Unraveling the Kinetics of the 10–23 RNA-Cleaving DNAzyme
Abstract
:1. Introduction
2. Results and Discussion
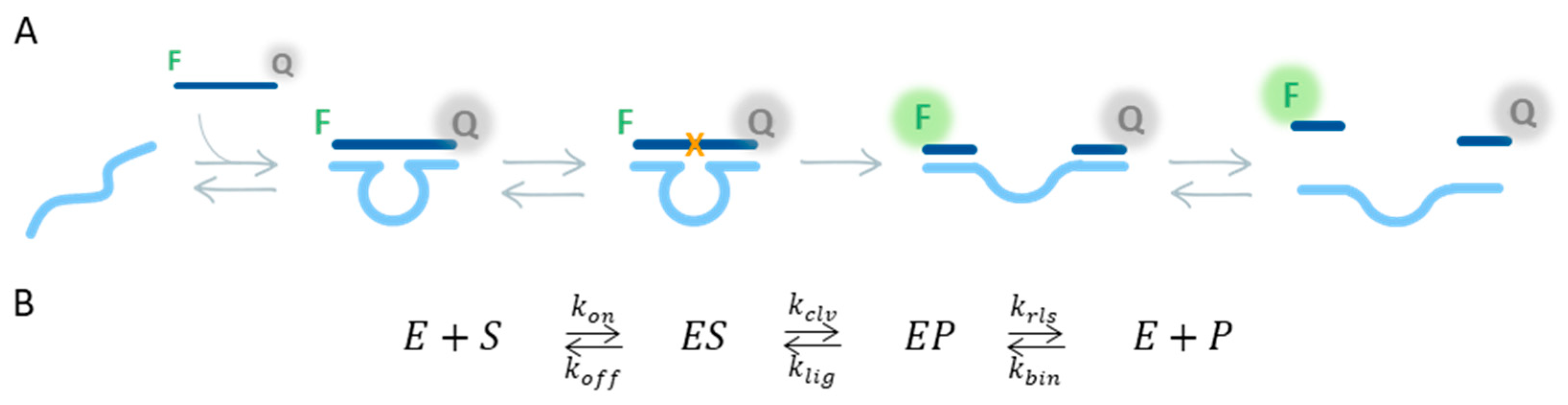
2.1. Activity Measurements
2.2. Model Optimization
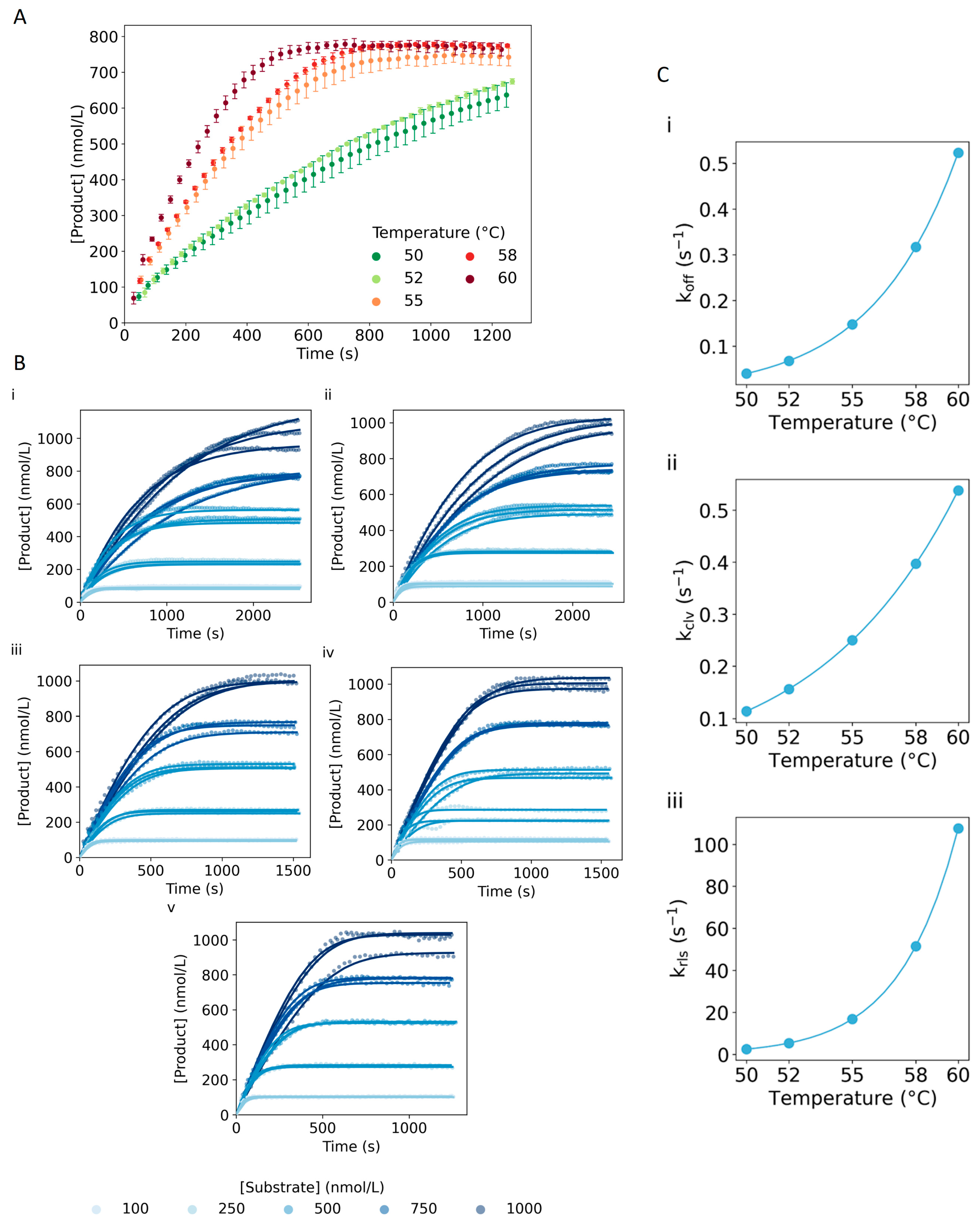
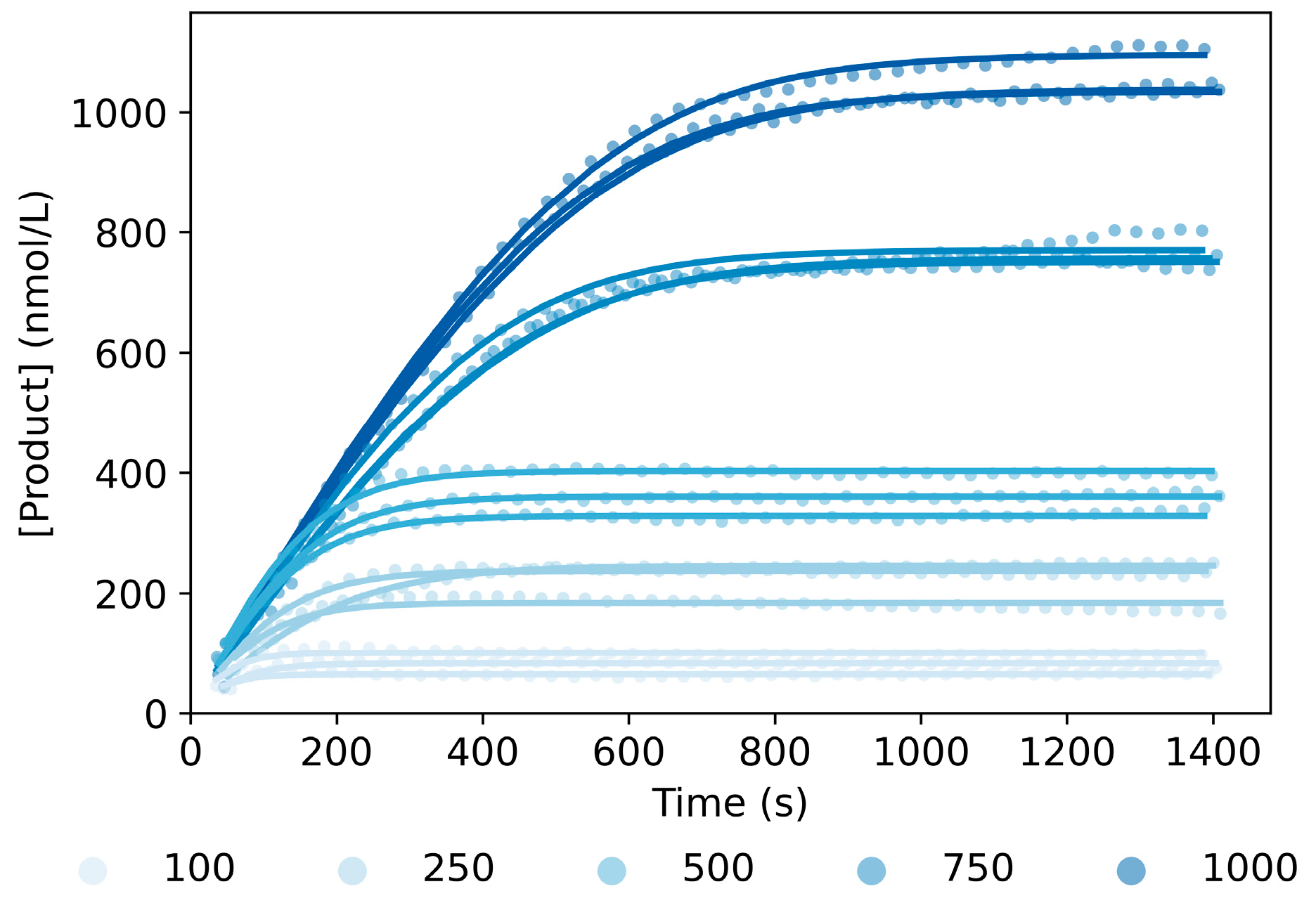
2.3. Model Validation
2.4. Temperature Dependence
2.5. Magnesium Dependence
2.6. Model Implementation for DNAzyme with Shorter Substrate-Binding Arm Sequences
2.7. Implementation of the Model for New Sequences
3. Materials and Methods
3.1. Reagents
3.2. Activity Measurements
3.3. Derivation of the ODEs
3.4. Model Optimization
3.4.1. Model Selection and Optimization
3.4.2. CI Determination
3.4.3. Monte Carlo Simulation of the Model Output
3.5. Temperature Dependence
3.6. Magnesium Dependence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Copeland, R.A. Enzymes. A Practical Introduction to Structure, Mechanism, and Data Analysis, 2nd ed.; Wiley-VCH: New York, NY, USA, 2000; ISBN 0471359297. [Google Scholar]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA Enzyme That Cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Joyce, G.F. Directed Evolution of Nucleic Acid Enzymes. Annu. Rev. Biochem. 2004, 73, 791–836. [Google Scholar] [CrossRef]
- Zhu, X.; Gao, X.; Liu, Q.; Lin, Z.; Qiu, B.; Chen, G. Pb2+-Introduced Activation of Horseradish Peroxidase (HRP)-Mimicking DNAzyme. Chem. Commun. 2011, 47, 7437–7439. [Google Scholar] [CrossRef]
- Yin, H.; Kuang, H.; Liu, L.; Xu, L.; Ma, W.; Wang, L.; Xu, C. A Ligation Dnazyme-Induced Magnetic Nanoparticles Assembly for Ultrasensitive Detection of Copper Ions. ACS Appl. Mater. Interfaces 2014, 6, 4752–4757. [Google Scholar] [CrossRef]
- Camden, A.J.; Walsh, S.M.; Suk, S.H.; Silverman, S.K. DNA Oligonucleotide 3′-Phosphorylation by a DNA Enzyme. Biochemistry 2016, 55, 2671–2676. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. A General Purpose RNA-Cleaving DNA Enzyme. Biochemistry 1997, 94, 4262–4266. [Google Scholar] [CrossRef]
- Safdar, S.; Lammertyn, J.; Spasic, D. RNA-Cleaving NAzymes: The Next Big Thing in Biosensing? Trends Biotechnol. 2020, 38, 1343–1359. [Google Scholar] [CrossRef]
- Silverman, S.K. Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes. Trends Biochem. Sci. 2016, 41, 595–609. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. Mechanism and Utility of an RNA-Cleaving DNA Enzyme. Biochemistry 1998, 37, 13330–13342. [Google Scholar] [CrossRef]
- Ven, K.; Safdar, S.; Dillen, A.; Spasic, D.; Lammertyn, J. Re-Engineering 10–23 Core DNA- and MNAzymes for Applications at Standard Room Temperature. Anal. Bioanal. Chem. 2018, 411, 205–215. [Google Scholar] [CrossRef] [PubMed]
- McGhee, C.E.; Loh, K.Y.; Lu, Y. DNAzyme Sensors for Detection of Metal Ions in the Environment and Imaging Them in Living Cells. Curr. Opin. Biotechnol. 2017, 45, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Safdar, S.; Ven, K.; van Lent, J.; Pavie, B.; Rutten, I.; Dillen, A.; Munck, S.; Lammertyn, J.; Spasic, D. DNA-Only, Microwell-Based Bioassay for Multiplex Nucleic Acid Detection with Single Base-Pair Resolution Using MNAzymes. Biosens. Bioelectron. 2020, 152. [Google Scholar] [CrossRef] [PubMed]
- Peeters, B.; Safdar, S.; Daems, D.; Goos, P.; Spasic, D.; Lammertyn, J. Solid-Phase PCR-Amplified DNAzyme Activity for Real-Time FO-SPR Detection of the MCR-2 Gene. Anal. Chem. 2020, 92, 10783–10791. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiao, L.; Xiang, Y.; Zhou, J.; Tong, A. A General Approach for Rational Design of Fluorescent DNA Aptazyme Sensors Based on Target-Induced Unfolding of DNA Hairpins. Anal. Chim. Acta 2015, 889, 179–186. [Google Scholar] [CrossRef]
- Huang, J.; He, Y.; Yang, X.H.; Quan, K.; Wang, K.M. Inhibited Aptazyme-Based Catalytic Molecular Beacon for Amplified Detection of Adenosine. Chin. Chem. Lett. 2014, 25, 1211–1214. [Google Scholar] [CrossRef]
- Zhang, L.; Guo, S.; Zhu, J.; Zhou, Z.; Li, T.; Li, J.; Dong, S.; Wang, E. Engineering DNA Three-Way Junction with Multifunctional Moieties: Sensing Platform for Bioanalysis. Anal. Chem. 2015, 87, 11295–11300. [Google Scholar] [CrossRef]
- Montserrat Pagès, A.; Safdar, S.; Ven, K.; Lammertyn, J.; Spasic, D. DNA-Only Bioassay for Simultaneous Detection of Proteins and Nucleic Acids. Anal. Bioanal. Chem. 2021, 413, 4925–4937. [Google Scholar] [CrossRef]
- Singh, N.; Ranjan, A.; Sur, S.; Chandra, R.; Tandon, V. Inhibition of HIV-1 Integrase Gene Expression by 10–23 DNAzyme. Proc. J. Biosci. 2012, 37, 493–502. [Google Scholar] [CrossRef]
- Fokina, A.A.; Stetsenko, D.A.; François, J.C. DNA Enzymes as Potential Therapeutics: Towards Clinical Application of 10–23 DNAzymes. Expert. Opin. Biol. Ther. 2015, 15, 689–711. [Google Scholar] [CrossRef]
- Zhang, J. RNA-Cleaving DNAzymes: Old Catalysts with New Tricks for Intracellular and in Vivo Applications. Catalysts 2018, 8, 550. [Google Scholar] [CrossRef]
- Victor, J.; Steger, G.; Riesner, D. Inability of DNAzymes to Cleave RNA in Vivo Is Due to Limited Mg2+ Concentration in Cells. Eur. Biophys. J. 2018, 47, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Ota, N.; Warashina, M.; Hirano, K.N.I.; Hatanaka, K.; Taira, K. Effects of Helical Structures Formed by the Binding Arms of DNAzymes and Their Substrates on Catalytic Activity. Nucleic Acids Res. 1998, 26, 3385–3391. [Google Scholar] [CrossRef] [PubMed]
- Nowakowski, J.; Shim, P.J.; Prasad, G.S.; Stout, C.D.; Joyce, G.F. Crystal Structure of an 82-Nucleotide RNA-DNA Complex Formed by the 10–23 DNA Enzyme. Nat. Struct. Biol. 1999, 6, 151–156. [Google Scholar] [CrossRef]
- Jung, J.; Han, K.Y.; Koh, H.R.; Lee, J.; Choi, Y.M.; Kim, C.; Kim, S.K. Effect of Single-Base Mutation on Activity and Folding of 10–23 Deoxyribozyme Studied by Three-Color Single-Molecule ALEX FRET. J. Phys. Chem. B 2012, 116, 3007–3012. [Google Scholar] [CrossRef]
- Dolot, R.; Sobczak, M.; Mikołajczyk, B.; Nawrot, B. Synthesis, Crystallization and Preliminary Crystallographic Analysis of a 52-Nucleotide DNA/2′-OMe-RNA Oligomer Mimicking 10–23 DNAzyme in the Complex with a Substrate. Nucleosides Nucleotides Nucleic Acids 2017, 36, 292–301. [Google Scholar] [CrossRef]
- Borggräfe, J.; Victor, J.; Rosenbach, H.; Viegas, A.; Gertzen, C.G.W.W.; Wuebben, C.; Kovacs, H.; Gopalswamy, M.; Riesner, D.; Steger, G.; et al. Time-Resolved Structural Analysis of an RNA-Cleaving DNA Catalyst. Nature 2022, 601, 144–149. [Google Scholar] [CrossRef]
- Zaborowska, Z.; Schubert, S.; Kurreck, J.; Erdmann, V.A. Deletion Analysis in the Catalytic Region of the 10–23 DNA Enzyme. FEBS Lett. 2005, 579, 554–558. [Google Scholar] [CrossRef]
- Zaborowska, Z.; Fürste, J.P.; Erdmann, V.A.; Kurreck, J. Sequence Requirements in the Catalytic Core of the “10–23” DNA Enzyme. J. Biol. Chem. 2002, 277, 40617–40622. [Google Scholar] [CrossRef]
- Li, P.; Du, S.; Li, Y.; He, J. Studies on the Two Thymine Residues in the Catalytic Core of 10–23 DNAzyme: The Impact on the Catalysis of Their 5-Substituted Functional Groups. Molecules 2017, 22, 1011. [Google Scholar] [CrossRef]
- Cairins, M.J.; Hopkins, T.M.; Witherington, C.; Sun, L.-Q. The Influence of Arm Length Asymmetry and Base Substitution on the Activity of the 10–23 DNA Enzyme. Antisense Nucleic Acid Drug Dev. 2000, 10, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ho, C.M. Aptamer-Based Optical Probes with Separated Molecular Recognition and Signal Transduction Modules. J. Am. Chem. Soc. 2008, 130, 2380–2381. [Google Scholar] [CrossRef]
- Wu, D.; Gao, T.; Lei, L.; Yang, D.; Mao, X.; Li, G. Colorimetric Detection of Proteins Based on Target-Induced Activation of Aptazyme. Anal. Chim. Acta 2016, 942, 68–73. [Google Scholar] [CrossRef]
- Warashina, M.; Takagi, Y.; Sawata, S.; Zhou, D.M.; Kuwabara, T.; Taira, K. Entropically Driven Enhancement of Cleavage Activity of a DNA-Armed Hammerhead Ribozyme: Mechanism of Action of Hammerhead Ribozymes. J. Org. Chem. 1997, 62, 9138–9147. [Google Scholar] [CrossRef]
- Kenward, M.; Dorfman, K.D. Coarse-Grained Brownian Dynamics Simulations of the 10–23 DNAzyme. Biophys. J. 2009, 97, 2785–2793. [Google Scholar] [CrossRef]
- Meng, X.X.; Yang, X.H.; Wang, K.M.; Tan, W.H.; Guo, Q.P. Real-Time Monitoring of DNAzyme Cleavage Process Using Fluorescent Assay. Chin. Chem. Lett. 2009, 20, 990–994. [Google Scholar] [CrossRef]
- Bone, S.M.; Hasick, N.J.; Lima, N.E.; Erskine, S.M.; Mokany, E.; Todd, A.V. DNA-Only Cascade: A Universal Tool for Signal Amplification, Enhancing the Detection of Target Analytes. Anal. Chem. 2014, 86, 9106–9113. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, J.E.; Neath, A.A. The Akaike Information Criterion: Background, Derivation, Properties, Application, Interpretation, and Refinements. Wiley Interdiscip. Rev. Comput. Stat. 2019, 11, e1460. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C.; Chen, C.C. Bootstrap Techniques for Error Estimation. IEEE Trans. Pattern Anal. Mach. Intell. 1987, PAMI-9, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Breaker, R.R.; Emilsson, G.M.; Lazarev, D.; Nakamura, S.; Puskarz, I.J.; Roth, A.; Sudarsan, N. A Common Speed Limit for RNA-Cleaving Ribozymes and Deoxyribozymes. RNA 2003, 9, 949–957. [Google Scholar] [CrossRef]
- Kroese, D.P.; Brereton, T.; Taimre, T.; Botev, Z.I. Why the Monte Carlo Method Is So Important Today. Wiley Interdiscip. Rev. Comput. Stat. 2014, 6, 386–392. [Google Scholar] [CrossRef]
- Bisswanger, H. Enzyme Kinetics: Principles and Methods; John Wiley & Sons: Hoboken, NJ, USA, 2009; Volume 46, ISBN 9783527342518. [Google Scholar]
- Ouldridge, T.E.; Šulc, P.; Romano, F.; Doye, J.P.K.; Louis, A.A. DNA Hybridization Kinetics: Zippering, Internal Displacement and Sequence Dependence. Nucleic Acids Res. 2013, 41, 8886–8895. [Google Scholar] [CrossRef]
- Sorgenfrei, S.; Chiu, C.Y.; Gonzalez, R.L.; Yu, Y.J.; Kim, P.; Nuckolls, C.; Shepard, K.L. Label-Free Single-Molecule Detection of DNA-Hybridization Kinetics with a Carbon Nanotube Field-Effect Transistor. Nat. Nanotechnol. 2011, 6, 126–132. [Google Scholar] [CrossRef]
- Chen, C.; Wang, W.; Wang, Z.; Wei, F.; Zhao, X.S. Influence of Secondary Structure on Kinetics and Reaction Mechanism of DNA Hybridization. Nucleic Acids Res. 2007, 35, 2875–2884. [Google Scholar] [CrossRef]
- Peleg, M.; Normand, M.D.; Corradini, M.G. The Arrhenius Equation Revisited. Crit. Rev. Food Sci. Nutr. 2012, 52, 830–851. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.J.; Chen, S.J. Nucleic Acid Helix Stability: Effects of Salt Concentration, Cation Valence and Size, and Chain Length. Biophys. J. 2006, 90, 1175–1190. [Google Scholar] [CrossRef]
- Hertog, M.; Verlinden, B.E.; Lammertyn, J.; Nicolaï, B.M. OptiPa, an Essential Primer to Develop Models in the Postharvest Area. Comput. Electron. Agric. 2007, 57, 99–106. [Google Scholar] [CrossRef]
- Egea, J.A.; Vazquez, E.; Banga, J.R.; Martí, R. Improved Scatter Search for the Global Optimization of Computationally Expensive Dynamic Models. J. Glob. Optim. 2009, 43, 175–190. [Google Scholar] [CrossRef]




| Parameter | 95% CI |
|---|---|
| kon ((nmol/L)−1·s−1) | 4.32 × 10−3–4.73 × 10−3 |
| koff (s−1) | 1.46 × 10−1–1.65 × 10−1 |
| kclv (s−1) | 2.10 × 10−1–2.34 × 10−1 |
| krls (s−1) | 1.91 × 101–2.22 × 101 |
| kbin ((nmol/L)−1·s−1) | 1.07 × 10−1–1.19 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montserrat Pagès, A.; Hertog, M.; Nicolaï, B.; Spasic, D.; Lammertyn, J. Unraveling the Kinetics of the 10–23 RNA-Cleaving DNAzyme. Int. J. Mol. Sci. 2023, 24, 13686. https://doi.org/10.3390/ijms241813686
Montserrat Pagès A, Hertog M, Nicolaï B, Spasic D, Lammertyn J. Unraveling the Kinetics of the 10–23 RNA-Cleaving DNAzyme. International Journal of Molecular Sciences. 2023; 24(18):13686. https://doi.org/10.3390/ijms241813686
Chicago/Turabian StyleMontserrat Pagès, Aida, Maarten Hertog, Bart Nicolaï, Dragana Spasic, and Jeroen Lammertyn. 2023. "Unraveling the Kinetics of the 10–23 RNA-Cleaving DNAzyme" International Journal of Molecular Sciences 24, no. 18: 13686. https://doi.org/10.3390/ijms241813686
APA StyleMontserrat Pagès, A., Hertog, M., Nicolaï, B., Spasic, D., & Lammertyn, J. (2023). Unraveling the Kinetics of the 10–23 RNA-Cleaving DNAzyme. International Journal of Molecular Sciences, 24(18), 13686. https://doi.org/10.3390/ijms241813686








