between two variables were assessed using the non-parametric Spearman’s rank correlation test. A value of p < 0.05 was considered statistically significant (* p < 0.05, ** p < 0.01, *** p < 0.001).
5. Conclusions
In summary, the present study indicated that immunohistochemical MUC3A and MUC3B expressions were localized to ciliated cells in human large airway epithelium. The results also suggested that MUC3A mRNA and MUC3B mRNA levels in samples acquired from large airways by bronchial brushing were higher in current smokers than in never-smokers. Furthermore, MUC3A and MUC3B mRNA levels correlated positively with exhaled carbon monoxide and negatively with expiratory air flows, which indicates that these mucins may be associated with smoking. Thus, MUC3A and MUC3B may present either a surrogate marker of lung function or play a causal role in the pathophysiology of lung diseases. Future research should focus on exploring the potential roles of MUC3A and MUC3B in human airways in different states of health and disease.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241713546/s1.
Author Contributions
M.C.S. and Å.M.W. designed the study; R.K. (Reza Karimi) and M.C.S. characterized study subjects, performed bronchoscopies, and generated clinical data; M.C.S., R.K. (Riitta Kaarteenaho) and Å.M.W. provided financial support and essential infrastructure; H.M., R.K. (Riitta Kaarteenaho) and E.L.-B. processed bronchial biopsies, evaluated biopsy quality, and performed initial stainings; R.K. (Reza Karimi) and S.N. evaluated HRCT; H.F. and M.M. performed flow cytometric analysis of bronchoalveolar lavage fluid; H.M. and R.K. (Riitta Kaarteenaho) analyzed immunohistochemical stainings; mRNA analyses were performed at the SABRE microarray facility at the University of California, San Francisco; Å.M.W., T.P., H.M., I.K. and C.-X.L. performed data analyses; T.P. and H.M. interpreted data; T.P. drafted the manuscript; H.M. and Å.M.W. contributed to the Methods section of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Jalmari and Rauha Ahokas Foundation, the Research Foundation of the Pulmonary Diseases, the Finnish Anti-Tuberculosis Association, a state subsidy of Oulu University Hospital, the Swedish Heart-Lung Foundation (MCS 20160418, 20180235, 20190320) and King Gustaf V’s and Queen Victoria’s Freemasons’ Foundation (MCS 2010, 2012, 2014), Karolinska Institutet, The Swedish Research Council (2018-00520), and through the Regional Agreement on Medical Training and Clinical Research (ALF) between Stockholm County Council and Karolinska Institutet (MCS 20170393, 20200287). Chuan-Xing Li is the recipient of an ERS-EU RESPIRE2 MCF [7214]-2013.
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Stockholm Regional Ethical Board (Case no 2006-959-31/1; 27 October 2006).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Acknowledgments
We thank Benita Engvall, Margitha Dahl, Helene Blomqvist, Gunnel de Forest, and Riitta Vuento for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| BALF | bronchoalveolar lavage fluid |
| COPD | chronic obstructive pulmonary disease |
| COSMIC | Clinical & Systems Medicine Investigations of Smoking-related Chronic Obstructive Pulmonary Disease |
| CT | computed tomography |
| EGF | epidermal growth factor |
| EGFR | epidermal growth factor receptor |
| FEV1 | forced expiratory volume in one second |
| VC | vital capacity |
| FVC | forced vital capacity |
| mRNA | messenger RNA |
| MUC | mucin |
| RV | residual volume |
References
- Caramori, G.; Casolari, P.; Barczyk, A.; Durham, A.L.; di Stefano, A.; Adcock, I. COPD immunopathology. Semin. Immunopathol. 2016, 38, 497–515. [Google Scholar] [PubMed]
- Shin, S.H.; Kwon, S.O.; Kim, V.; Silverman, E.K.; Kim, T.H.; Kim, D.K.; Hwang, Y.I.; Yoo, K.H.; Kim, W.J.; Park, H.Y. Association of body mass index and COPD exacerbation among patients with chronic bronchitis. Respir. Res. 2022, 23, 52. [Google Scholar]
- Lahousse, L.; Seys, L.J.M.; Joos, G.F.; Franco, O.H.; Stricker, B.H.; Brusselle, G.G. Epidemiology and impact of chronic bronchitis in chronic obstructive pulmonary disease. Eur. Respir. J. 2017, 50, 1602470. [Google Scholar] [PubMed]
- Marshall, D.C.; Al Omari, O.; Goodall, R.; Shalhoub, J.; Adcock, I.M.; Chung, K.F.; Salciccioli, J.D. Trends in prevalence, mortality, and disability-adjusted life-years relating to chronic obstructive pulmonary disease in Europe: An observational study of the global burden of disease database, 2001–2019. BMC Pulm. Med. 2022, 22, 289. [Google Scholar]
- Institute for Health Metrics and Evaluation. Global Burden of Disease Study 2019 (GBD 2019) Results; Institute for Health Metrics and Evaluation: Seattle, WA, USA, 2019. [Google Scholar]
- Terry, P.D.; Dhand, R. The 2023 GOLD Report: Updated Guidelines for Inhaled Pharmacological Therapy in Patients with Stable COPD. Pulm. Ther. 2023, 9, 345–357. [Google Scholar]
- Adeloye, D.; Song, P.; Zhu, Y.; Campbell, H.; Sheikh, A.; Rudan, I.; NIHR RESPIRE Global Respiratory Health Unit. Global, regional, and national prevalence of, and risk factors for, chronic obstructive pulmonary disease (COPD) in 2019: A systematic review and modelling analysis. Lancet Respir. Med. 2022, 10, 447–458. [Google Scholar]
- Vij, N.; Chandramani-Shivalingappa, P.; Van Westphal, C.; Hole, R.; Bodas, M. Cigarette smoke-induced autophagy impairment accelerates lung aging, COPD-emphysema exacerbations and pathogenesis. Am. J. Physiol. Cell Physiol. 2018, 314, C73–C87. [Google Scholar] [PubMed]
- Rathnayake, S.N.H.; Ditz, B.; van Nijnatten, J.; Sadaf, T.; Hansbro, P.M.; Brandsma, C.A.; Timens, W.; van Schadewijk, A.; Hiemstra, P.S.; Ten Hacken, N.H.T.; et al. Smoking induces shifts in cellular composition and transcriptome within the bronchial mucus barrier. Respirology 2023, 28, 132–142. [Google Scholar] [PubMed]
- Kim, V.; Jeong, S.; Zhao, H.; Kesimer, M.; Boucher, R.C.; Wells, J.M.; Christenson, S.A.; Han, M.K.; Dransfield, M.; Paine, R., 3rd; et al. Current smoking with or without chronic bronchitis is independently associated with goblet cell hyperplasia in healthy smokers and COPD subjects. Sci. Rep. 2020, 10, 20133. [Google Scholar]
- Alsharairi, N.A. Antioxidant Intake and Biomarkers of Asthma in Relation to Smoking Status—A Review. Curr. Issues Mol. Biol. 2023, 45, 5099–5117. [Google Scholar]
- Bradicich, M.; Schuurmans, M.M. Smoking status and second-hand smoke biomarkers in COPD, asthma and healthy controls. ERJ Open Res. 2020, 6, 00192–02019. [Google Scholar] [PubMed]
- Rabe, K.F.; Watz, H. Chronic obstructive pulmonary disease. Lancet 2017, 389, 1931–1940. [Google Scholar] [PubMed]
- Fermont, J.M.; Masconi, K.L.; Jensen, M.T.; Ferrari, R.; Di Lorenzo, V.A.P.; Marott, J.M.; Schuetz, P.; Watz, H.; Waschki, B.; Müllerova, H.; et al. Biomarkers and clinical outcomes in COPD: A systematic review and meta-analysis. Thorax 2019, 74, 439–446. [Google Scholar] [PubMed]
- Vivodtzev, I.; Tamisier, R.; Baguet, J.P.; Borel, J.C.; Levy, P.; Pépin, J.L. Arterial stiffness in COPD. Chest 2014, 145, 861–875. [Google Scholar]
- Papaioannou, A.I.; Mazioti, A.; Kiropoulos, T.; Tsilioni, I.; Koutsokera, A.; Tanou, K.; Nikoulis, D.J.; Georgoulias, P.; Zakynthinos, E.; Gourgoulianis, K.I.; et al. Systemic and airway inflammation and the presence of emphysema in patients with COPD. Respir. Med. 2010, 104, 275–282. [Google Scholar] [PubMed]
- Zhang, Y.; Tedrow, J.; Nouraie, M.; Li, X.; Chandra, D.; Bon, J.; Kass, D.J.; Fuhrman, C.R.; Leader, J.K.; Duncan, S.R.; et al. Elevated plasma level of Pentraxin 3 is associ-ated with emphysema and mortality in smokers. Thorax 2021, 76, 335–342. [Google Scholar] [PubMed]
- Ma, J.; Rubin, B.K.; Voynow, J.A. Mucins, Mucus, and Goblet Cells. Chest 2018, 154, 169–176. [Google Scholar]
- O’Donnell, R.A.; Richter, A.; Ward, J.; Angco, G.; Mehta, A.; Rousseau, K.; Swallow, D.M.; Holgate, S.T.; Djukanovic, R.; Davies, D.E.; et al. Expression of ErbB receptors and mucins in the airways of long term current smokers. Thorax 2004, 59, 1032–1040. [Google Scholar]
- Merikallio, H.; Kaarteenaho, R.; Lindén, S.; Padra, M.; Karimi, R.; Li, C.X.; Lappi-Blanco, E.; Wheelock, Å.M.; Sköld, C.M. Smoking-associated increase in mucins 1 and 4 in human airways. Respir. Res. 2020, 21, 239. [Google Scholar]
- Kato, K.; Chang, E.H.; Chen, Y.; Lu, W.; Kim, M.M.; Niihori, M.; Hecker, L.; Kim, K.C. MUC1 contributes to goblet cell metaplasia and MUC5AC expression in response to cigarette smoke in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L82–L90. [Google Scholar]
- Khorasani, A.M.; Mohammadi, B.; Saghafi, M.R.; Mohammadi, S.; Ghaffari, S.; Mirsadraee, M.; Khakzad, M.R. The association between MUC5AC and MUC5B genes expression and remodeling progression in severe neutrophilic asthma: A direct relationship. Respir. Med. 2023, 213, 107260. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Zhang, X.; Xu, Y.; Liu, L.; Sun, X.; Wang, B.; Yu, K.; Zhang, H.; Zhao, X.; Wang, X. BMAL1 regulates MUC1 overexpression in ovalbumin-induced asthma. Mol. Immunol. 2023, 156, 77–84. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Q.; Lu, H.; Wang, C.; Xia, L. Suppression of lncRNA MALAT1 Reduces LPS- or IL-17A-Induced Inflammatory Response in Human Middle Ear Epithelial Cells via the NF-κB Signaling Pathway. BioMed Res. Int. 2021, 2021, 8844119. [Google Scholar] [CrossRef]
- Jono, H.; Xu, H.; Kai, H.; Lim, D.J.; Kim, Y.S.; Feng, X.H.; Li, J.D. Transforming growth factor-beta-Smad signaling pathway negatively regulates nontypeable Haemophilus influenzae-induced MUC5AC mucin transcription via mitogen-activated protein kinase (MAPK) phosphatase-1-dependent inhibition of p38 MAPK. J. Biol. Chem. 2003, 278, 27811–27819. [Google Scholar] [CrossRef]
- Fischer, B.; Voynow, J. Neutrophil elastase induces MUC5AC messenger RNA expression by an oxidant-dependent mechanism. Chest 2000, 117, 317S–320S. [Google Scholar] [CrossRef]
- Wu, M.; Lai, T.; Jing, D.; Yang, S.; Wu, Y.; Li, Z.; Wu, Y.; Zhao, Y.; Zhou, L.; Chen, H.; et al. Epithelium-derived IL17A promotes cigarette smoke-induced inflammation and mucus hyperproduction. Am. J. Respir. Cell Mol. Biol. 2021, 65, 581–592. [Google Scholar] [PubMed]
- Nishida, Y.; Yagi, H.; Ota, M.; Tanaka, A.; Sato, K.; Inoue, T.; Yamada, S.; Arakawa, N.; Ishige, T.; Kobayashi, Y.; et al. Oxidative stress induces MUC5AC expression through mitochondrial damage-dependent STING signaling in human bronchial epithelial cells. FASEB BioAdvances 2023, 5, 171–181. [Google Scholar] [CrossRef]
- Deng, J.; Tang, H.; Zhang, Y.; Yuan, X.; Ma, N.; Hu, H.; Wang, X.; Liu, C.; Xu, G.; Li, Y.; et al. House dust mite-induced endo-plasmic reticulum stress mediates MUC5AC hypersecretion via TBK1 in airway epithelium. Exp. Lung Res. 2023, 49, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Huang, L.Z.X.; Mykytyn, A.Z.; Wang, C.; Lamers, M.M.; Westendorp, B.; Wubbolts, R.W.; van Putten, J.P.M.; Bosch, B.J.; Haagmans, B.L.; et al. Glycosylated extracellu-lar mucin domains protect against SARS-CoV-2 infection at the respiratory surface. PLoS Pathog. 2023, 19, e1011571. [Google Scholar] [CrossRef]
- Arruda, B.L.; Kanefsky, R.A.; Hau, S.; Janzen, G.M.; Anderson, T.K.; Vincent Baker, A.L. Mucin 4 is a cellular biomarker of necrotizing bronchiolitis in influenza A virus infection. Microbes Infect. 2023, 105169. [Google Scholar] [CrossRef]
- Pratt, W.S.; Crawley, S.; Hicks, J.; Ho, J.; Nash, M.; Kim, Y.S.; Gum, J.R.; Swallow, D.M. Multiple transcripts of MUC3: Evidence for two genes, MUC3A and MUC3B. Biochem. Biophys. Res. Commun. 2000, 275, 916–923. [Google Scholar] [CrossRef]
- Kyo, K.; Muto, T.; Nagawa, H.; Lathrop, G.M.; Nakamura, Y. Associations of distinct variants of the intestinal mucin gene MUC3A with ulcerative colitis and Crohn’s disease. J. Hum. Genet. 2001, 46, 5–20. [Google Scholar] [CrossRef]
- Dohrman, A.; Tsuda, T.; Escudier, E.; Cardone, M.; Jany, B.; Gum, J.; Kim, Y.; Basbaum, C. Distribution of lysozyme and mucin (MUC2 and MUC3) mRNA in human bronchus. Exp. Lung Res. 1994, 20, 367–380. [Google Scholar] [CrossRef] [PubMed]
- López-Ferrer, A.; Curull, V.; Barranco, C.; Garrido, M.; Lloreta, J.; Real, F.X.; de Bolós, C. Mucins as Differentiation Markers in Bronchial Epithelium. Am. J. Respir. Cell Mol. Biol. 2001, 24, 22–29. [Google Scholar] [CrossRef]
- Copin, M.C.; Devisme, L.; Buisine, M.P.; Marquette, C.H.; Wurtz, A.; Aubert, J.P.; Gosselin, B.; Porchet, N. From normal respiratory mucosa to epidermoid carcinoma: Expression of human mucin genes. Int. J. Cancer 2000, 86, 162–168. [Google Scholar] [CrossRef]
- Copin, M.C.; Buisine, M.P.; Devisme, L.; Leroy, X.; Escande, F.; Gosselin, B.; Aubert, J.P.; Porchet, N. Normal respiratory mucosa, precursor lesions and lung carcinomas: Differential expression of human mucin genes. Front. Biosci. 2001, 6, D1264–D1275. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, S.; Dorofieiev, A. The Content of Mucin MUC-2, -3 and -4 Antigens in the Bronchial Mucosa Membrane of Chronic Obstructive Pulmonary Disease Patients dur-ing Acute Exacerbation—Initial Report. Adv. Respir. Med. 2017, 85, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Gum, J.R.; Hicks, J.W.; Crawley, S.C.; Dahl, C.M.; Yang, S.C.; Roberton, A.M.; Kim, Y.S. Initiation of transcription of the MUC3A human intestinal mucin from a TATA-less promoter and comparison with the MUC3B amino terminus. J. Biol. Chem. 2003, 278, 49600–49609. [Google Scholar] [CrossRef]
- Weidner, J.; Jogdand, P.; Jarenbäck, L.; Åberg, I.; Helihel, D.; Ankerst, J.; Westergren-Thorsson, G.; Bjermer, L.; Erjefält, J.S.; Tufvesson, E. Expression, activity and localization of lysosomal sulfatases in Chronic Obstructive Pulmonary Disease. Sci. Rep. 2019, 9, 1991. [Google Scholar] [CrossRef]
- Perez, T.A.; Castillo, E.G.; Ancochea, J.; Pastor Sanz, M.T.; Almagro, P.; Martínez-Camblor, P.; Miravitlles, M.; Rodríguez-Carballeira, M.; Navarro, A.; Lamprecht, B.; et al. Sex differences between women and men with COPD: A new analysis of the 3CIA study. Respir. Med. 2020, 171, 106105. [Google Scholar] [CrossRef]
- Machado, G.C.; Ferrer, V.P. MUC17 mutations and methylation are associated with poor prognosis in adult-type diffuse glioma patients. J. Neurol. Sci. 2023, 452, 120762. [Google Scholar] [CrossRef]
- Czogalla, B.; Dötzer, K.; Sigrüner, N.; von Koch, F.E.; Brambs, C.E.; Anthuber, S.; Frangini, S.; Burges, A.; Werner, J.; Mahner, S.; et al. Combined Expression of HGFR with Her2/neu, EGFR, IGF1R, Mucin-1 and Integrin α2β1 Is Associated with Aggressive Epithelial Ovarian Cancer. Biomedicines 2022, 10, 2694. [Google Scholar] [CrossRef]
- Kumar, A.R.; Devan, A.R.; Nair, B.; Nair, R.R.; Nath, L.R. Biology, Significance and Immune Signaling of Mucin 1 in Hepatocellular Carcinoma. Curr. Cancer Drug Targets 2022, 22, 725–740. [Google Scholar]
- Lin, S.; Tian, C.; Li, J.; Liu, B.; Ma, T.; Chen, K.; Gong, W.; Wang, J.M.; Huang, J. Differential MUC22 expression by epigenetic alterations in human lung squamous cell carcinoma and adenocarcinoma. Oncol. Rep. 2021, 45, 78. [Google Scholar] [CrossRef] [PubMed]
- Taverna, C.; Maggiore, G.; Cannavicci, A.; Bonomo, P.; Santucci, M.; Franchi, A. Immunohistochemical profiling of mucins in sinonasal adenocarcinomas. Pathol. Res. Pract. 2019, 215, 152439. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, T.; Okudela, K.; Nakashima, Y.; Mitsui, H.; Matsumura, M.; Umeda, S.; Arai, H.; Baba, T.; Suzuki, T.; Koike, C.; et al. Unique expression profiles of mucin proteins in interstitial pneumonia-associated lung adenocarcinomas. Histol. Histopathol. 2019, 34, 1243–1254. [Google Scholar]
- Tu, J.; Tang, M.; Li, G.; Chen, L.; Wang, Y.; Huang, Y. Expression of Mucin Family Proteins in Non-Small-Cell Lung Cancer and its Role in Evaluation of Prognosis. J. Oncol. 2022, 2022, 4181658. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Matsushima, M.; Carreras, J.; Hirabayashi, K.; Kikuti, Y.; Ueda, T.; Kaneko, M.; Fujimoto, R.; Sano, M.; Teramura, E.; et al. Whole-genome copy number and immunohistochemical analyses on surgically resected intracholecystic papillary neoplasms. Pathol. Int. 2021, 71, 823–830. [Google Scholar] [CrossRef]
- Robinson, L.; van Heerden, M.B.; Ker-Fox, J.G.; Hunter, K.D.; van Heerden, W.F.P. Expression of Mucins in Salivary Gland Mucoepidermoid Carcinoma. Head Neck Pathol. 2021, 15, 491–502. [Google Scholar] [CrossRef]
- Yokoyama, S.; Hamada, T.; Higashi, M.; Matsuo, K.; Maemura, K.; Kurahara, H.; Horinouchi, M.; Hiraki, T.; Sugimoto, T.; Akahane, T.; et al. Predicted Prognosis of Patients with Pancreatic Cancer by Machine Learning. Clin. Cancer Res. 2020, 26, 2411–2421. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zuo, D.; Liu, T.; Yin, L.; Li, C.; Wang, L. Prognostic and Clinicopathological Significance of MUC Family Members in Colorectal Cancer: A Systematic Review and Meta-Analysis. Gastroenterol. Res. Pract. 2019, 2019, 2391670. [Google Scholar] [CrossRef] [PubMed]
- Ratan, C.; Cicily, K.D.D.; Nair, B.; Nath, L.R. MUC Glycoproteins: Potential Biomarkers and Molecular Targets for Cancer Therapy. Curr. Cancer Drug Targets 2021, 21, 132–152. [Google Scholar] [CrossRef] [PubMed]
- Behl, A.; Sarwalia, P.; Kumar, S.; Behera, C.; Mintoo, M.J.; Datta, T.K.; Gupta, P.N.; Chhillar, A.K. Codelivery of Gemcita-bine and MUC1 Inhibitor Using PEG-PCL Nanoparticles for Breast Cancer Therapy. Mol. Pharm. 2022, 19, 2429–2440. [Google Scholar] [CrossRef]
- Lee, D.H.; Choi, S.; Park, Y.; Jin, H.S. Mucin1 and Mu-cin16: Therapeutic Targets for Cancer Therapy. Pharmaceuticals 2021, 14, 1053. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.; Chaudhary, S.; Lakshmanan, I.; Aithal, A.; Kisling, S.G.; Sorrell, C.; Marimuthu, S.; Gautam, S.K.; Rauth, S.; Kshirsagar, P.; et al. Chi-meric antibody targeting unique epitope on onco-mucin16 reduces tumor burden in pancreatic and lung malignancies. NPJ Precis. Oncol. 2023, 7, 74. [Google Scholar] [CrossRef]
- Schicht, M.; Reichle, A.; Schapher, M.; Garreis, F.; Kleinsasser, B.; Aydin, M.; Sahin, A.; Iro, H.; Paulsen, F. The Translational Role of MUC8 in Salivary Glands: A Potential Biomarker for Salivary Stone Disease? Diagnostics 2021, 11, 2330. [Google Scholar] [CrossRef]
- Lewandowska, K.B.; Szturmowicz, M.; Lechowicz, U.; Franczuk, M.; Błasińska, K.; Falis, M.; Błaszczyk, K.; Sobiecka, M.; Wyrostkiewicz, D.; Siemion-Szcześniak, I.; et al. The Presence of T Allele (rs35705950) of the MUC5B Gene Predicts Lower Baseline Forced Vital Capacity and Its Subsequent Decline in Patients with Hypersensitivity Pneumonitis. Int. J. Mol. Sci. 2023, 24, 10748. [Google Scholar] [CrossRef]
- Packer, R.J.; Shrine, N.; Hall, R.; Melbourne, C.A.; Thompson, R.; Williams, A.T.; Paynton, M.L.; Guyatt, A.L.; Allen, R.J.; Lee, P.H.; et al. Genome-wide association study of chronic sputum production implicates loci in-volved in mucus production and infection. Eur. Respir. J. 2023, 61, 2201667. [Google Scholar] [CrossRef]
- Tajiri, T.; Matsumoto, H.; Jinnai, M.; Kanemitsu, Y.; Nagasaki, T.; Iwata, T.; Inoue, H.; Nakaji, H.; Oguma, T.; Ito, I.; et al. Pathophysiological relevance of sputum MUC5AC and MUC5B levels in patients with mild asthma. Allergol. Int. 2022, 71, 193–199. [Google Scholar] [CrossRef]
- Shah, B.K.; Singh, B.; Wang, Y.; Xie, S.; Wang, C. Mucus Hypersecretion in Chronic Obstruc-tive Pulmonary Disease and Its Treatment. Mediat. Inflamm. 2023, 2023, 8840594. [Google Scholar] [CrossRef]
- Kumar, S.S.; Binu, A.; Devan, A.R.; Nath, L.R. Mucus targeting as a plausible approach to improve lung function in COVID-19 patients. Med. Hypotheses 2021, 156, 110680. [Google Scholar] [CrossRef]
- Kesimer, M.; Ford, A.A.; Ceppe, A.; Radicioni, G.; Cao, R.; Davis, C.W.; Doerschuk, C.M.; Alexis, N.E.; Anderson, W.H.; Henderson, A.G.; et al. Airway Mucin Concentration as a Marker of Chronic Bronchitis. N. Engl. J. Med. 2017, 377, 911–922. [Google Scholar] [CrossRef]
- Radicioni, G.; Ceppe, A.; Ford, A.A.; Alexis, N.E.; Barr, R.G.; Bleecker, E.R.; Christenson, S.A.; Cooper, C.B.; Han, M.K.; Hansel, N.N.; et al. Airway mucin MUC5AC and MUC5B concentrations and the initiation and progression of chronic obstructive pulmonary disease: An analysis of the SPIROMICS cohort. Lancet Respir. Med. 2021, 9, 1241–1254. [Google Scholar] [CrossRef]
- Forsslund, H.; Yang, M.; Mikko, M.; Karimi, R.; Nyrén, S.; Engvall, B.; Grunewald, J.; Merikallio, H.; Kaarteenaho, R.; Wahlström, J.; et al. Gender differences in the T-cell profiles of the airways in COPD patients associated with clinical phenotypes. Int. J. Chronic Obstr. Pulm. Dis. 2016, 12, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Balgoma, D.; Yang, M.; Sjödin, M.; Snowden, S.; Karimi, R.; Levänen, B.; Merikallio, H.; Kaarteenaho, R.; Palmberg, L.; Larsson, K.; et al. Linoleic acid-derived lipid mediators increase in a female-dominated subphenotype of COPD. Eur. Respir. J. 2016, 47, 1645–1656. [Google Scholar] [CrossRef] [PubMed]
- Karimi, R.; Tornling, G.; Forsslund, H.; Mikko, M.; Wheelock, Å.M.; Nyrén, S.; Sköld, C.M. Differences in regional air trapping in current smokers with normal spirometry. Eur. Respir. J. 2017, 49, 1600345. [Google Scholar] [CrossRef]
- Karimi, R.; Tornling, G.; Forsslund, H.; Mikko, M.; Wheelock, Å.M.; Nyrén, S.; Sköld, C.M. Lung density on high resolution computer tomography (HRCT) reflects degree of inflammation in smokers. Respir. Res. 2014, 15, 23. [Google Scholar] [CrossRef]
- Kohler, M.; Sandberg, A.; Kjellqvist, S.; Thomas, A.; Karimi, R.; Nyrén, S.; Eklund, A.; Thevis, M.; Sköld, C.M.; Wheelock, Å.M. Gender differences in the bronchoalveolar lavage cell proteome of patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2013, 131, 743–751. [Google Scholar] [CrossRef]
- Sandberg, A.S.; Sköld, C.M.; Grunewald, J.; Eklund, A.; Wheelock, Å.M. Assessing recent smoking status by measuring exhaled carbon monoxide levels. PLoS ONE 2011, 6, e28864. [Google Scholar] [CrossRef] [PubMed]
- Karimi, R.; Tornling, G.; Grunewald, J.; Eklund, A.; Sköld, C.M. Cell recovery in bronchoalveolar lavage fluid in smokers is dependent on cumulative smoking history. PLoS ONE 2012, 7, e34232. [Google Scholar] [CrossRef]
- Forsslund, H.; Mikko, M.; Karimi, R.; Grunewald, J.; Wheelock, Å.M.; Wahlström, J.; Sköld, C.M. Distribution of T-cell subsets in BAL fluid of patients with mild to moderate COPD depends on current smoking status and not airway obstruction. Chest 2014, 145, 711–722. [Google Scholar] [CrossRef]
- Löfdahl, J.M.; Cederlund, K.; Nathell, L.; Eklund, A.; Sköld, C.M. Bronchoalveolar lavage in COPD: Fluid recovery correlates with the degree of emphysema. Eur. Respir. J. 2005, 25, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Kohler, M.; Heyder, T.; Forsslund, H.; Garberg, H.K.; Karimi, R.; Grunewald, J.; Berven, F.S.; Nyrén, S.; Sköld, C.M. Proteomic profiling of lung immune cells reveals dysregulation of phagocytotic pathways in female-dominated molecular COPD phenotype. Respir. Res. 2018, 19, 39. [Google Scholar] [CrossRef]
- Lee, H.J.; Xu, X.; Choe, G.; Chung, D.H.; Seo, J.W.; Lee, J.H.; Lee, C.T.; Jheon, S.; Sung, S.W.; Chung, J.H. Protein overexpression and gene amplification of epidermal growth factor receptor in nonsmall cell lung carcinomas: Comparison of four commercially available antibodies by immunohistochemistry and fluorescence in situ hybridization study. Lung Cancer 2010, 68, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Lappi-Blanco, E.; Mäkinen, J.M.; Lehtonen, S.; Karvonen, H.; Sormunen, R.; Laitakari, K.; Johnson, S.; Mäkitaro, R.; Bloigu, R.; Kaarteenaho, R. Mucin-1 correlates with survival, smoking status, and growth patterns in lung adenocarcinoma. Tumor Biol. 2016, 37, 13811–13820. [Google Scholar] [CrossRef]
- Li, C.X.; Wheelock, C.E.; Sköld, C.M.; Wheelock, Å.M. Integration of multi-omics datasets enables molecular classification of COPD. Eur. Respir. J. 2018, 51, 1701930. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Åstrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).