Whole-Genome Sequencing Identified New Structural Variations in the DMD Gene That Cause Duchenne Muscular Dystrophy in Two Girls
Abstract
1. Introduction
2. Results
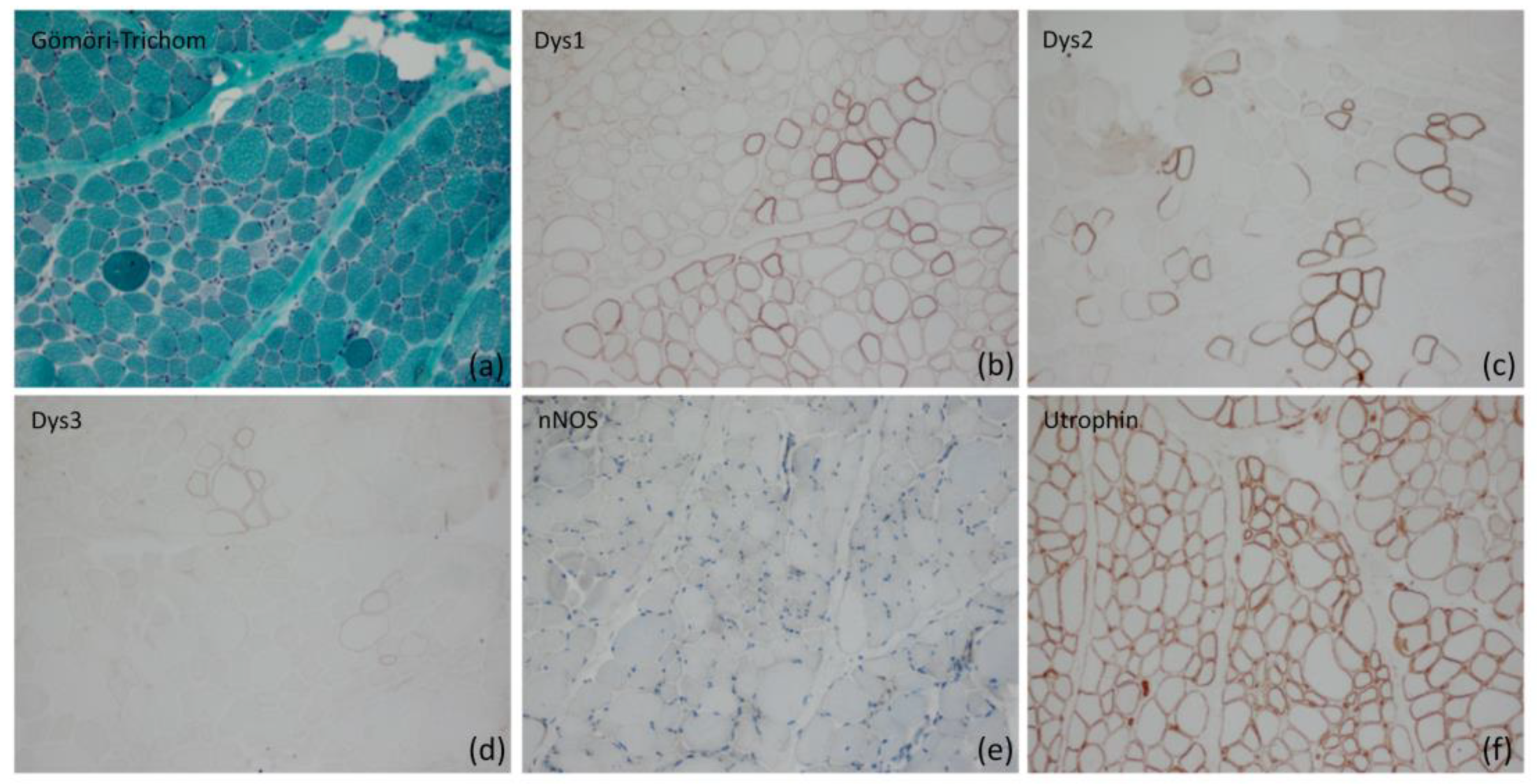
2.1. Morphological Analysis of Skeletal Muscle
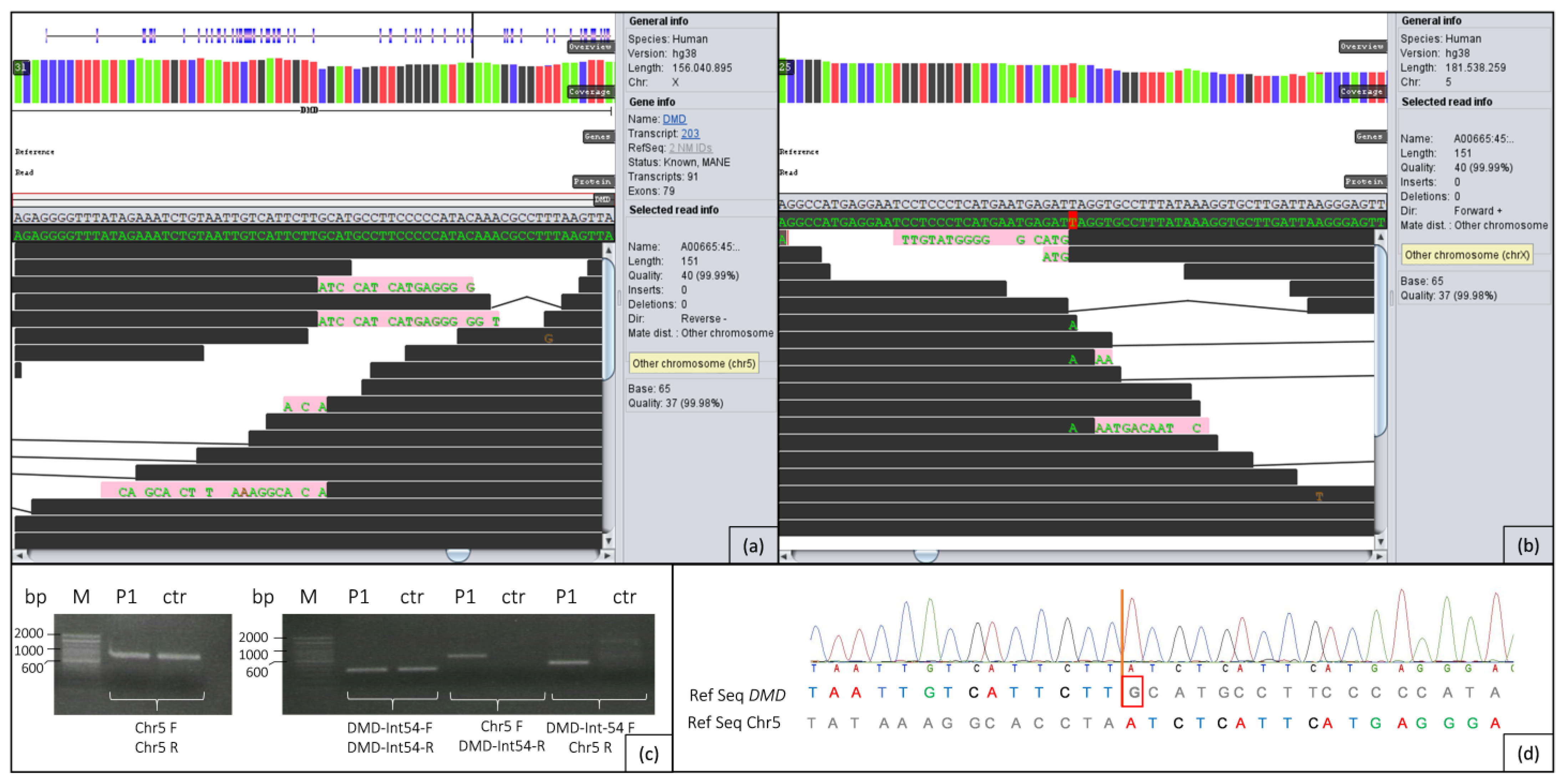
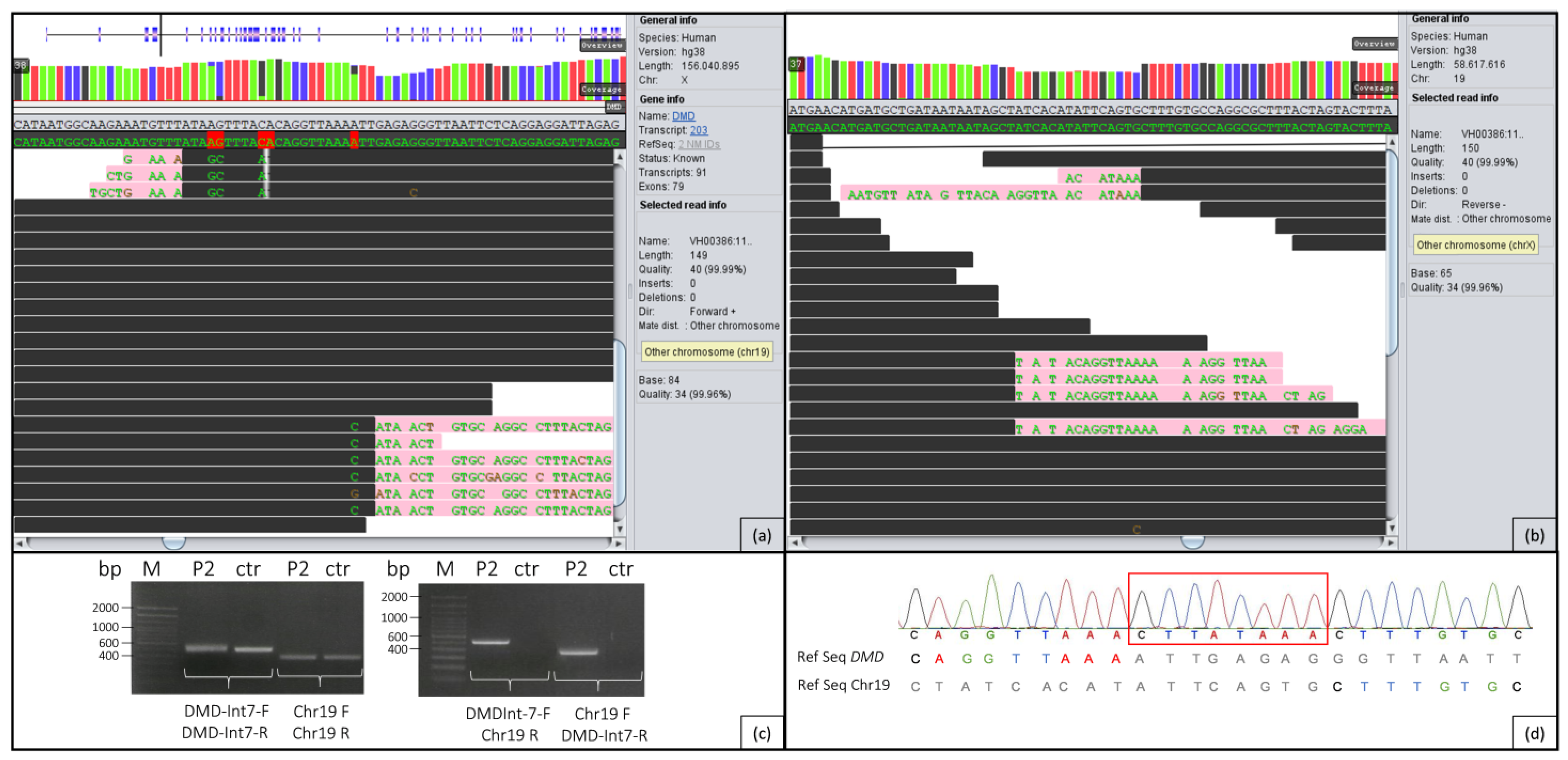
2.2. Genome Analysis
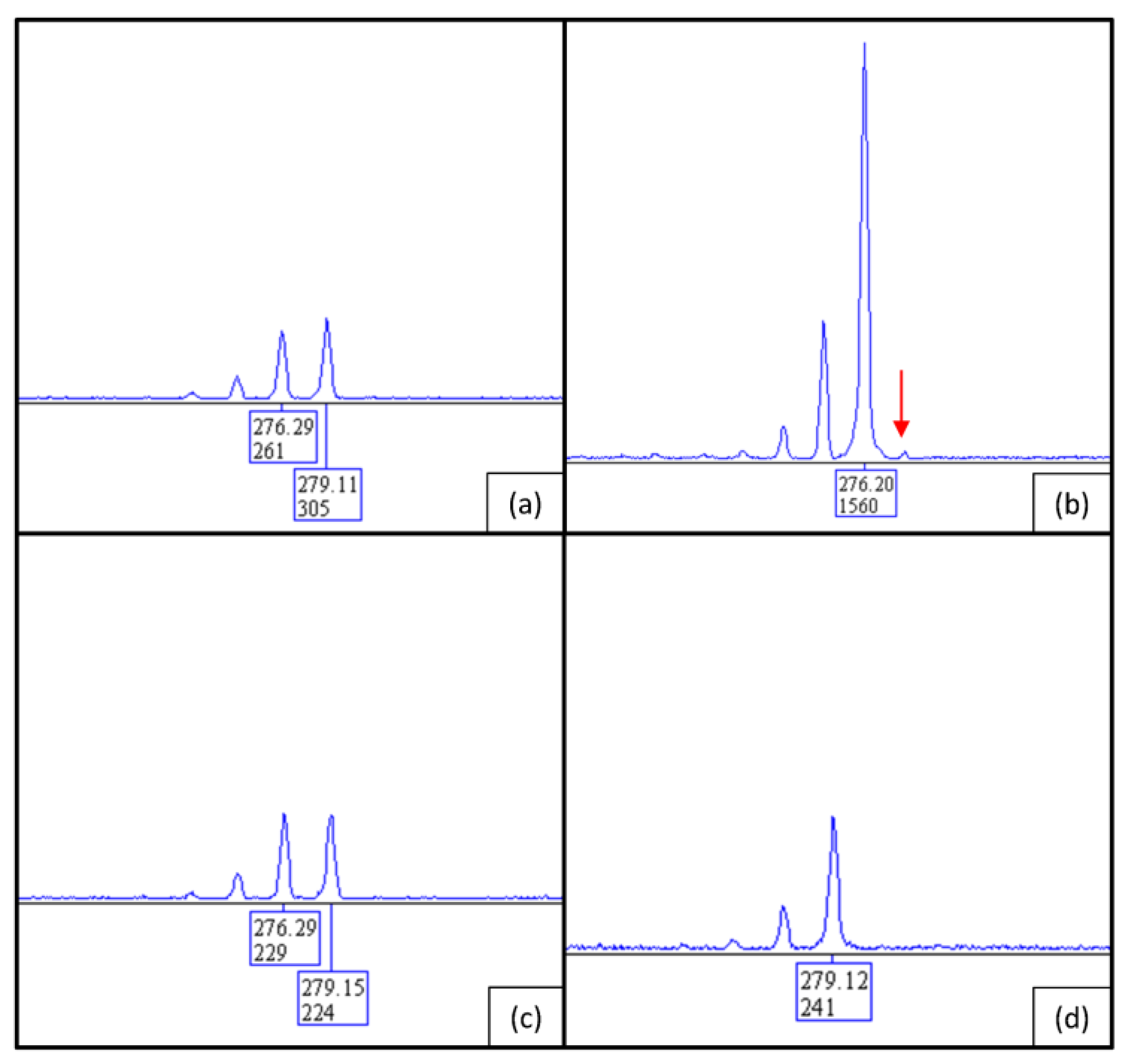
2.3. PCR/Sanger Analysis
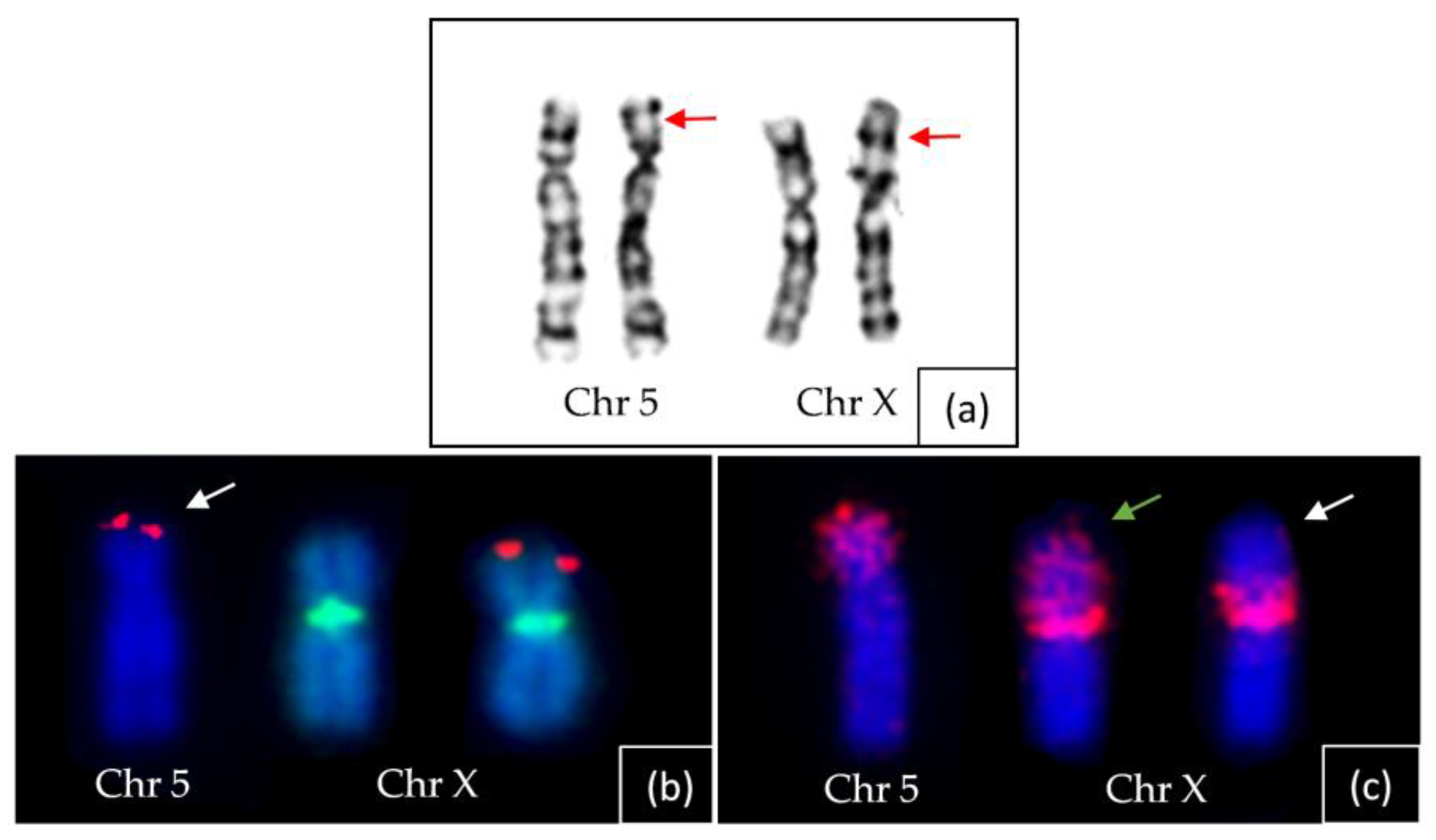
2.4. Chromosome Analysis
2.5. X-Inactivation Analysis
3. Discussion
4. Materials and Methods
4.1. Case Reports
4.2. X-Inactivation Analysis
4.3. DMD Analysis by MLPA and NGS
4.4. Sanger Sequencing and Cytogenetics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bushby, K.; Bourke, J.; Bullock, R.; Eagle, M.; Gibson, M.; Quinby, J. The multidisciplinary management of Duchenne muscular dystrophy. Curr. Paediatr. 2005, 15, 292–300. [Google Scholar] [CrossRef]
- Verma, S.; Anziska, Y.; Cracco, J. Review of Duchenne muscular dystrophy (DMD) for the pediatricians in the community. Clin. Pediatr. 2010, 49, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Hoogerwaard, E.; Bakker, E.; Ippel, P.; Oosterwijk, J.; Majoor-Krakauer, D.; Leschot, N.; Van Essen, A.; Brunner, H.; van der Wouw, P.; Wilde, A.; et al. Signs and symptoms of Duchenne muscular dystrophy and Becker muscular dystrophy among carriers in the Netherlands: A cohort study. Lancet 1999, 353, 2116–2119. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Xie, Y.; Bhandari, V.; Chen, G.; Dang, Y.; Liao, H.; Zhang, J.; Lan, D. Clinical and genetic characteristics of female dystrophinopathy carriers. Mol. Med. Rep. 2019, 19, 3035–3044. [Google Scholar] [CrossRef] [PubMed]
- Brioschi, S.; Gualandi, F.; Scotton, C.; Armaroli, A.; Bovolenta, M.; Falzarano, M.S.; Sabatelli, P.; Selvatici, R.; D’amico, A.; Pane, M.; et al. Genetic characterization in symptomatic female DMD carriers: Lack of relationship between X-inactivation, transcriptional DMD allele balancing and phenotype. BMC Med. Genet. 2012, 13, 73. [Google Scholar] [CrossRef]
- Sahashi, K.; Ibi, T.; Suoh, H.; Nakao, N.; Tashiro, M.; Marui, K.; Arahata, K.; Sugita, H. Immunostaining of dystrophin and utrophin in skeletal muscle of dystrophinopathies. Intern. Med. 1994, 33, 277–283. [Google Scholar] [CrossRef]
- Falzarano, M.S.; Scotton, C.; Passarelli, C.; Ferlini, A. Duchenne muscular dystrophy: From diagnosis to therapy. Molecules 2015, 20, 18168–18184. [Google Scholar] [CrossRef]
- Fratter, C.; Dalgleish, R.; Allen, S.K.; Santos, R.; Abbs, S.; Tuffery-Giraud, S.; Ferlini, A. EMQN best practice guidelines for genetic testing in dystrophinopathies. Eur. J. Hum. Genet. 2020, 28, 1141–1159. [Google Scholar] [CrossRef]
- Dent, K.; Dunn, D.; von Niederhausern, A.; Aoyagi, A.; Kerr, L.; Bromberg, M.; Hart, K.; Tuohy, T.; White, S.; Dunnen, J.D.; et al. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am. J. Med. Genet. A 2005, 134, 295–298. [Google Scholar] [CrossRef]
- Okubo, M.; Minami, N.; Goto, K.; Noguchi, S.; Mitsuhashi, S.; Nishino, I. Genetic diagnosis of Duchenne/Becker muscular dystrophy using next-generation sequencing: Validation analysis of DMD mutations. Neuromuscul. Disord. 2016, 26, S96. [Google Scholar] [CrossRef]
- Muntoni, F.; Torelli, S.; Ferlini, A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003, 2, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Zaum, A.-K.; Stüve, B.; Gehrig, A.; Kölbel, H.; Schara, U.; Kress, W.; Rost, S. Deep intronic variants introduce DMD pseudoexon in patient with muscular dystrophy. Neuromuscul. Disord. 2017, 27, 631–634. [Google Scholar] [CrossRef]
- Xie, Z.; Sun, C.; Liu, Y.; Yu, M.; Zheng, Y.; Meng, L.; Wang, G.; Cornejo-Sanchez, D.M.; Bharadwaj, T.; Yan, J.; et al. Practical approach to the genetic diagnosis of unsolved dystrophinopathies: A stepwise strategy in the genomic era. J. Med. Genet. 2020, 58, 743–751. [Google Scholar] [CrossRef]
- Xie, Z.; Tang, L.; Xie, Z.; Sun, C.; Shuai, H.; Zhou, C.; Liu, Y.; Yu, M.; Zheng, Y.; Meng, L.; et al. Splicing characteristics of dystrophin pseudoexons and identification of a novel pathogenic intronic variant in the DMD gene. Genes 2020, 11, 1180. [Google Scholar] [CrossRef]
- Gonorazky, H.; Liang, M.; Cummings, B.; Lek, M.; Micallef, J.; Hawkins, C.; Basran, R.; Cohn, R.; Wilson, M.D.; MacArthur, D.; et al. RNAseq analysis for the diagnosis of muscular dystrophy. Ann. Clin. Transl. Neurol. 2016, 3, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Saito-Ohara, F.; Fukuda, Y.; Ito, M.; Agarwala, K.L.; Hayashi, M.; Matsuo, M.; Imoto, I.; Yamakawa, K.; Nakamura, Y.; Inazawa, J. The Xq22 inversion breakpoint interrupted a novel Ras-like GTPase gene in a patient with Duchenne muscular dystrophy and profound mental retardation. Am. J. Hum. Genet. 2002, 71, 637–645. [Google Scholar] [CrossRef]
- Zaum, A.; Nanda, I.; Kress, W.; Rost, S. Detection of pericentric inversion with breakpoint in DMD by whole genome sequencing. Mol. Genet. Genomic Med. 2022, 10, e2028. [Google Scholar] [CrossRef]
- Bach, E.J.; Wolf, B.; Oldenburg, J.; Müller, C.R.; Rost, S. Identification of deep intronic variants in 15 haemophilia A patients by next generation sequencing of the whole factor VIII gene. Thromb. Haemost. 2015, 114, 757–767. [Google Scholar]
- Nallamilli, B.R.R.; Chaubey, A.; Valencia, C.A.; Stansberry, L.; Behlmann, A.M.; Ma, Z.; Mathur, A.; Shenoy, S.; Ganapathy, V.; Jagannathan, L.; et al. A single NGS-based assay covering the entire genomic sequence of the DMD gene facilitates diagnostic and newborn screening confirmatory testing. Hum. Mutat. 2021, 42, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Sun, C.; Zhang, S.; Liu, Y.; Yu, M.; Zheng, Y.; Meng, L.; Acharya, A.; Cornejo-Sanchez, D.M.; Wang, G.; et al. Long-read whole-genome sequencing for the genetic diagnosis of dystrophinopathies. Ann. Clin. Transl. Neurol. 2020, 7, 2041–2046. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.; Hubley, R.; Green, P. 2013–2015. RepeatMasker Open-4.0. Available online: http://www.repeatmasker.org (accessed on 25 May 2021).
- Shaffer, L.G.; Schultz, R.A.; Ballif, B.C. The use of new technologies in the detection of balanced translocations in hematologic disorders. Curr. Opin. Genet. Dev. 2012, 22, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, A.A.; Meloni-Ehrig, A.M. Cytogenetics and genetics of human cancer: Methods and accomplishments. Cancer Genet. Cytogenet. 2010, 203, 102–126. [Google Scholar] [CrossRef] [PubMed]
- Van Bakel, I.; Holt, S.; Craig, I.; Boyd, Y. Sequence analysis of the breakpoint regions of an X;5 translocation in a female with Duchenne muscular dystrophy. Am. J. Hum. Genet. 1995, 57, 329–336. [Google Scholar]
- Nevin, N.C.; Hughes, A.E.; Calwell, M.; Lim, J.H. Duchenne muscular dystrophy in a female with a translocation involving Xp21. J. Med. Genet. 1986, 23, 171–173. [Google Scholar] [CrossRef][Green Version]
- Zatz, M.; Vianna-Morgante, A.M.; Campos, P.; Diament, A.J. Translocation (X;6) in a female with Duchenne muscular dystrophy: Implications for the localisation of the DMD locus. J. Med. Genet. 1981, 18, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Trippe, H.; Wieczorek, S.; Kötting, J.; Kress, W.; Schara, U. Xp21/A translocation: A rarely considered genetic cause for manifesting carriers of duchenne muscular dystrophy. Neuropediatrics 2014, 45, 333–335. [Google Scholar]
- Szűcs, Z.; Pinti, É.; Haltrich, I.; Szén, O.P.; Nagy, T.; Barta, E.; Méhes, G.; Bidiga, L.; Török, O.; Ujfalusi, A.; et al. An ultra-rare manifestation of an X-linked recessive disorder: Duchenne muscular dystrophy in a female patient. Int. J. Mol. Sci. 2022, 23, 13076. [Google Scholar] [CrossRef]
- Meitinger, T.; Boyd, Y.; Anand, R.; Craig, I. Mapping of Xp21 translocation breakpoints in and around the DMD gene by pulsed field gel electrophoresis. Genomics 1988, 3, 315–322. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Liu, J.; Chen, X.C.; Liu, X.; Wang, C.Z.; He, X.Y. Whole dystrophin gene analysis by next-generation sequencing: A comprehensive genetic diagnosis of Duchenne and Becker muscular dystrophy. Mol. Genet. Genom. 2014, 289, 1013–1021. [Google Scholar] [CrossRef]
- Fujii, K.; Minami, N.; Hayashi, Y.; Nishino, I.; Nonaka, I.; Tanabe, Y.; Takanashi, J.-I.; Kohno, Y. Homozygous female Becker muscular dystrophy. Am. J. Med. Genet. A 2009, 149A, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Quan, F.; Janas, J.; Toth-Fejel, S.; Johnson, D.B.; Wolford, J.K.; Popovich, B.W. Uniparental disomy of the entire X chromosome in a female with Duchenne muscular dystrophy. Am. J. Hum. Genet. 1997, 60, 160–165. [Google Scholar] [PubMed]
- Chelly, J.; Marlhens, F.; Le Marec, B.; Jeanpierre, M.; Lambert, M.; Hamard, G.; Dutrillaux, B.; Kaplan, J.-C. De novo DNA microdeletion in a girl with Turner syndrome and Duchenne muscular dystrophy. Hum. Genet. 1986, 74, 193–196. [Google Scholar] [CrossRef]
- Katayama, Y.; Tran, V.K.; Hoan, N.T.; Zhang, Z.; Goji, K.; Yagi, M.; Takeshima, Y.; Saiki, K.; Nhan, N.T.; Matsuo, M. Co-occurrence of mutations in both dystrophin- and androgen-receptor genes is a novel cause of female Duchenne muscular dystrophy. Human. Genet. 2006, 119, 516–519. [Google Scholar] [CrossRef] [PubMed]
- Verellen-Dumoulin, C.; Freund, M.; De Meyer, R.; Laterre, C.; Frédéric, J.; Thompson, M.W.; Markovic, V.D.; Worton, R.G. Expression of an X-linked muscular-dystrophy in a female due to translocation involving Xp21 and non-random inactivation of the normal X-chromosome. Human. Genet. 1984, 67, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, M.; Yorifuji, T.; Mituyoshi, I. Skewed X inactivation in manifesting carriers of Duchenne muscular dystrophy. Clin. Genet. 1998, 53, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Van den Veyver, I.B. Skewed X inactivation in X-linked disorders. Semin. Reprod. Med. 2001, 19, 183–191. [Google Scholar] [CrossRef]
- Mattei, M.G.; Mattei, J.F.; Ayme, S.; Giraud, F. X-Autosome Translocations—Cytogenetic Characteristics and Their Consequences. Human. Genet. 1982, 61, 295–309. [Google Scholar] [CrossRef]
- Sisdelli, L.; Vidi, A.C.; Moysés-Oliveira, M.; Di Battista, A.; Bortolai, A.; Moretti-Ferreira, D.; Dias da Silva, M.R.; Melaragno, M.I.; Carvalheira, G. Incorporation of 5-ethynyl-2′-deoxyuridine (EdU) as a novel strategy for identification of the skewed X inactivation pattern in balanced and unbalanced X-rearrangements. Hum. Genet. 2016, 135, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Bodrug, S.; Holden, J.; Ray, P.; Worton, R. Molecular analysis of X-autosome translocations in females with Duchenne muscular dystrophy. EMBO J. 1991, 10, 3931–3939. [Google Scholar] [CrossRef]
- Robinson, D.O.; Boyd, Y.; Cockburn, D.; Collinson, M.N.; Craig, I.; Jacobs, P.A. The parental origin of de novo X-autosome translocations in females with Duchenne muscular dystrophy revealed by M27 beta methylation analysis. Genet. Res. 1990, 56, 135–140. [Google Scholar] [CrossRef]
- Allen, R.C.; Zoghbi, H.Y.; Moseley, A.B.; Rosenblatt, H.M.; Belmont, J.W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am. J. Hum. Genet. 1992, 51, 1229–1239. [Google Scholar] [PubMed]
- Wolf, B.; Kuonen, P.; Dandekar, T.; Atlan, D. DNAseq Workflow in a diagnostic context and an example of a user friendly implementation. Biomed. Res. Int. 2015, 2015, 403497. [Google Scholar] [CrossRef] [PubMed]



| Patient 1 | Patient 2 | |
|---|---|---|
| age (years) | 9 | 14 |
| age of onset (months) | 6 | 6 |
| first symptoms | motor developmental delay, hypotonia | motor developmental delay, hypotonia, failure to thrive |
| independent walking (months) | 24 | 20 |
| kardiac involvement | no | no |
| mental involvement | yes | no |
| CK levels | 12,000–20,000 U/L | 7000–12,000 U/L |
| muscle biopsy | dystrophic pattern; reduction of dystrophin using dys1–3 antibodies, upregulation of utrophin | dystrophic picture with fibrous necrosis, connective tissue proliferation, absent staining of dystrophin with dys1–3 antibodies |
| clinical findings | proximal weakness, positive Gowers sign; walking distance, reduced; impaired cognitive skills, IQ 61 | proximal weakness, walking distance 100 m; |
| Primer Name 1 | Sequence | Primer Name 2 | Sequence | Fragment Size (bp) |
|---|---|---|---|---|
| DMD-Int54-F | 5′-GGTTTGTCTCAAATTTGGCAGT-3′, | DMD-Int54-R | 5′-TGGTGCACCTAGTGAACTCC-3′, | 362 |
| Chr5-F | 5′-TTAGGTGGGAACATGGCATT-3′ | Chr5-R | 5′-TGATCTCACAGGCTCACAGC-3′ | 798 |
| DMD-Int54-F | Ibid. | Chr5-R | Ibid. | 472 |
| Chr5-F | Ibid. | DMD-Int54-R | Ibid. | 687 |
| DMD-Int7-F | 5′-GAGTGAATGCTTTCAGACTTGG-3′ | DMD- Int7-R | 5′-ATTTTCAACTGCAGAGTTTGACT-3′ | 488 |
| Chr19-F | 5′-GGGTTACTAATGTGTTTATTCATCTG-3′ | Chr19-R | 5′-CTGACCCTTTGAGCCTTGTC-3′ | 372 |
| DMD-Int7-F | Ibid. | Chr19-R | Ibid. | 511 |
| Chr19-F | Ibid. | DMD- Int7-R | Ibid. | 357 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pluta, N.; von Moers, A.; Pechmann, A.; Stenzel, W.; Goebel, H.-H.; Atlan, D.; Wolf, B.; Nanda, I.; Zaum, A.-K.; Rost, S. Whole-Genome Sequencing Identified New Structural Variations in the DMD Gene That Cause Duchenne Muscular Dystrophy in Two Girls. Int. J. Mol. Sci. 2023, 24, 13567. https://doi.org/10.3390/ijms241713567
Pluta N, von Moers A, Pechmann A, Stenzel W, Goebel H-H, Atlan D, Wolf B, Nanda I, Zaum A-K, Rost S. Whole-Genome Sequencing Identified New Structural Variations in the DMD Gene That Cause Duchenne Muscular Dystrophy in Two Girls. International Journal of Molecular Sciences. 2023; 24(17):13567. https://doi.org/10.3390/ijms241713567
Chicago/Turabian StylePluta, Natalie, Arpad von Moers, Astrid Pechmann, Werner Stenzel, Hans-Hilmar Goebel, David Atlan, Beat Wolf, Indrajit Nanda, Ann-Kathrin Zaum, and Simone Rost. 2023. "Whole-Genome Sequencing Identified New Structural Variations in the DMD Gene That Cause Duchenne Muscular Dystrophy in Two Girls" International Journal of Molecular Sciences 24, no. 17: 13567. https://doi.org/10.3390/ijms241713567
APA StylePluta, N., von Moers, A., Pechmann, A., Stenzel, W., Goebel, H.-H., Atlan, D., Wolf, B., Nanda, I., Zaum, A.-K., & Rost, S. (2023). Whole-Genome Sequencing Identified New Structural Variations in the DMD Gene That Cause Duchenne Muscular Dystrophy in Two Girls. International Journal of Molecular Sciences, 24(17), 13567. https://doi.org/10.3390/ijms241713567






