The Intergeneration Long-Lasting Consequences of Pre-Conceptional Exposure to Sofosbuvir on the Ovarian Tissues of F1 Offspring: Experimental Study on Rats
Abstract
:1. Introduction
2. Results
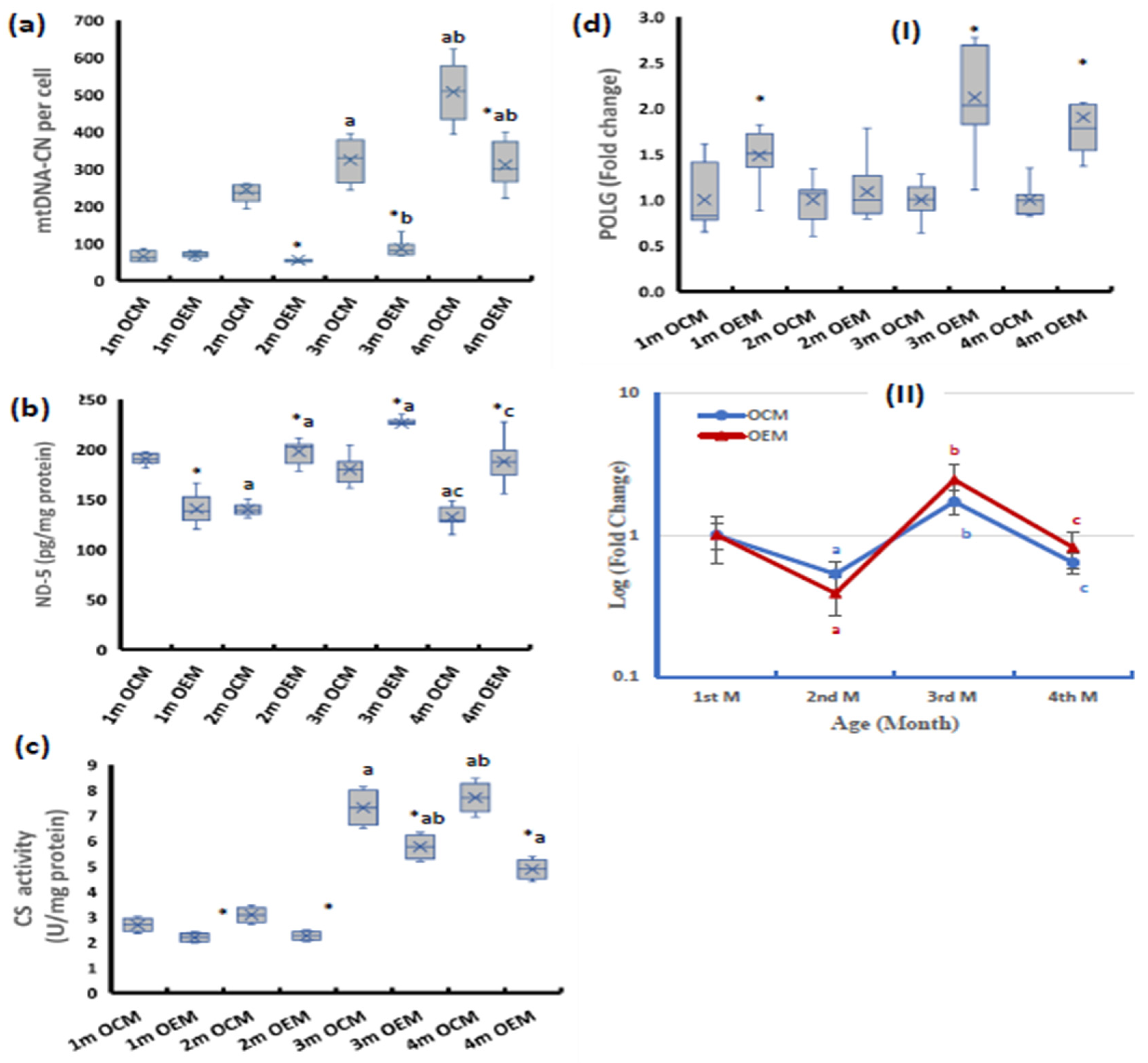
2.1. Ovarian Mitochondrial Parameters
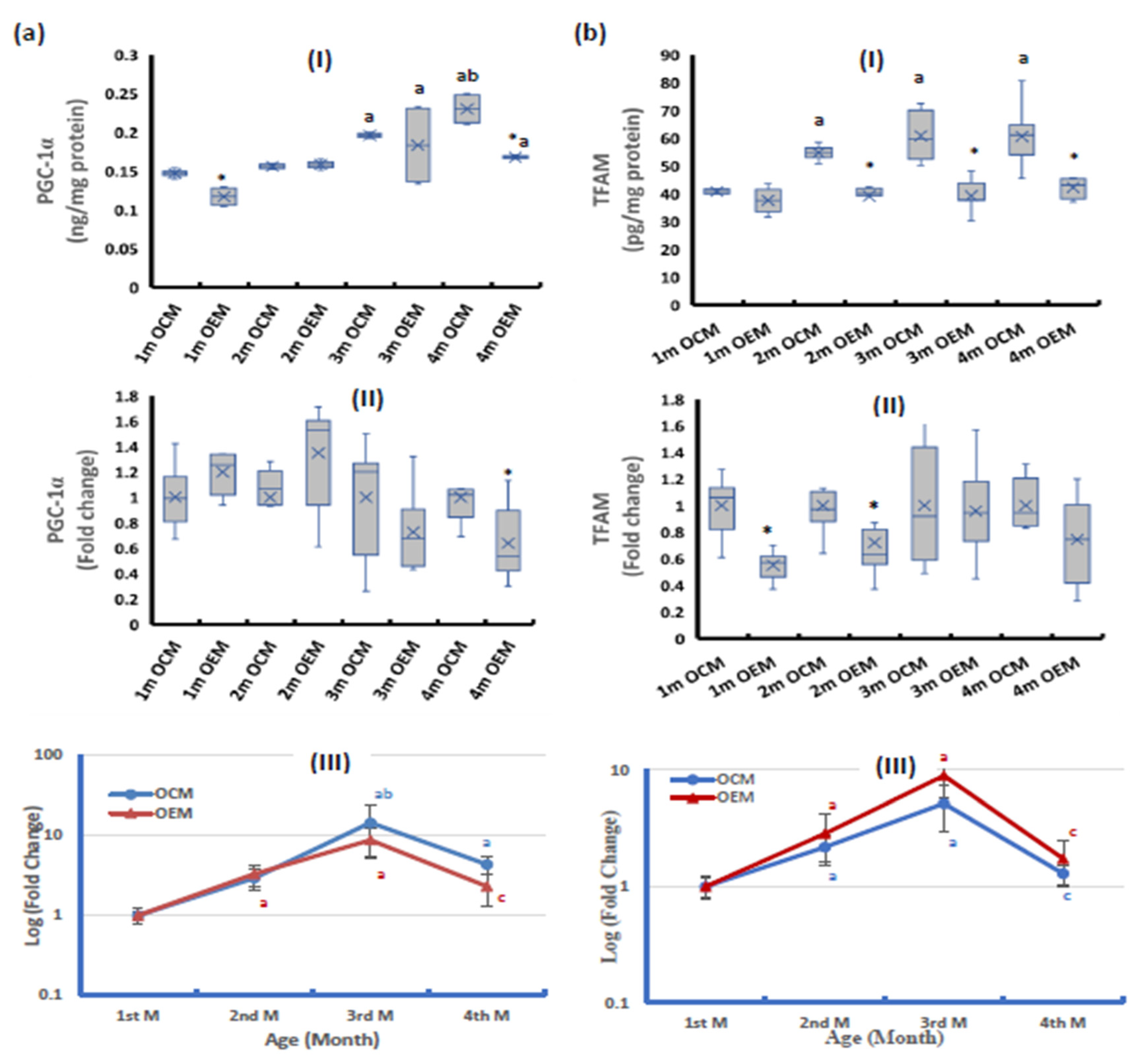
2.2. Parameters of Mitochondrial Biogenesis Pathway
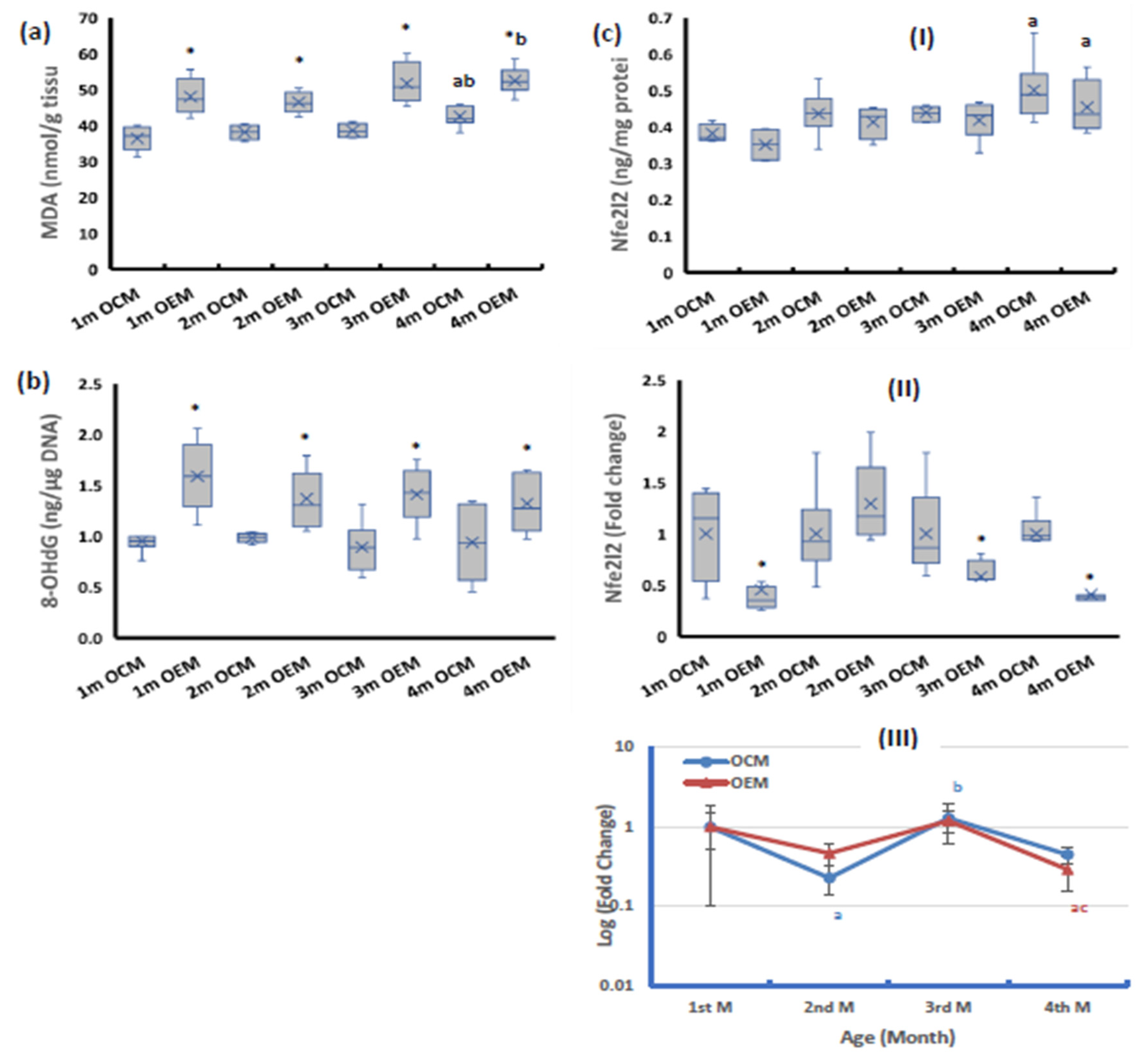
2.3. Redox Parameters
2.4. The Ovarian Expression of Nuclear Factor kappa-B (NF-κB)
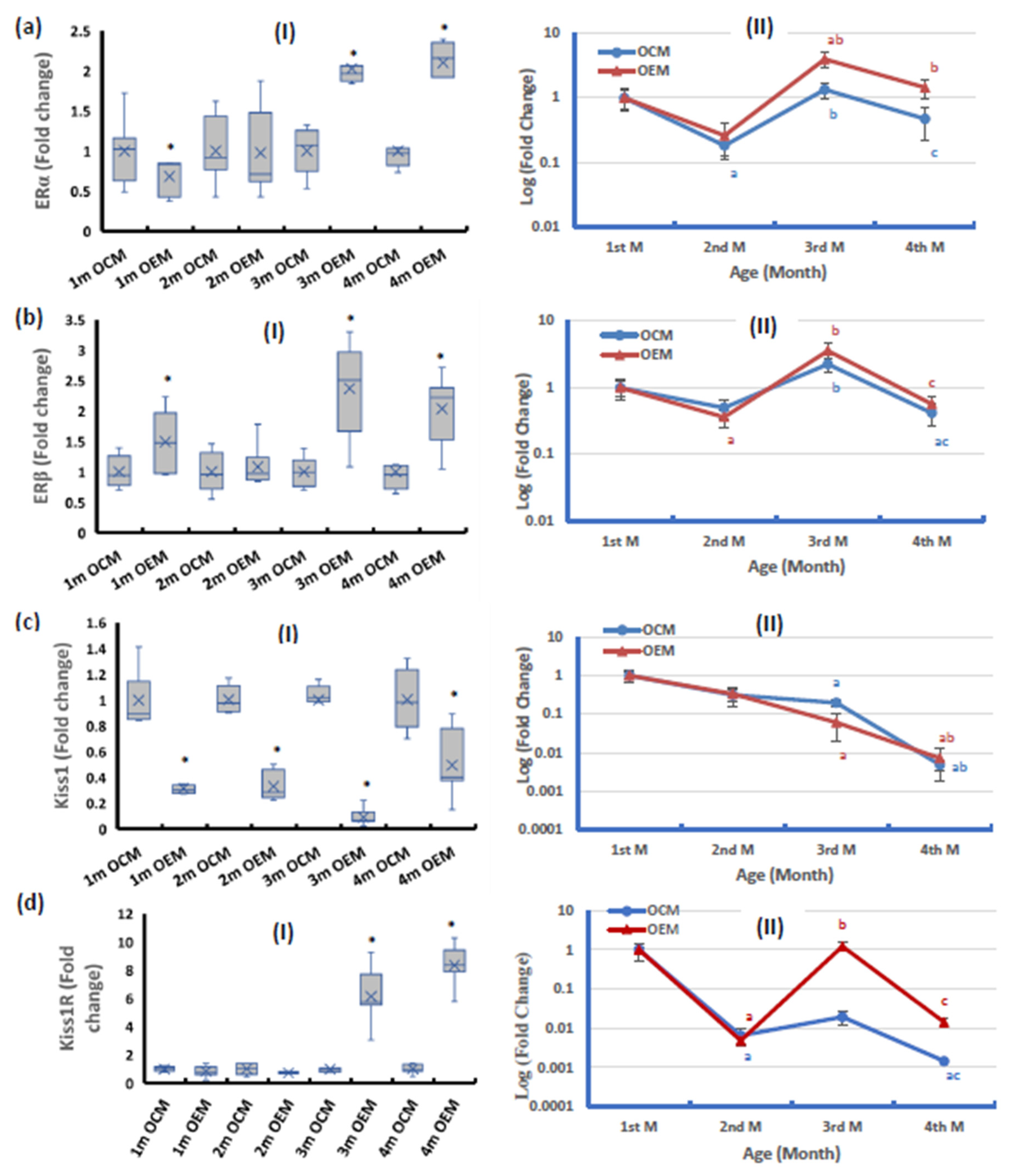
2.5. Ovarian Expression of the Components of the Estrogen Pathway
2.6. Correlation Studies
3. Discussion
4. Materials and Methods
4.1. Experimental Animals
4.2. Ethical Statement
4.3. Drug
4.4. Experimental Design
4.5. Tissue Preparation
4.6. Determination of Malondialdehyde as a Marker for Lipid Peroxidation
4.7. Elisa Measurements
4.8. Ovarian Citrate Synthase (CS) Activity
4.9. Determination of Total Protein
4.10. Gene Expression Analysis Using Quantitative Real-Time Polymerase Chain Reaction (qPCR)
4.11. Determination of Mitochondrial DNA Copy Number per Cell
4.12. Determination of 8-Hydroxy Deoxyguanosine (8-OHdG)
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization [WHO]. Accelerating Access to Hepatitis C Diagnostics and Treatment: Overcoming Barriers in Low-and Middle-Income Countries: Global Progress Report 2020; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- World Health Organization [WHO]. Updated Recommendations on Simplified Service Delivery and Disagnostics for Hepititis C Infection: Policy Brief; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Asselah, T.; Boyer, N.; Saadoun, D.; Martinot-Peignoux, M.; Marcellin, P. Direct-acting antivirals for the treatment of hepatitis C virus infection: Optimizing current IFN-free treatment and future perspectives. Liver Int. 2016, 36, 47–57. [Google Scholar] [CrossRef]
- Kim, S.J.; Jang, J.Y.; Kim, E.J.; Cho, E.K.; Ahn, D.G.; Kim, C.; Park, H.S.; Jeong, S.W.; Lee, S.H.; Kim, S.G. Ginsenoside Rg3 restores hepatitis C virus–induced aberrant mitochondrial dynamics and inhibits virus propagation. Hepatology 2017, 66, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Stoker, M.L.; Newport, E.; Hulit, J.C.; West, A.P.; Morten, K.J. Impact of pharmacological agents on mitochondrial function: A growing opportunity? Biochem. Soc. Trans. 2019, 47, 1757–1772. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Ward, W.F. PGC-1α: A key regulator of energy metabolism. Adv. Physiol. Educ. 2006, 30, 145–151. [Google Scholar] [CrossRef]
- Copeland, W.C. The mitochondrial DNA polymerase in health and disease. Genome Stab. Hum. Dis. 2010, 50, 211–222. [Google Scholar]
- Morrison, H. Enzyme Active Sites and Their Reaction Mechanisms; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Li, X.; Zhang, W.; Cao, Q.; Wang, Z.; Zhao, M.; Xu, L.; Zhuang, Q. Mitochondrial dysfunction in fibrotic diseases. Cell Death Discov. 2020, 6, 80. [Google Scholar] [CrossRef]
- Zhang, D.D.; Lo, S.C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 2004, 24, 10941–10953. [Google Scholar] [CrossRef] [PubMed]
- Mattingly, K.A.; Ivanova, M.M.; Riggs, K.A.; Wickramasinghe, N.S.; Barch, M.J.; Klinge, C.M. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol. Endocrinol. 2008, 22, 609–622. [Google Scholar] [CrossRef]
- Chen, J.Q.; Cammarata, P.R.; Baines, C.P.; Yager, J.D. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2009, 1793, 1540–1570. [Google Scholar] [CrossRef]
- Chakravarthi, V.P.; Ghosh, S.; Housami, S.M.; Wang, H.; Roby, K.F.; Wolfe, M.W.; Kinsey, W.H.; Rumi, M.K. ERβ regulated ovarian kisspeptin plays an important role in oocyte maturation. Mol. Cell. Endocrinol. 2021, 527, 111208. [Google Scholar] [CrossRef]
- D’Occhio, M.J.; Campanile, G.; Baruselli, P.S. Peripheral action of kisspeptin at reproductive tissues—Role in ovarian function and embryo implantation and relevance to assisted reproductive technology in livestock: A review. Biol. Reprod. 2020, 103, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Kamel, M.A.; Mahmoud, S.A.; Elfetooh, H.A. In-Utero Exposure to Maternal Diabetes Increase the Risk of Vascular Diseases in the F1 Offspring in Rats. Am. J. Biomed. Sci. 2014, 6, 201–216. [Google Scholar] [CrossRef]
- Nagaty, A.; Abd El-Wahab, E.W. Real-life results of sofosbuvir based therapy in chronic hepatitis C-naïve and-experienced patients in Egypt. Public Libr. Sci. One 2017, 12, e0184654. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.A.; Abdel-Aziz, M.M.; Khafaga, R.H.M.; Hafez, H.A.; Kamel, M.A.; Shaker, S.A. The pre-conception maternal exposure to Sofosbuvir affects the mitochondrial biogenesis in prenatal fetal tissues: Experimental study on rats. Mol. Med. 2023, 29, 71. [Google Scholar] [CrossRef] [PubMed]
- Hoque, S.M.; Kawai, T.; Zhu, Z.; Shimada, M. Mitochondrial protein turnover is critical for granulosa cell proliferation and differentiation in antral follicles. J. Endocr. Soc. 2019, 3, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.R.; Murray, A.; Day, F.R.; Ong, K.K. Molecular insights into the aetiology of female reproductive ageing. Nat. Rev. Endocrinol. 2015, 11, 725–734. [Google Scholar] [CrossRef]
- Miyamoto, K.; Sato, E.F.; Kasahara, E.; Jikumaru, M.; Hiramoto, K.; Tabata, H.; Katsuragi, M.; Odo, S.; Utsumi, K.; Inoue, M. Effect of oxidative stress during repeated ovulation on the structure and functions of the ovary, oocytes, and their mitochondria. Free Radic. Biol. Med. 2010, 49, 674–681. [Google Scholar] [CrossRef]
- Boucret, L.; Chao De La Barca, J.; Morinière, C.; Desquiret, V.; Ferré-L’Hôtellier, V.; Descamps, P.; Marcaillou, C.; Reynier, P.; Procaccio, V.; May-Panloup, P. Relationship between diminished ovarian reserve and mitochondrial biogenesis in cumulus cells. Hum. Reprod. 2015, 30, 1653–1664. [Google Scholar] [CrossRef]
- Zhang, G.; Wan, Y.; Zhang, Y.; Lan, S.; Jia, R.; Wang, Z.; Fan, Y.; Wang, F. Expression of Mitochondria-Associated Genes (PPARGC 1A, NRF-1, BCL-2 and BAX) in Follicular Development and Atresia of Goat Ovaries. Reprod. Domest. Anim. 2015, 50, 465–473. [Google Scholar] [CrossRef]
- Zhang, G.M.; Deng, M.T.; Lei, Z.H.; Wan, Y.J.; Nie, H.T.; Wang, Z.Y.; Fan, Y.X.; Wang, F.; Zhang, Y.L. Effects of NRF1 on steroidogenesis and apoptosis in goat luteinized granulosa cells. Reproduction 2017, 154, 111–122. [Google Scholar] [CrossRef]
- Kaufman, B.A.; Durisic, N.; Mativetsky, J.M.; Costantino, S.; Hancock, M.A.; Grutter, P.; Shoubridge, E.A. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell 2007, 18, 3225–3236. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Ao, A.; Zhang, X.; Cyr, D.; Dufort, D.; Shoubridge, E.A. The role of mitochondrial DNA copy number in mammalian fertility. Biol. Reprod. 2010, 83, 52–62. [Google Scholar] [CrossRef]
- Sharma, L.K.; Lu, J.; Bai, Y. Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr. Med. Chem. 2009, 16, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Atef, M.M.; Abd-Ellatif, R.N.; Emam, M.N.; Amer, A.I.; Hafez, Y.M. Therapeutic potential of sodium selenite in letrozole induced polycystic ovary syndrome rat model: Targeting mitochondrial approach (selenium in PCOS). Arch. Biochem. Biophys. 2019, 671, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhao, M.; Liu, X.; Wang, X.; Nie, Y.; Li, P.; Liu, T.; Ge, R.; Han, F. Reduced expression of citrate synthase leads to excessive superoxide formation and cell apoptosis. Biochem. Biophys. Res. Commun. 2017, 485, 388–394. [Google Scholar] [CrossRef]
- Alcendor, R.R.; Gao, S.; Zhai, P.; Zablocki, D.; Holle, E.; Yu, X.; Tian, B.; Wagner, T.; Vatner, S.F.; Sadoshima, J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007, 100, 1512–1521. [Google Scholar] [CrossRef]
- Zhang, M.; Yu, X.; Li, D.; Ma, N.; Wei, Z.; Ci, X.; Zhang, S. Nrf2 Signaling Pathway Mediates the Protective Effects of Daphnetin Against D-Galactose Induced-Premature Ovarian Failure. Front. Pharmacol. 2022, 13, 810524. [Google Scholar] [CrossRef]
- Sun, T.C.; Liu, X.C.; Yang, S.H.; Song, L.L.; Zhou, S.J.; Deng, S.L.; Tian, L.; Cheng, L.Y. Melatonin Inhibits Oxidative Stress and Apoptosis in Cryopreserved Ovarian Tissues via Nrf2/HO-1 Signaling Pathway. Front. Mol. Biosci. 2020, 7, 163. [Google Scholar] [CrossRef]
- Navarro, E.; Gonzalez-Lafuente, L.; Pérez-Liébana, I.; Buendia, I.; López-Bernardo, E.; Sánchez-Ramos, C.; Prieto, I.; Cuadrado, A.; Satrustegui, J.; Cadenas, S. Heme-oxygenase I and PCG-1α regulate mitochondrial biogenesis via microglial activation of alpha7 nicotinic acetylcholine receptors using PNU282987. Antioxid. Redox Signal. 2017, 27, 93–105. [Google Scholar] [CrossRef]
- Cherry, A.D.; Suliman, H.B.; Bartz, R.R.; Piantadosi, C.A. Peroxisome proliferator-activated receptor γ co-activator 1-α as a critical co-activator of the murine hepatic oxidative stress response and mitochondrial biogenesis in Staphylococcus aureus sepsis. J. Biol. Chem. 2014, 289, 41–52. [Google Scholar] [CrossRef]
- Aquilano, K.; Baldelli, S.; Pagliei, B.; Cannata, S.M.; Rotilio, G.; Ciriolo, M.R. p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid. Redox Signal. 2013, 18, 386–399. [Google Scholar] [CrossRef]
- Paciolla, M.; Boni, R.; Fusco, F.; Pescatore, A.; Poeta, L.; Ursini, M.; Lioi, M.; Miano, M.G. Nuclear factor-kappa-B-inhibitor alpha (NFKBIA) is a developmental marker of NF- B/p65 activation during in vitro oocyte maturation and early embryogenesis. Hum. Reprod. Oxf. Engl. 2011, 26, 1191–1201. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Z.; Min, W. Mitochondria, oxidative stress and innate immunity. Front. Physiol. 2018, 9, 1487. [Google Scholar] [CrossRef]
- Khristi, V.; Chakravarthi, V.P.; Singh, P.; Ghosh, S.; Pramanik, A.; Ratri, A.; Borosha, S.; Roby, K.F.; Wolfe, M.W.; Rumi, M.K. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 2018, 474, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Marcondes, R.R.; Carvalho, K.C.; Giannocco, G.; Duarte, D.C.; Garcia, N.; Soares-Junior, J.M.; da Silva, I.; Maliqueo, M.; Baracat, E.C.; Maciel, G.A.R. Hypothalamic transcriptional expression of the kisspeptin system and sex steroid receptors differs among polycystic ovary syndrome rat models with different endocrine phenotypes. Clinics 2017, 72, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, K.J.; Arao, Y.; Korach, K.S. Estrogen hormone physiology: Reproductive findings from estrogen receptor mutant mice. Reprod. Biol. 2014, 14, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Wall, E.H.; Hewitt, S.C.; Case, L.K.; Lin, C.Y.; Korach, K.S.; Teuscher, C. The role of genetics in estrogen responses: A critical piece of an intricate puzzle. Fed. Am. Soc. Exp. Biol. 2014, 28, 5042. [Google Scholar] [CrossRef] [PubMed]
- Hu, K.-L.; Zhao, H.; Chang, H.M.; Yu, Y.; Qiao, J. Kisspeptin/kisspeptin receptor system in the ovary. Front. Endocrinol. 2018, 8, 365. [Google Scholar] [CrossRef]
- Issa, N.; El-Sherif, N. Histological and Immunohistochemical Studies on the Cornea and Retina of Sofosbuvir Treated Rats. Austin J. Anat. 2017, 4, 1068. [Google Scholar]
- Draper, H.H.; Hadley, M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990, 186, 421–431. [Google Scholar]
- Shepherd, D.; Garland, P. [2] Citrate synthase from rat liver: [EC 4.1. 3.7 Citrate oxaloacetage-lyase (CoA-acetylating)]. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1969; Volume 13, pp. 11–16. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Gowayed, M.A.; Mahmoud, S.A.; El-Sayed, Y.; Abu-Samra, N.; Kamel, M.A. Enhanced mitochondrial biogenesis is associated with the ameliorative action of creatine supplementation in rat soleus and cardiac muscles. Exp. Ther. Med. 2020, 19, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hagen, S. SPSS in Practice. Nurse Res. 2002, 10, 86–88. [Google Scholar] [CrossRef]

| ND-5 Protein | POLG mRNA | PGC-1α Protein | TFAM Protein | MDA | 8-OHdG | mtDNA-CN | ERα mRNA | ERβ mRNA | |
|---|---|---|---|---|---|---|---|---|---|
| ND-5 protein | − | 0.338 * | −0.006 | −0.406 * | 0.258 * | 0.218 | −0.393 * | 0.422 * | 0.296 * |
| TFAM protein | −0.406 * | −0.282 * | 0.510 * | − | −0.419 * | −0.439 * | 0.685 * | −0.038 | −0.349 * |
| MDA | 0.258 * | 0.519 * | −0.045 | −0.419 * | − | 0.615 * | −0.079 | 0.372 * | 0.623 * |
| 8-OHdG | 0.218 | 0.413 * | −0.288 * | −0.439 * | 0.615 * | − | −0.294 * | 0.138 | 0.556 * |
| Nfe2l2 protein | −0.066 | 0.055 | 0.536 * | 0.423 * | 0.074 | −0.204 | 0.490 * | 0.096 | 0.036 |
| NF-κB protein | 0.373 * | 0.542 * | 0.208 | −0.420 * | 0.833 * | 0.545 * | −0.025 | 0.400 * | 0.533 * |
| mtDNA-CN | −0.393 * | 0.015 | 0.636 * | 0.685 * | −0.079 | −0.294 * | − | 0.170 | −0.034 |
| Kiss1 mRNA | −0.484 * | −0.521 * | 0.139 | 0.593 * | −0.571 * | −0.610 * | 0.420 * | −0.221 | −0.510 * |
| Kiss1R mRNA | 0.284 * | 0.371 * | 0.244 | −0.103 | 0.355 * | 0.079 | 0.285 * | 0.559 * | 0.323 * |
| CS activity | −0.049 | 0.038 | 0.789 * | 0.588 * | −0.093 | −0.327 * | 0.766 * | 0.309 * | −0.039 |
| Gene | Accession Number | Primer Sequence | |
|---|---|---|---|
| 18S rRNA (Reference gene) | NR_046237.2 | F: | GTAACCCGTTGAACCCCATT |
| R: | CAAGCTTATGACCCGCACTT | ||
| PGC-1α | NM_031347.1 | F: | GTGCAGCCAAGACTCTGTATGG |
| R: | GTCCAGGTCATTCACATCAAGTTC | ||
| TFAM | NM_031326.2 | F: | CCCACAGAGAACAGAAACAG |
| R: | CCCTGGAAGCTTTCAGATACG | ||
| POLG | NM_053528.1 | F: | GGACCTCCCTTAGAGAGGGA |
| R: | AGCATGCCAGCCAGAGTCACT | ||
| Nfe2l2 | NM_017008.4 | F: | CGAGATATACGCAGCAGGAGAGGTAAG |
| R: | GCTCGACAATGTTCTCCAGCTT | ||
| NF-κB (P65) | NM_199267.2 | F: | CAGGACCAGGAACAGTTCGAA |
| R: | CCAGGTTCTGGAAGCTATGGAT | ||
| ERα | NM_012689.1 | F: | ATGAGAGCTGCCAACCTT |
| R: | AACAAGGCACTGACCATC | ||
| ERβ | NM_012754 | F: | AGGTGCTAATGGTGGGACTG |
| R: | ACTTTCTGCCTCCTGGTTTG | ||
| Kiss1 | NM_181692.1 | F: | GCTGCTGCTTCTCCTCTGTGT |
| R: | CTGTTGGCCTGTGGGTTCA | ||
| Kiss1R | NM_023992.2 | F: | AGCACATGCAGACCGTCACC |
| R: | GACGAATTTGCACATGAAGTCTCC | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hafez, H.A.; Mahmoud, S.A.; Alhmoud, J.F.; Khafaga, R.H.M.; Kamel, M.A.; Shaker, S.A. The Intergeneration Long-Lasting Consequences of Pre-Conceptional Exposure to Sofosbuvir on the Ovarian Tissues of F1 Offspring: Experimental Study on Rats. Int. J. Mol. Sci. 2023, 24, 13675. https://doi.org/10.3390/ijms241813675
Hafez HA, Mahmoud SA, Alhmoud JF, Khafaga RHM, Kamel MA, Shaker SA. The Intergeneration Long-Lasting Consequences of Pre-Conceptional Exposure to Sofosbuvir on the Ovarian Tissues of F1 Offspring: Experimental Study on Rats. International Journal of Molecular Sciences. 2023; 24(18):13675. https://doi.org/10.3390/ijms241813675
Chicago/Turabian StyleHafez, Hala A., Shimaa A. Mahmoud, Jehad F. Alhmoud, Rana H.M. Khafaga, Maher A. Kamel, and Sara A. Shaker. 2023. "The Intergeneration Long-Lasting Consequences of Pre-Conceptional Exposure to Sofosbuvir on the Ovarian Tissues of F1 Offspring: Experimental Study on Rats" International Journal of Molecular Sciences 24, no. 18: 13675. https://doi.org/10.3390/ijms241813675





