Imprinted Long Non-Coding RNAs in Mammalian Development and Disease
Abstract
1. Introduction
2. Regulatory lncRNAs at Developmental Imprinted Gene Domains
2.1. The Igf2-H19 Imprinted Domain
2.2. The Igf2r Imprinted Domain
2.3. The Gnas Imprinted Domain
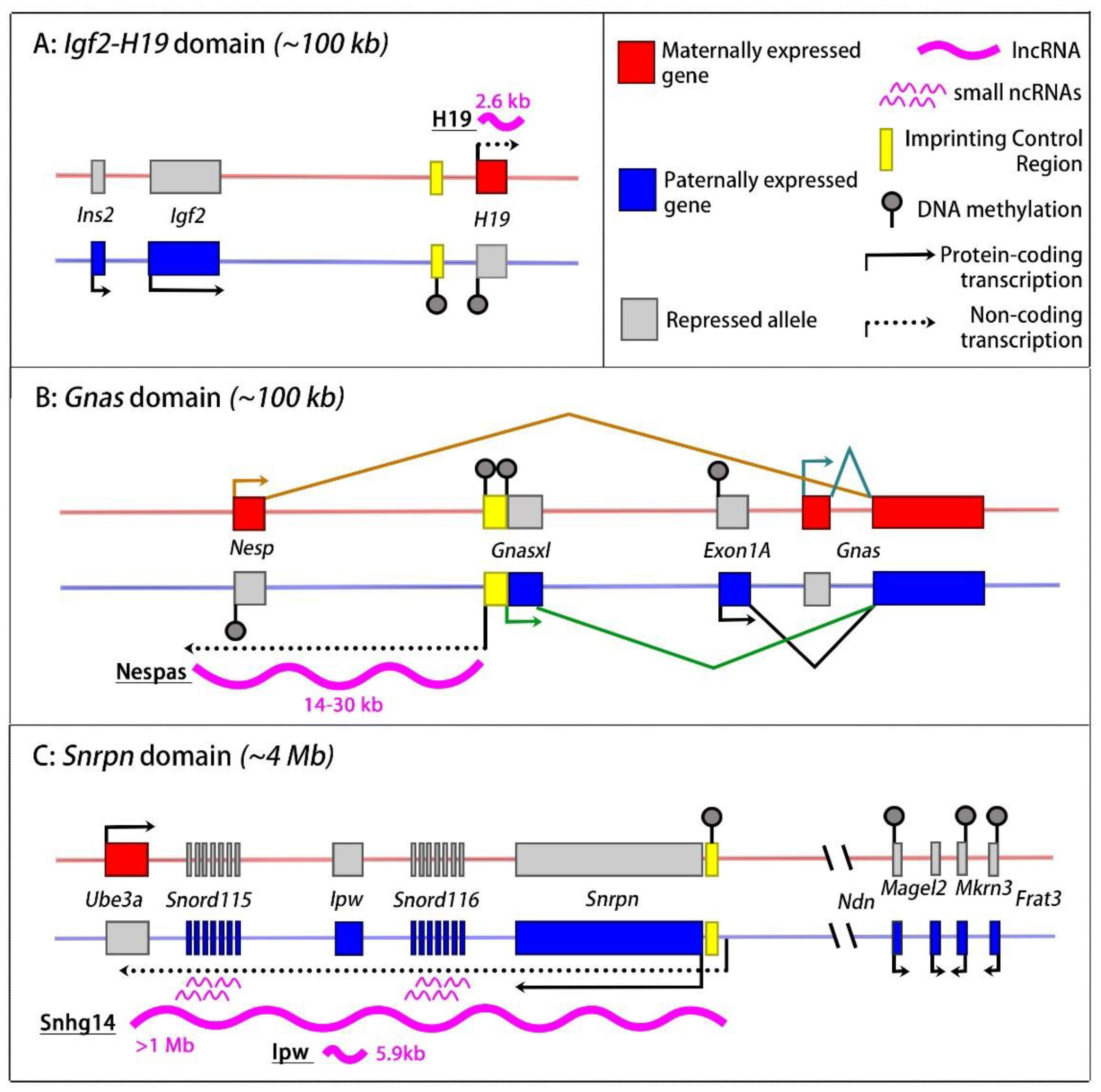
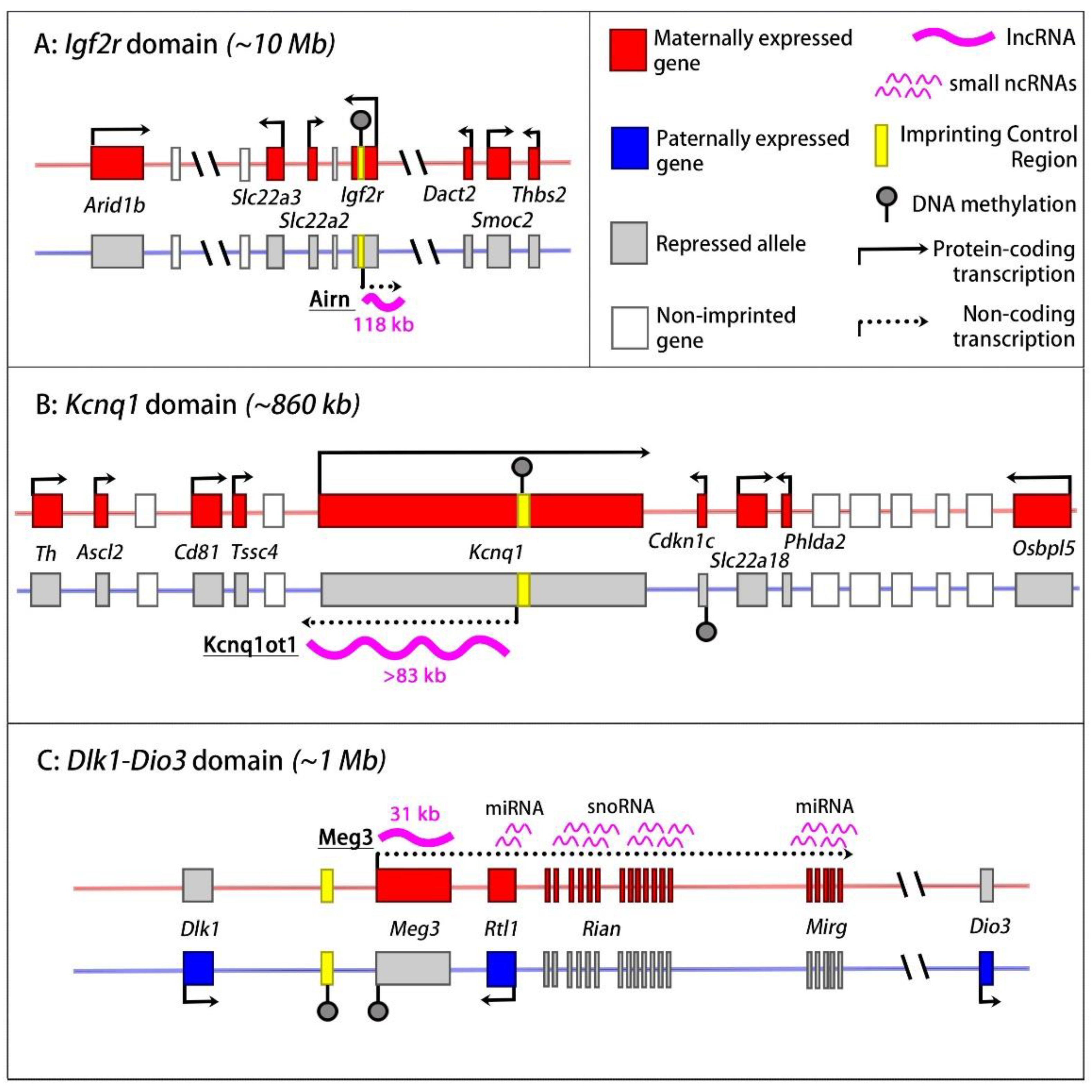
2.4. The Kcnq1 Domain
2.5. The Dlk1-Dio3 Imprinted Domain
2.6. The Snrpn Domain
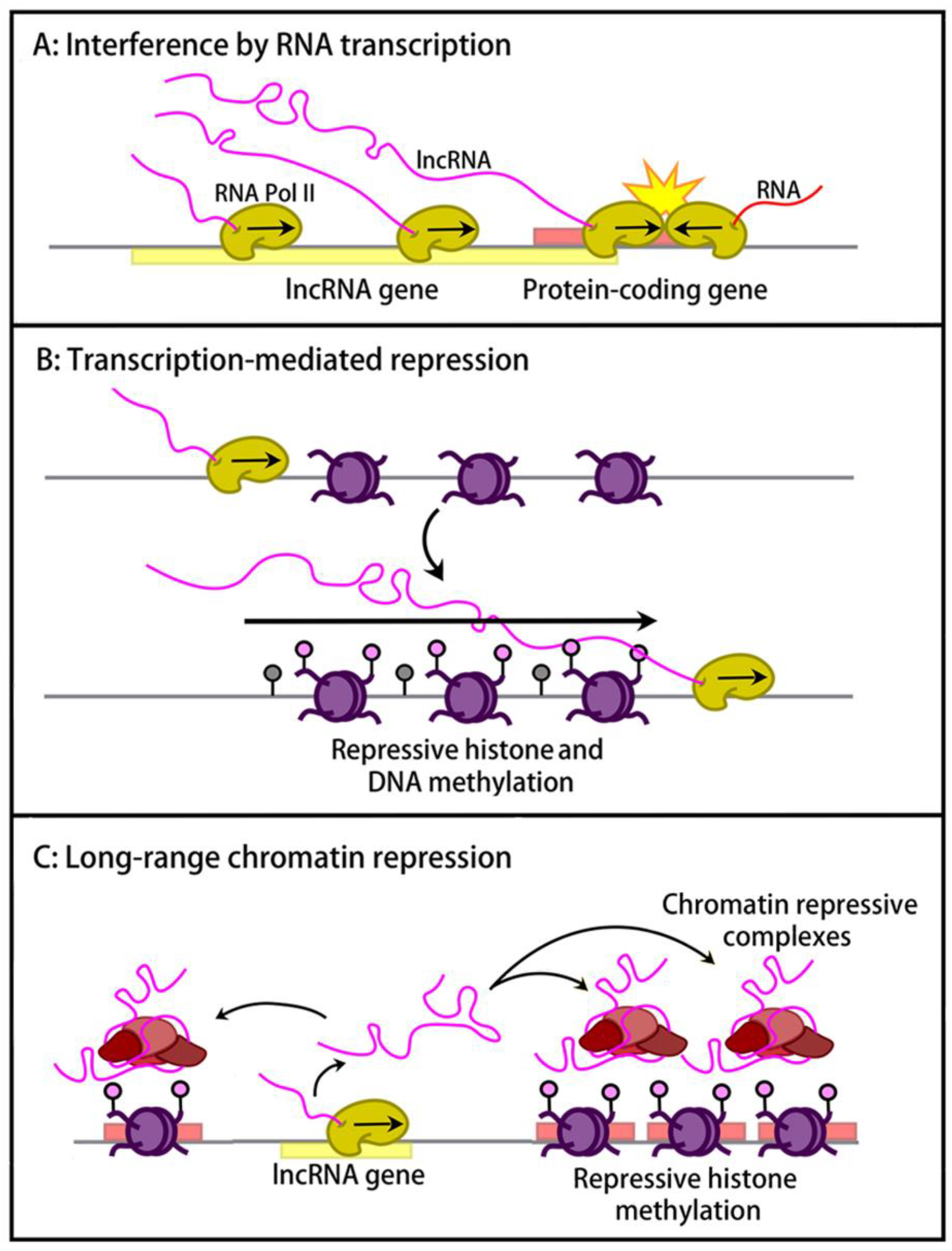
3. Cis-Regulatory Effects of lncRNAs at Imprinted Domains
3.1. lncRNA-Transcription-Mediated Interference and Chromatin Repression
3.2. lncRNA-Mediated Long-Range Chromatin Repression
3.3. Putative Structural Roles in cis of Imprinted lncRNAs
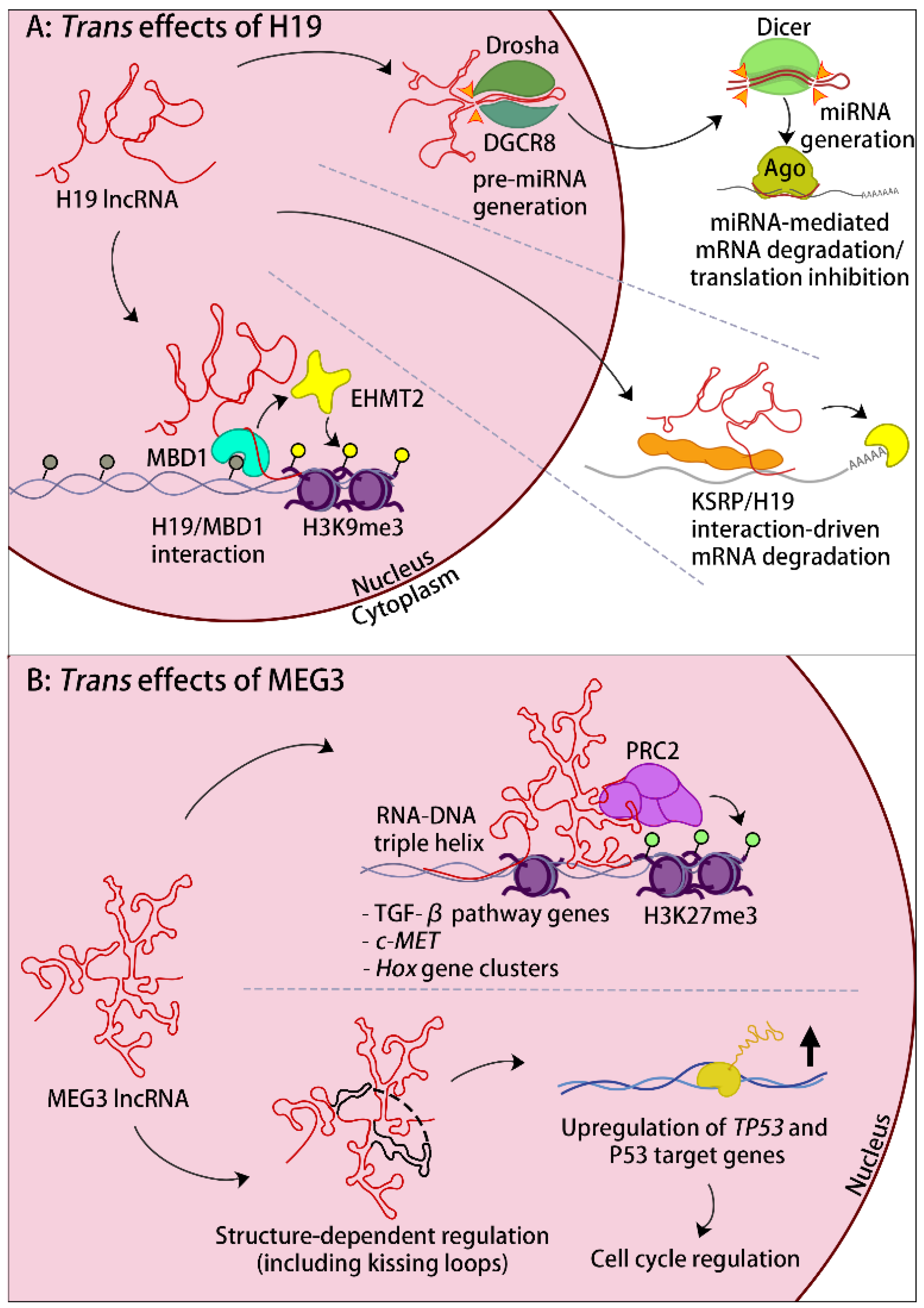
4. Emerging trans-Regulatory Roles
4.1. lncRNAs That Influence Other Imprinted Domains
4.2. Non-Imprinted trans Targets of Imprinted lncRNAs
5. Perspectives
| Imprinted Domain | lncRNA Name | lncRNA Gene Location 1 | Transcript(s) Size (nt) 2 | Expressed Allele | Action | lncRNA Function and Molecular Mechanism |
|---|---|---|---|---|---|---|
| Dlk1-Dio3 | Meg3 | Chr12: 109506879-109538163 | 31,285 (unspliced), 11,488 (v1), 11,476 (v2), 1924 (v3) | Maternal | Cis | Silencing of Dlk1, probably by PRC2 scaffolding and histone methylation deposition [78,84] |
| Trans | Enhances Hox gene repression by H3K27me3, by facilitating the interaction between EZH2 (PRC2 complex) and JARID2 [152] | |||||
| In humans, activation of a p53 target gene subset by an unknown mechanism [158] | ||||||
| In humans, activation of a TGF-beta pathway target gene subset by formation of RNA–DNA triplex structures at distal regulatory elements [145] | ||||||
| Igf2-H19 | H19 | Chr7: 142129267-142131883 | 2625 (unspliced), 2288 (v1, spliced), 2284 (v2, spliced) | Maternal | Trans | Influences an imprinted gene network (including Igf2) by MBD1 recruitment and subsequent histone KMT interaction at specific genes [142] |
| Promotes decaying of unstable mRNAs through interaction with the KSRP protein [159] | ||||||
| Hosting and processing regulation of the microRNA precursor miR-675 to control Igf1r expression in placenta [41] | ||||||
| In humans, functions as a tumour suppressor through 4E-BP1 binding and mTORC1 inhibition in pituitary tumours [172] | ||||||
| Kcnq1 | Kcnq1ot1 | Chr7: 142766848-142850284 | >83,437 (unspliced) | Paternal | Cis | Silencing of Kcnq1, Cdkn1c, Slc22a18, and Phlda2 in the embryo and the placenta [68,74,75] Silencing of the Ascl2, Cd81, Tssc4, and Osbpl5 genes in the placenta through recruitment of PRC complexes and KMT EHMT2 [50,68,72,74,126] |
| Trans | In human cells, this lncRNA contributes to retrotransposon repression by influencing HP1 binding [149] | |||||
| Igf2r | Airn | Chr17: 12960198-13079023 | 118,574 (unspliced), 1176 (v1), 413 (v2), 604 (v3), 1399 (v4) | Paternal | Cis | Silencing of Igf2r through transcriptional interference [47] |
| Silencing of Slc222a3 and several other genes in the placenta through recruitment of PRC complexes and KMT EHMT2 [46,49,50,51] | ||||||
| Gnas | Nespas | Chr2: 174123030-174137229, complement | 14,200 (unspliced), 2248 (v1) | Paternal | Cis | Silencing of Nesp, likely through a transcription-mediated process [60,107] |
| Trans | Modulation of IKBKE and Tmed9 expression levels by hosting the miR-296 microRNA [162] | |||||
| Snrpn | Snhg14 (Ube3a-ATS) | Chr7: 58922485-60099925, complement | 117,7441 (unspliced), 24,206 (v1) and >13 variants with diff. 5′ and 3′ ends | Paternal | Cis | Regulation of Ube3a expression by transcriptional interference [95,104] |
| IPW 3 | Chr15: 25116545-25122476 | 5932 (unspliced), 4498 (v1) | Paternal | Trans | In humans, downregulation of the Meg3 ncRNA polycistron by mediating repressive histone methylation [143] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33 (Suppl. S3), 245–254. [Google Scholar] [CrossRef] [PubMed]
- Feil, R.; Fraga, M.F. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Khamlichi, A.A.; Feil, R. Parallels between Mammalian Mechanisms of Monoallelic Gene Expression. Trends Genet. 2018, 34, 954–971. [Google Scholar] [CrossRef]
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Tucci, V.; Isles, A.R.; Kelsey, G.; Ferguson-Smith, A.C.; Grp, E.I. Genomic Imprinting and Physiological Processes in Mammals. Cell 2019, 176, 952. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Ferguson-Smith, A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011, 3, a002592. [Google Scholar] [CrossRef]
- Morison, I.M.; Ramsay, J.P.; Spencer, H.G. A census of mammalian imprinting. Trends Genet. 2005, 21, 457–465. [Google Scholar] [CrossRef]
- Andergassen, D.; Dotter, C.P.; Wenzel, D.; Sigl, V.; Bammer, P.C.; Muckenhuber, M.; Mayer, D.; Kulinski, T.M.; Theussl, H.C.; Penninger, J.M.; et al. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. Elife 2017, 6, e25125. [Google Scholar] [CrossRef]
- Surani, M.A.; Barton, S.C.; Norris, M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984, 308, 548–550. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.; Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef]
- Cattanach, B.M.; Kirk, M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature 1985, 315, 496–498. [Google Scholar] [CrossRef] [PubMed]
- Peters, J. The role of genomic imprinting in biology and disease: An expanding view. Nat. Rev. Genet. 2014, 15, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Eggermann, T.; Perez de Nanclares, G.; Maher, E.R.; Temple, I.K.; Tumer, Z.; Monk, D.; Mackay, D.J.; Gronskov, K.; Riccio, A.; Linglart, A.; et al. Imprinting disorders: A group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenet. 2015, 7, 123. [Google Scholar] [CrossRef]
- Hirasawa, R.; Feil, R. Genomic imprinting and human disease. Essays Biochem. 2010, 48, 187–200. [Google Scholar]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef]
- Kota, S.K.; Feil, R. Epigenetic transitions in germ cell development and meiosis. Dev. Cell 2010, 19, 675–686. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Sanli, I.; Feil, R. Chromatin mechanisms in the developmental control of imprinted gene expression. Int. J. Biochem. Cell Biol. 2015, 67, 139–147. [Google Scholar] [CrossRef]
- Zhang, Y.; Guan, D.G.; Yang, J.H.; Shao, P.; Zhou, H.; Qu, L.H. ncRNAimprint: A comprehensive database of mammalian imprinted noncoding RNAs. RNA 2010, 16, 1889–1901. [Google Scholar] [CrossRef]
- Girardot, M.; Cavaille, J.; Feil, R. Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenet. Off. J. DNA Methylation Soc. 2012, 7, 1341–1348. [Google Scholar] [CrossRef]
- Malnou, E.C.; Umlauf, D.; Mouysset, M.; Cavaille, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2018, 9, 706. [Google Scholar] [CrossRef]
- Royo, H.; Cavaille, J. Non-coding RNAs in imprinted gene clusters. Biol. Cell/Under Auspices Eur. Cell Biol. Organ. 2008, 100, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Ghousein, A.; Feil, R. Imprinted Small RNAs Unraveled: Maternal MicroRNAs Antagonize a Paternal-Genome-Driven Gene Expression Network. Mol. Cell 2020, 78, 3–5. [Google Scholar] [CrossRef]
- Cavaille, J. Box C/D small nucleolar RNA genes and the Prader-Willi syndrome: A complex interplay. Wiley Interdiscip. Rev. RNA 2017, 8, e1417. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Marcia, M. The multiple molecular dimensions of long noncoding RNAs that regulate gene expression and tumorigenesis. Curr. Opin. Oncol. 2022, 34, 141–147. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The Product of the H19 Gene May Function as an Rna. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [PubMed]
- MacDonald, W.A.; Mann, M.R.W. Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet. 2020, 16, e1008930. [Google Scholar] [CrossRef] [PubMed]
- Kanduri, C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim. Biophys. Acta 2016, 1859, 102–111. [Google Scholar] [CrossRef]
- Pauler, F.M.; Barlow, D.P.; Hudson, Q.J. Mechanisms of long range silencing by imprinted macro non-coding RNAs. Curr. Opin. Genet. Dev. 2012, 22, 283–289. [Google Scholar] [CrossRef]
- Latos, P.A.; Barlow, D.P. Regulation of imprinted expression by macro non-coding RNAs. RNA Biol. 2009, 6, 100–106. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef]
- Eggenschwiler, J.; Ludwig, T.; Fisher, P.; Leighton, P.A.; Tilghman, S.M.; Efstratiadis, A. Mouse mutant embryos overexpressing IGF-II exhibit phenotypic features of the Beckwith-Wiedemann and Simpson-Golabi-Behmel syndromes. Genes. Dev. 1997, 11, 3128–3142. [Google Scholar] [CrossRef] [PubMed]
- Louvi, A.; Accili, D.; Efstratiadis, A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev. Biol. 1997, 189, 33–48. [Google Scholar] [CrossRef]
- Weksberg, R.; Smith, A.C.; Squire, J.; Sadowski, P. Beckwith-Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum. Mol. Genet. 2003, 12, R61–R68. [Google Scholar] [CrossRef]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and lgf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Brioude, F.; Toutain, A.; Giabicani, E.; Cottereau, E.; Cormier-Daire, V.; Netchine, I. Overgrowth syndromes—Clinical and molecular aspects and tumour risk. Nat. Rev. Endocrinol. 2019, 15, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Lyle, R.; Watanabe, D.; te Vruchte, D.; Lerchner, W.; Smrzka, O.W.; Wutz, A.; Schageman, J.; Hahner, L.; Davies, C.; Barlow, D.P. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000, 25, 19–21. [Google Scholar] [CrossRef]
- Ludwig, T.; Eggenschwiler, J.; Fisher, P.; DErcole, A.J.; Davenport, M.L.; Efstratiadis, A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev. Biol. 1996, 177, 517–535. [Google Scholar] [CrossRef]
- Blanchard, F.; Duplomb, L.; Raher, S.; Vusio, P.; Hoflack, B.; Jacques, Y.; Godard, A. Mannose 6-Phosphate/Insulin-like growth factor II receptor mediates internalization and degradation of leukemia inhibitory factor but not signal transduction. J. Biol. Chem. 1999, 274, 24685–24693. [Google Scholar] [CrossRef]
- Sleutels, F.; Zwart, R.; Barlow, D.P. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002, 415, 810–813. [Google Scholar] [CrossRef]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Zwart, R.; Sleutels, F.; Wutz, A.; Schinkel, A.H.; Barlow, D.P. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 2001, 15, 2361–2366. [Google Scholar] [CrossRef]
- Andergassen, D.; Muckenhuber, M.; Bammer, P.C.; Kulinski, T.M.; Theussl, H.C.; Shimizu, T.; Penninger, J.M.; Pauler, F.M.; Hudson, Q.J. The Airn lncRNA does not require any DNA elements within its locus to silence distant imprinted genes. PLoS Genet. 2019, 15, e1008268. [Google Scholar] [CrossRef]
- Schertzer, M.D.; Braceros, K.C.A.; Starmer, J.; Cherneyt, R.E.; Lee, D.M.; Salazar, G.; Justice, M.; Bischoff, S.R.; Cowley, D.O.; Ariel, P.; et al. lncRNA-Induced Spread of Polycomb Controlled by Genome Architecture, RNA Abundance, and CpG Island DNA. Mol. Cell 2019, 75, 523. [Google Scholar] [CrossRef]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef]
- Killian, J.K.; Nolan, C.M.; Wylie, A.A.; Li, T.; Vu, T.H.; Hoffman, A.R.; Jirtle, R.L. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum. Mol. Genet. 2001, 10, 1721–1728. [Google Scholar] [CrossRef]
- Yotova, I.Y.; Vlatkovic, I.M.; Pauler, F.M.; Warczok, K.E.; Ambros, P.F.; Oshimura, M.; Theussl, H.C.; Gessler, M.; Wagner, E.F.; Barlow, D.P. Identification of the human homolog of the imprinted mouse Air non-coding RNA. Genomics 2008, 92, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, T.A.; Brizel, D.M.; Killian, J.K.; Oka, Y.; Jang, H.S.; Fu, X.; Clough, R.W.; Vollmer, R.T.; Anscher, M.S.; Jirtle, R.L. M6P/IGF2R loss of heterozygosity in head and neck cancer associated with poor patient prognosis. BMC Cancer 2003, 3, 4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Souza, A.T.; Hankins, G.R.; Washington, M.K.; Orton, T.C.; Jirtle, R.L. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nat. Genet. 1995, 11, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Plagge, A.; Kelsey, G.; Germain-Lee, E.L. Physiological functions of the imprinted Gnas locus and its protein variants Galpha(s) and XLalpha(s) in human and mouse. J. Endocrinol. 2008, 196, 193–214. [Google Scholar] [CrossRef]
- Peters, J.; Wroe, S.F.; Wells, C.A.; Miller, H.J.; Bodle, D.; Beechey, C.V.; Williamson, C.M.; Kelsey, G. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl. Acad. Sci. USA 1999, 96, 3830–3835. [Google Scholar] [CrossRef]
- Kelsey, G. Epigenetics and imprinted genes: Insights from the imprinted Gnas locus. Horm. Res. 2009, 71 (Suppl. S2), 22–29. [Google Scholar] [CrossRef]
- Coombes, C.; Arnaud, P.; Gordon, E.; Dean, W.; Coar, E.A.; Williamson, C.M.; Feil, R.; Peters, J.; Kelsey, G. Epigenetic properties and identification of an imprint mark in the Nesp-Gnasxl domain of the mouse Gnas imprinted locus. Mol. Cell Biol. 2003, 23, 5475–5488. [Google Scholar] [CrossRef]
- Tibbit, C.J.; Williamson, C.M.; Mehta, S.; Ball, S.T.; Chotalia, M.; Nottingham, W.T.; Eaton, S.A.; Quwailid, M.M.; Teboul, L.; Kelsey, G.; et al. Antisense Activity across the Nesp Promoter is Required for Nespas-Mediated Silencing in the Imprinted Gnas Cluster. Noncoding RNA 2015, 1, 246–265. [Google Scholar] [CrossRef]
- Wroe, S.F.; Kelsey, C.; Skinner, J.A.; Bodle, D.; Ball, S.T.; Beechey, C.V.; Peters, J.; Williamson, C.M. An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl. Acad. Sci. USA 2000, 97, 3342–3346. [Google Scholar] [CrossRef]
- Williamson, C.M.; Turner, M.D.; Ball, S.T.; Nottingham, W.T.; Glenister, P.; Fray, M.; Tymowska-Lalanne, Z.; Plagge, A.; Powles-Glover, N.; Kelsey, G.; et al. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 2006, 38, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitz, S.; Kaufman, Y.; Ludwig, G.; Razin, A.; Shemer, R. Mechanisms of activation of the paternally expressed genes by the Prader-Willi imprinting center in the Prader-Willi/Angelman syndromes domains. Proc. Natl. Acad. Sci. USA 2012, 109, 7403–7408. [Google Scholar] [CrossRef]
- Kelsey, G. Imprinting on chromosome 20: Tissue-specific imprinting and imprinting mutations in the GNAS locus. Am. J. Med. Genet. C Semin. Med. Genet. 2010, 154C, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, G.; Bastepe, M.; Monk, D.; de Sanctis, L.; Thiele, S.; Usardi, A.; Ahmed, S.F.; Bufo, R.; Choplin, T.; De Filippo, G.; et al. Diagnosis and management of pseudohypoparathyroidism and related disorders: First international Consensus Statement. Nat. Rev. Endocrinol. 2018, 14, 476–500. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Jaritz, M.; Guenzl, P.; Vlatkovic, I.; Sommer, A.; Tamir, I.M.; Marks, H.; Klampfl, T.; Kralovics, R.; Stunnenberg, H.G.; et al. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS ONE 2011, 6, e27288. [Google Scholar] [CrossRef]
- Redrup, L.; Branco, M.R.; Perdeaux, E.R.; Krueger, C.; Lewis, A.; Santos, F.; Nagano, T.; Cobb, B.S.; Fraser, P.; Reik, W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 2009, 136, 525–530. [Google Scholar] [CrossRef]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Golding, M.C.; Magri, L.S.; Zhang, L.; Lalone, S.A.; Higgins, M.J.; Mann, M.R. Depletion of Kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development 2011, 138, 3667–3678. [Google Scholar] [CrossRef]
- Fitzpatrick, G.V.; Soloway, P.D.; Higgins, M.J. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002, 32, 426–431. [Google Scholar] [CrossRef]
- Shin, J.Y.; Fitzpatrick, G.V.; Higgins, M.J. Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 2008, 27, 168–178. [Google Scholar] [CrossRef]
- Umlauf, D.; Goto, Y.; Cao, R.; Cerqueira, F.; Wagschal, A.; Zhang, Y.; Feil, R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004, 36, 1296–1300. [Google Scholar] [CrossRef]
- Lewis, A.; Mitsuya, K.; Umlauf, D.; Smith, P.; Dean, W.; Walter, J.; Higgins, M.; Feil, R.; Reik, W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004, 36, 1291–1295. [Google Scholar] [CrossRef]
- Terranova, R.; Yokobayashi, S.; Stadler, M.B.; Otte, A.P.; van Lohuizen, M.; Orkin, S.H.; Peters, A.H. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 2008, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Mancini-DiNardo, D.; Steele, S.J.S.; Levorse, J.M.; Ingram, R.S.; Tilghman, S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes. Dev. 2006, 20, 1268–1282. [Google Scholar] [CrossRef]
- da Rocha, S.T.; Edwards, C.A.; Ito, M.; Ogata, T.; Ferguson-Smith, A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008, 24, 306–316. [Google Scholar] [CrossRef]
- Lin, S.P.; Youngson, N.; Takada, S.; Seitz, H.; Reik, W.; Paulsen, M.; Cavaille, J.; Ferguson-Smith, A.C. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003, 35, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Kota, S.K.; Lleres, D.; Bouschet, T.; Hirasawa, R.; Marchand, A.; Begon-Pescia, C.; Sanli, I.; Arnaud, P.; Journot, L.; Girardot, M.; et al. ICR noncoding RNA expression controls imprinting and DNA replication at the Dlk1-Dio3 domain. Dev. Cell 2014, 31, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Lin, C.; Woodfin, A.R.; Bartom, E.T.; Gao, X.; Smith, E.R.; Shilatifard, A. Regulation of the imprinted Dlk1-Dio3 locus by allele-specific enhancer activity. Genes. Dev. 2016, 30, 92–101. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, Y.; Dai, Q.; Yang, Q.; Zhang, Y.; Wang, X.; Xie, W.; Luo, Z.; Lin, C. A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 2017, 45, 1177–1185. [Google Scholar] [CrossRef]
- Das, P.P.; Hendrix, D.A.; Apostolou, E.; Buchner, A.H.; Canver, M.C.; Beyaz, S.; Ljuboja, D.; Kuintzle, R.; Kim, W.; Karnik, R.; et al. PRC2 Is Required to Maintain Expression of the Maternal Gtl2-Rian-Mirg Locus by Preventing De Novo DNA Methylation in Mouse Embryonic Stem Cells. Cell Rep. 2015, 12, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Gossler, K.; Bilinski, P.; Sado, T.; Ferguson-Smith, A.; Gossler, A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1998, 212, 214–228. [Google Scholar]
- Kagami, M.; O’Sullivan, M.J.; Green, A.J.; Watabe, Y.; Arisaka, O.; Masawa, N.; Matsuoka, K.; Fukami, M.; Matsubara, K.; Kato, F.; et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: Hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010, 6, e1000992. [Google Scholar] [CrossRef]
- Sanli, I.; Lalevee, S.; Cammisa, M.; Perrin, A.; Rage, F.; Lleres, D.; Riccio, A.; Bertrand, E.; Feil, R. Meg3 Non-coding RNA Expression Controls Imprinting by Preventing Transcriptional Upregulation in cis. Cell Rep. 2018, 23, 337–348. [Google Scholar] [CrossRef]
- Renfree, M.B.; Suzuki, S.; Kaneko-Ishino, T. The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120151. [Google Scholar] [CrossRef]
- Habib, W.A.; Brioude, F.; Azzi, S.; Rossignol, S.; Linglart, A.; Sobrier, M.L.; Giabicani, E.; Steunou, V.; Harbison, M.D.; Le Bouc, Y.; et al. Transcriptional profiling at the DLK1/MEG3 domain explains clinical overlap between imprinting disorders. Sci. Adv. 2019, 5, eaau9425. [Google Scholar] [CrossRef] [PubMed]
- Beygo, J.; Elbracht, M.; de Groot, K.; Begemann, M.; Kanber, D.; Platzer, K.; Gillessen-Kaesbach, G.; Vierzig, A.; Green, A.; Heller, R.; et al. Novel deletions affecting the MEG3-DMR provide further evidence for a hierarchical regulation of imprinting in 14q32. Eur. J. Hum. Genet. 2015, 23, 180–188. [Google Scholar] [CrossRef]
- van der Werf, I.M.; Buiting, K.; Czeschik, C.; Reyniers, E.; Vandeweyer, G.; Vanhaesebrouck, P.; Ludecke, H.J.; Wieczorek, D.; Horsthemke, B.; Mortier, G.; et al. Novel microdeletions on chromosome 14q32.2 suggest a potential role for non-coding RNAs in Kagami-Ogata syndrome. Eur. J. Hum. Genet. 2016, 24, 1724–1729. [Google Scholar] [CrossRef]
- Prasasya, R.; Grotheer, K.V.; Siracusa, L.D.; Bartolomei, M.S. Temple syndrome and Kagami-Ogata syndrome: Clinical presentations, genotypes, models and mechanisms. Hum. Mol. Genet. 2020, 29, R108–R117. [Google Scholar] [CrossRef]
- Sanchez-Delgado, M.; Riccio, A.; Eggermann, T.; Maher, E.R.; Lapunzina, P.; MacKay, D.; Monk, D. Causes and Consequences of Multi-Locus Imprinting Disturbances in Humans. Trends Genet. 2016, 32, 444–455. [Google Scholar] [CrossRef]
- Horsthemke, B.; Wagstaff, J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am. J. Med. Genet. A 2008, 146A, 2041–2052. [Google Scholar] [CrossRef]
- Mendiola, A.J.P.; LaSalle, J.M. Epigenetics in Prader-Willi Syndrome. Front. Genet. 2021, 12, 624581. [Google Scholar] [CrossRef] [PubMed]
- Buiting, K.; Williams, C.; Horsthemke, B. Angelman syndrome—Insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016, 12, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Landers, M.; Bancescu, D.L.; Le Meur, E.; Rougeulle, C.; Glatt-Deeley, H.; Brannan, C.; Muscatelli, F.; Lalande, M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res 2004, 32, 3480–3492. [Google Scholar] [CrossRef]
- Meng, L.Y.; Person, R.E.; Huang, W.; Zhu, P.J.; Costa-Mattioli, M.; Beaudet, A.L. Truncation of Ube3a-ATS Unsilences Paternal Ube3a and Ameliorates Behavioral Defects in the Angelman Syndrome Mouse Model. PLoS Genet. 2013, 9, e1004039. [Google Scholar] [CrossRef]
- Meng, L.Y.; Ward, A.J.; Chun, S.; Bennett, C.F.; Beaudet, A.L.; Rigo, F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature 2015, 518, 409–412. [Google Scholar] [CrossRef]
- Lewis, M.W.; Vargas-Franco, D.; Morse, D.A.; Resnick, J.L. A mouse model of Angelman syndrome imprinting defects. Hum. Mol. Genet. 2019, 28, 220–229. [Google Scholar] [CrossRef]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Wright, J.; Zheng, K.J.; Shalem, O.; Fontanillas, P.; Joung, J.; Cheng, C.; Regev, A.; Zhang, F. High-resolution interrogation of functional elements in the noncoding genome. Science 2016, 353, 1545–1549. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Kaikkonen, M.U.; Adelman, K. Emerging Roles of Non-Coding RNA Transcription. Trends Biochem. Sci. 2018, 43, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Orom, U.A.; Shiekhattar, R. Noncoding RNAs and enhancers: Complications of a long-distance relationship. Trends Genet. 2011, 27, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.S.; Germain, N.D.; Wilderman, A.; Stoddard, C.; Wojenski, L.A.; Villafano, G.J.; Core, L.; Cotney, J.; Chamberlain, S.J. A bipartite boundary element restricts UBE3A imprinting to mature neurons. Proc. Natl. Acad. Sci. USA 2019, 116, 2181–2186. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, J.M.; Reiter, L.T.; Chamberlain, S.J. Epigenetic regulation of UBE3A and roles in human neurodevelopmental disorders. Epigenomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Lalevee, S.; Feil, R. Long noncoding RNAs in human disease: Emerging mechanisms and therapeutic strategies. Epigenomics 2015, 7, 877–879. [Google Scholar] [CrossRef]
- Wolter, J.M.; Mao, H.; Fragola, G.; Simon, J.M.; Krantz, J.L.; Bazick, H.O.; Oztemiz, B.; Stein, J.L.; Zylka, M.J. Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature 2020, 587, 281–284. [Google Scholar] [CrossRef]
- Williamson, C.M.; Ball, S.T.; Dawson, C.; Mehta, S.; Beechey, C.V.; Fray, M.; Teboul, L.; Dear, T.N.; Kelsey, G.; Peters, J. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted Gnas cluster. PLoS Genet. 2011, 7, e1001347. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Williamson, C.M.; Ball, S.; Tibbit, C.; Beechey, C.; Fray, M.; Peters, J. Transcription Driven Somatic DNA Methylation within the Imprinted Gnas Cluster. PLoS ONE 2015, 10, e0117378. [Google Scholar] [CrossRef]
- Baubec, T.; Colombo, D.F.; Wirbelauer, C.; Schmidt, J.; Burger, L.; Krebs, A.R.; Akalin, A.; Schubeler, D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 2015, 520, 243–247. [Google Scholar] [CrossRef]
- Sendzikaite, G.; Hanna, C.W.; Stewart-Morgan, K.R.; Ivanova, E.; Kelsey, G. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019, 10, 1884. [Google Scholar] [CrossRef]
- Santoro, F.; Mayer, D.; Klement, R.M.; Warczok, K.E.; Stukalov, A.; Barlow, D.P.; Pauler, F.M. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 2013, 140, 1184–1195. [Google Scholar] [CrossRef]
- Latos, P.A.; Stricker, S.H.; Steenpass, L.; Pauler, F.M.; Huang, R.; Senergin, B.H.; Regha, K.; Koerner, M.V.; Warczok, K.E.; Unger, C.; et al. An in vitro ES cell imprinting model shows that imprinted expression of the Igf2r gene arises from an allele-specific expression bias. Development 2009, 136, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Sakurai, T.; Sato, S.; Nakabayashi, K.; Hata, K.; Kono, T. Imprinted DNA methylation reprogramming during early mouse embryogenesis at the Gpr1-Zdbf2 locus is linked to long cis-intergenic transcription. FEBS Lett. 2012, 586, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Hiura, H.; Sugawara, A.; Ogawa, H.; John, R.M.; Miyauchi, N.; Miyanari, Y.; Horiike, T.; Li, Y.; Yaegashi, N.; Sasaki, H.; et al. A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res. 2010, 38, 4929–4945. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Glaser, J.; Borsos, M.; El Marjou, F.; Walter, M.; Teissandier, A.; Bourc’his, D. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat. Genet. 2017, 49, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Duffie, R.; Ajjan, S.; Greenberg, M.V.; Zamudio, N.; del Arenal, M.E.; Iranzo, J.; Okamoto, I.; Barbaux, S.; Fauque, P.; Bourc’his, D. The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev. 2014, 28, 463–478. [Google Scholar] [CrossRef]
- Greenberg, M.; Teissandier, A.; Walter, M.; Noordermeer, D.; Bourc’his, D. Dynamic enhancer partitioning instructs activation of a growth-related gene during exit from naive pluripotency. Elife 2019, 8, e44057. [Google Scholar] [CrossRef]
- Kobayashi, H.; Yanagisawa, E.; Sakashita, A.; Sugawara, N.; Kumakura, S.; Ogawa, H.; Akutsu, H.; Hata, K.; Nakabayashi, K.; Kono, T. Epigenetic and transcriptional features of the novel human imprined lncRNA GPR1AS suggest it is a functonal homolog of mouse Zdbf2linc. Epigenetics 2013, 8, 635–645. [Google Scholar] [CrossRef]
- Niemczyk, M.; Ito, Y.; Huddleston, J.; Git, A.; Abu-Amero, S.; Caldas, C.; Moore, G.E.; Stojic, L.; Murrell, A. Imprinted chromatin around DIRAS3 regulates alternative splicing of GNG12-AS1, a long noncoding RNA. Am. J. Hum. Genet. 2013, 93, 224–235. [Google Scholar] [CrossRef]
- Yu, Y.; Luo, R.; Lu, Z.; Feng, W.W.; Badgwell, D.; Issa, J.P.; Rosen, D.G.; Liu, J.; Bast, R.C., Jr. Biochemistry and biology of ARHI (DIRAS3), an imprinted tumor suppressor gene whose expression is lost in ovarian and breast cancers. Methods Enzymol. 2006, 407, 455–468. [Google Scholar]
- Stojic, L.; Niemczyk, M.; Orjalo, A.; Ito, Y.; Ruijter, A.E.; Uribe-Lewis, S.; Joseph, N.; Weston, S.; Menon, S.; Odom, D.T.; et al. Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 2016, 7, 10406. [Google Scholar] [CrossRef]
- Loda, A.; Collombet, S.; Heard, E. Gene regulation in time and space during X-chromosome inactivation. Nat. Rev. Mol. Cell Biol. 2022, 23, 231–249. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef]
- Kaneko, S.; Bonasio, R.; Saldana-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef]
- Mager, J.; Montgomery, N.D.; de Villena, F.P.; Magnuson, T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 2003, 33, 502–507. [Google Scholar] [CrossRef]
- Wagschal, A.; Sutherland, H.G.; Woodfine, K.; Henckel, A.; Chebli, K.; Schulz, R.; Oakey, R.J.; Bickmore, W.A.; Feil, R. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol. Cell Biol. 2008, 28, 1104–1113. [Google Scholar] [CrossRef]
- Pintacuda, G.; Wei, G.; Roustan, C.; Kirmizitas, B.A.; Solcan, N.; Cerase, A.; Castello, A.; Mohammed, S.; Moindrot, B.; Nesterova, T.B.; et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 2017, 68, 955–969.e910. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef]
- Kaneko, S.; Son, J.; Shen, S.S.; Reinberg, D.; Bonasio, R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013, 20, 1258–1264. [Google Scholar] [CrossRef]
- Monk, D.; Wagschal, A.; Arnaud, P.; Muller, P.S.; Parker-Katiraee, L.; Bourc’his, D.; Scherer, S.W.; Feil, R.; Stanier, P.; Moore, G.E. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 2008, 18, 1270–1281. [Google Scholar] [CrossRef]
- Lleres, D.; Imaizumi, Y.; Feil, R. Exploring chromatin structural roles of non-coding RNAs at imprinted domains. Biochem. Soc. Trans. 2021, 49, 1867–1879. [Google Scholar] [CrossRef]
- Noordermeer, D.; Feil, R. Differential 3D chromatin organization and gene activity in genomic imprinting. Curr. Opin. Genet. Dev. 2020, 61, 17–24. [Google Scholar] [CrossRef]
- Saldana-Meyer, R.; Rodriguez-Hernaez, J.; Escobar, T.; Nishana, M.; Jacome-Lopez, K.; Nora, E.P.; Bruneau, B.G.; Tsirigos, A.; Furlan-Magaril, M.; Skok, J.; et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol. Cell 2019, 76, 412–422. [Google Scholar] [CrossRef]
- Hansen, A.S.; Hsieh, T.H.S.; Cattoglio, C.; Pustova, I.; Saldana-Meyer, R.; Reinberg, D.; Darzacq, X.; Tjian, R. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol. Cell 2019, 76, 395. [Google Scholar] [CrossRef]
- Thakur, J.; Henikoff, S. Architectural RNA in chromatin organization. Biochem. Soc. Trans. 2020, 48, 1967–1978. [Google Scholar] [CrossRef]
- McCann, J.A.; Zheng, H.; Islam, A.; Goodyer, C.G.; Polychronakos, C. Evidence against GRB10 as the gene responsible for Silver-Russell syndrome. Biochem. Biophys. Res. Commun. 2001, 286, 943–948. [Google Scholar] [CrossRef]
- Monk, D.; Wakeling, E.L.; Proud, V.; Hitchins, M.; Abu-Amero, S.N.; Stanier, P.; Preece, M.A.; Moore, G.E. Duplication of 7p11.2-p13, including GRB10, in Silver-Russell syndrome. Am. J. Hum. Genet. 2000, 66, 36–46. [Google Scholar] [CrossRef]
- Patten, M.M.; Cowley, M.; Oakey, R.J.; Feil, R. Regulatory links between imprinted genes: Evolutionary predictions and consequences. Proc. Biol. Sci. 2016, 283, 20152760. [Google Scholar] [CrossRef]
- Varrault, A.; Gueydan, C.; Delalbre, A.; Bellmann, A.; Houssami, S.; Aknin, C.; Severac, D.; Chotard, L.; Kahli, M.; Le Digarcher, A.; et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell 2006, 11, 711–722. [Google Scholar] [CrossRef]
- Al Adhami, H.; Evano, B.; Le Digarcher, A.; Gueydan, C.; Dubois, E.; Parrinello, H.; Dantec, C.; Bouschet, T.; Varrault, A.; Journot, L. A systems-level approach to parental genomic imprinting: The imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 2015, 25, 353–367. [Google Scholar] [CrossRef]
- Gabory, A.; Ripoche, M.A.; Le Digarcher, A.; Watrin, F.; Ziyyat, A.; Forne, T.; Jammes, H.; Ainscough, J.F.; Surani, M.A.; Journot, L.; et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 2009, 136, 3413–3421. [Google Scholar] [CrossRef]
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, Y.; Sagi, I.; Yanuka, O.; Eiges, R.; Benvenisty, N. The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome. Nat. Genet. 2014, 46, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915.e2904. [Google Scholar] [CrossRef]
- Iyer, S.; Modali, S.D.; Agarwal, S.K. Long Noncoding RNA MEG3 Is an Epigenetic Determinant of Oncogenic Signaling in Functional Pancreatic Neuroendocrine Tumor Cells. Mol. Cell Biol. 2017, 37, e00278-17. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, Q.; Li, J.; Zhang, S.; Cao, Y.; Xia, X.; Cai, D.; Tan, J.; Chen, J.; Han, J.J. KCNQ1OT1 promotes genome-wide transposon repression by guiding RNA-DNA triplexes and HP1 binding. Nat. Cell Biol. 2022, 24, 1617–1629. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.P.; Hsieh, W.F.; Tsai, Y.Y.; Lu, Y.L.; Liau, E.S.; Hsu, H.C.; Chen, Y.C.; Liu, T.C.; Chang, M.; Li, J.; et al. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 2018, 7, e38080. [Google Scholar] [CrossRef] [PubMed]
- Higgs, M.J.; Hill, M.J.; John, R.M.; Isles, A.R. Systematic investigation of imprinted gene expression and enrichment in the mouse brain explored at single-cell resolution. BMC Genome 2022, 23, 754. [Google Scholar] [CrossRef] [PubMed]
- Gejman, R.; Batista, D.L.; Zhong, Y.; Zhou, Y.L.; Zhang, X.; Swearingen, B.; Stratakis, C.A.; Hedley-Whyte, E.T.; Klibanski, A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocr. Metab. 2008, 93, 4119–4125. [Google Scholar] [CrossRef] [PubMed]
- Buttarelli, M.; De Donato, M.; Raspaglio, G.; Babini, G.; Ciucci, A.; Martinelli, E.; Baccaro, P.; Pasciuto, T.; Fagotti, A.; Scambia, G.; et al. Clinical Value of lncRNA MEG3 in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 966. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheunsuchon, P.; Nakayama, Y.; Lawlor, M.W.; Zhong, Y.; Rice, K.A.; Zhang, L.; Zhang, X.; Gordon, F.E.; Lidov, H.G.; et al. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 2010, 137, 2643–2652. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Uroda, T.; Anastasakou, E.; Rossi, A.; Teulon, J.M.; Pellequer, J.L.; Annibale, P.; Pessey, O.; Inga, A.; Chillon, I.; Marcia, M. Conserved Pseudoknots in lncRNA MEG3 Are Essential for Stimulation of the p53 Pathway. Mol. Cell 2019, 75, 982. [Google Scholar] [CrossRef]
- Giovarelli, M.; Bucci, G.; Ramos, A.; Bordo, D.; Wilusz, C.J.; Chen, C.Y.; Puppo, M.; Briata, P.; Gherzi, R. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc. Natl. Acad. Sci. USA 2014, 111, E5023–E5028. [Google Scholar] [CrossRef]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef]
- Martinet, C.; Monnier, P.; Louault, Y.; Benard, M.; Gabory, A.; Dandolo, L. H19 controls reactivation of the imprinted gene network during muscle regeneration. Development 2016, 143, 962–971. [Google Scholar] [CrossRef] [PubMed]
- Robson, J.E.; Eaton, S.A.; Underhill, P.; Williams, D.; Peters, J. MicroRNAs 296 and 298 are imprinted and part of the GNAS/Gnas cluster and miR-296 targets IKBKE and Tmed9. RNA 2012, 18, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Shen, A.; Liu, A. Long non-coding RNA H19 and cancer: A competing endogenous RNA. Bull. Cancer 2019, 106, 1152–1159. [Google Scholar] [CrossRef]
- Muller, V.; Oliveira-Ferrer, L.; Steinbach, B.; Pantel, K.; Schwarzenbach, H. Interplay of lncRNA H19/miR-675 and lncRNA NEAT1/miR-204 in breast cancer. Mol. Oncol. 2019, 13, 1137–1149. [Google Scholar] [CrossRef]
- Li, J.; Wang, X.; Wang, Y.; Yang, Q. H19 promotes the gastric carcinogenesis by sponging miR-29a-3p: Evidence from lncRNA-miRNA-mRNA network analysis. Epigenomics 2020, 12, 989–1002. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, F.; Zhou, J.; Yan, L.; Jurczak, M.J.; Lee, H.Y.; Yang, L.; Mueller, M.; Zhou, X.B.; Dandolo, L.; et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014, 42, 13799–13811. [Google Scholar] [CrossRef]
- Park, K.S.; Rahat, B.; Lee, H.C.; Yu, Z.X.; Noeker, J.; Mitra, A.; Kean, C.M.; Knutsen, R.H.; Springer, D.; Gebert, C.M.; et al. Cardiac pathologies in mouse loss of imprinting models are due to misexpression of H19 long noncoding RNA. Elife 2021, 10, e67250. [Google Scholar] [CrossRef]
- Chu, H.P.; Minajigi, A.; Chen, Y.F.; Morris, R.; Guh, C.Y.; Hsieh, Y.H.; Boukhali, M.; Haas, W.; Lee, J.T. iDRiP for the systematic discovery of proteins bound directly to noncoding RNA. Nat. Protoc. 2021, 16, 3672. [Google Scholar] [CrossRef]
- Cao, H.; Kapranov, P. Methods to Analyze the Non-Coding RNA Interactome-Recent Advances and Challenges. Front. Genet. 2022, 13, 857759. [Google Scholar] [CrossRef]
- Frost, J.M.; Udayashankar, R.; Moore, H.D.; Moore, G.E. Telomeric NAP1L4 and OSBPL5 of the KCNQ1 cluster, and the DECORIN gene are not imprinted in human trophoblast stem cells. PLoS ONE 2010, 5, e11595. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.R.; Yan, L.; Liu, Y.T.; Cao, L.; Guo, Y.H.; Zhang, Y.; Yao, H.; Cai, L.; Shang, H.B.; Rui, W.W.; et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat. Commun. 2018, 9, 4624. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Michele, F.; Chillón, I.; Feil, R. Imprinted Long Non-Coding RNAs in Mammalian Development and Disease. Int. J. Mol. Sci. 2023, 24, 13647. https://doi.org/10.3390/ijms241713647
Di Michele F, Chillón I, Feil R. Imprinted Long Non-Coding RNAs in Mammalian Development and Disease. International Journal of Molecular Sciences. 2023; 24(17):13647. https://doi.org/10.3390/ijms241713647
Chicago/Turabian StyleDi Michele, Flavio, Isabel Chillón, and Robert Feil. 2023. "Imprinted Long Non-Coding RNAs in Mammalian Development and Disease" International Journal of Molecular Sciences 24, no. 17: 13647. https://doi.org/10.3390/ijms241713647
APA StyleDi Michele, F., Chillón, I., & Feil, R. (2023). Imprinted Long Non-Coding RNAs in Mammalian Development and Disease. International Journal of Molecular Sciences, 24(17), 13647. https://doi.org/10.3390/ijms241713647






