Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder
Abstract
:1. Introduction
2. Results
2.1. Prenatal EtOH Exposure Inhibits 18S rRNA Expression in Fetal Neurons and FB-Es
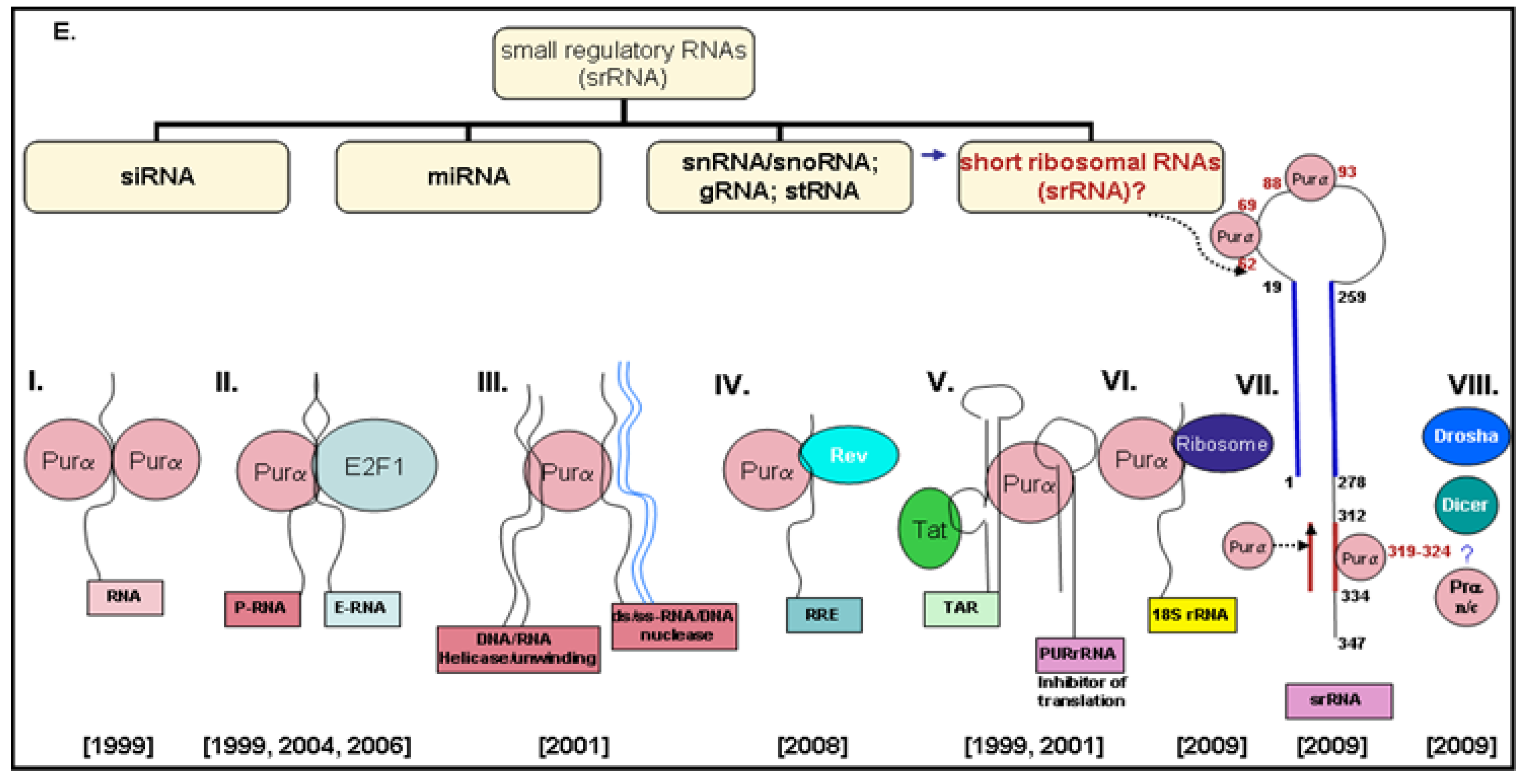
2.2. Generation of 18S-Homologous Small Ribosomal RNAs (srRNAs)
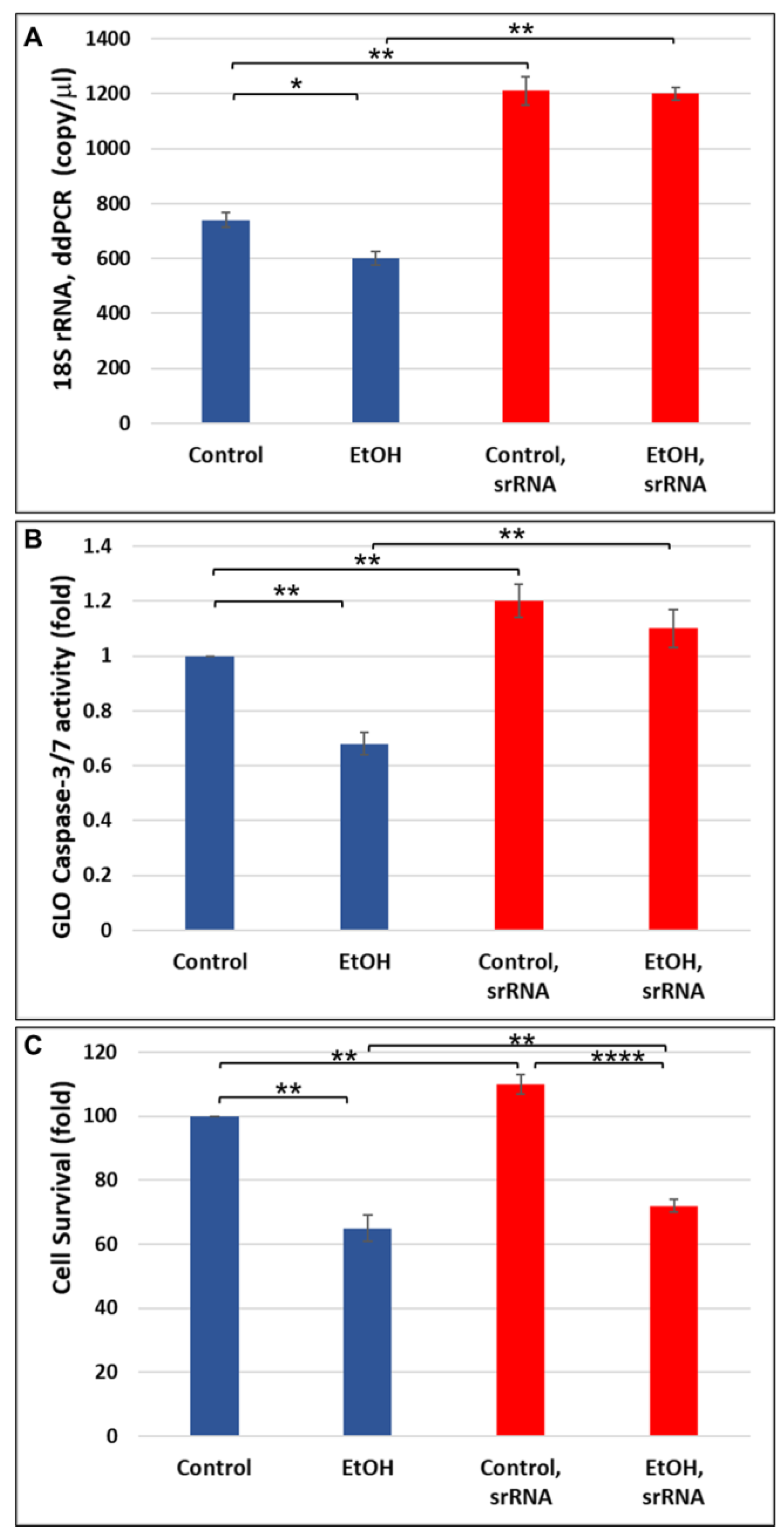
2.3. srRNA Reverses Inhibitory Effect of EtOH Exposure on 18S rRNA Expression in Human Neuronal Cells
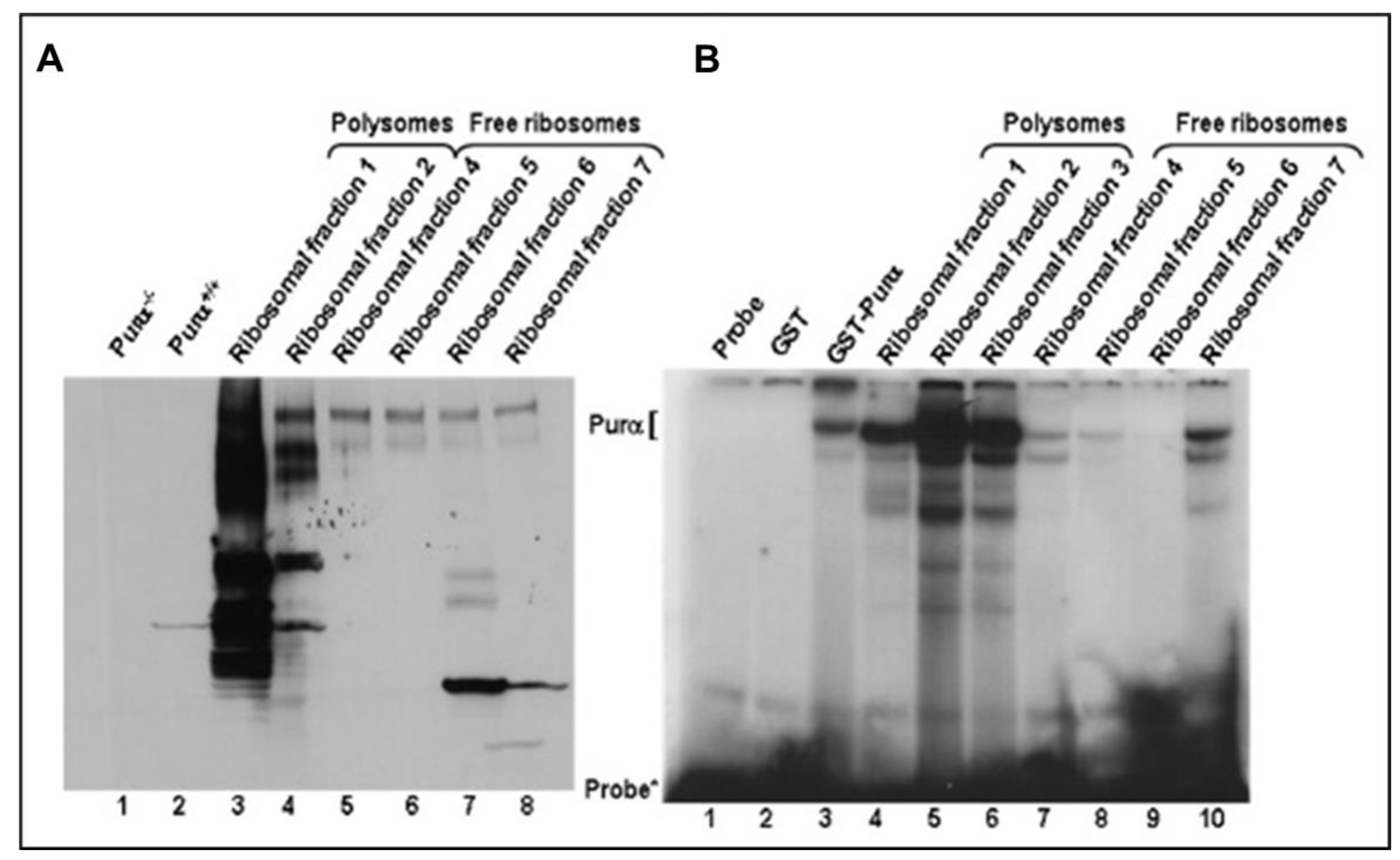
2.4. 18S Ribosomal RNA in Polyribosomes Is in the Complex with the Cellular Protein (Purα)
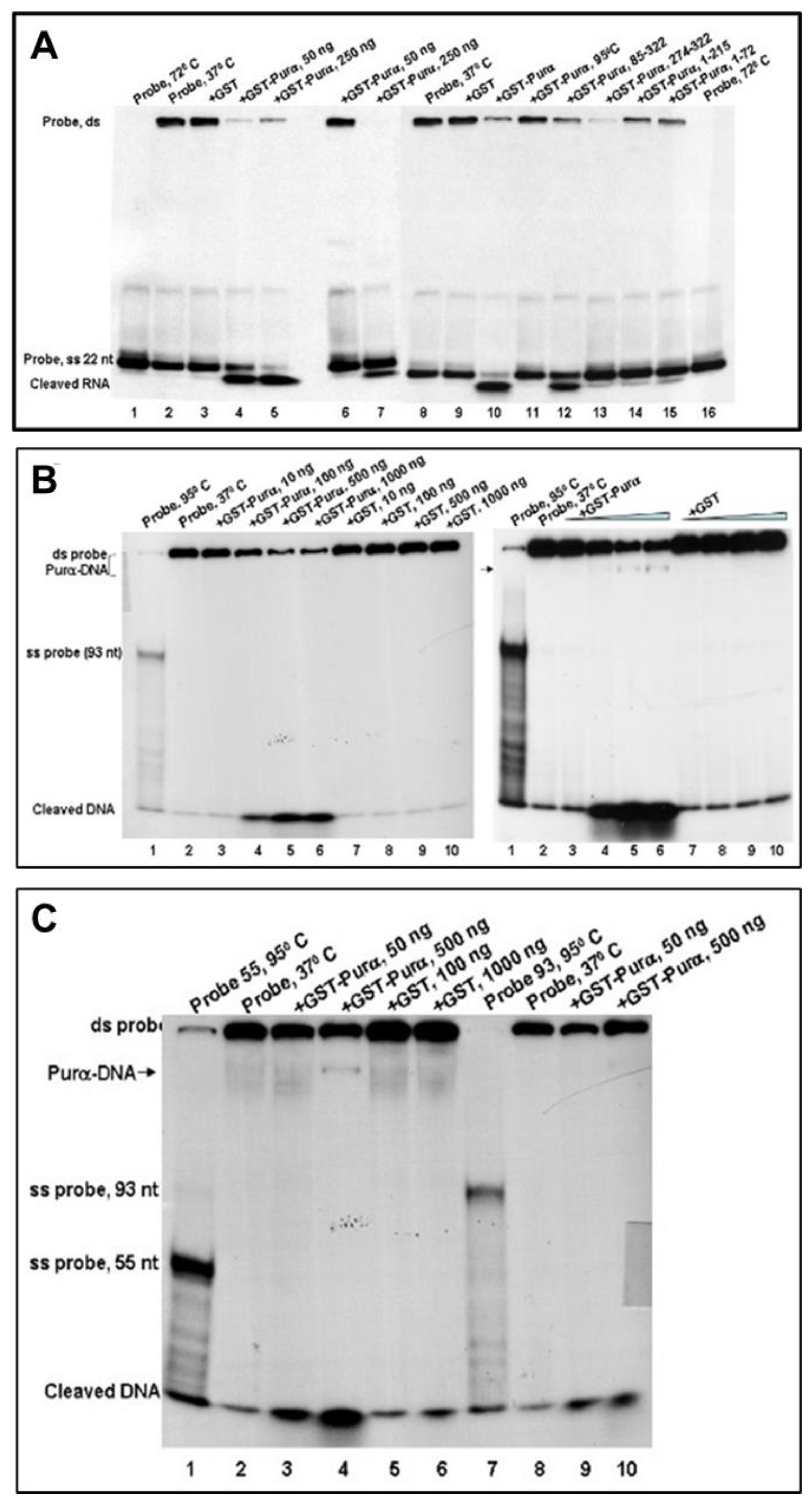
2.5. 18S Ribosomal RNA Is Cleaved by a Cellular Protein (Purα)
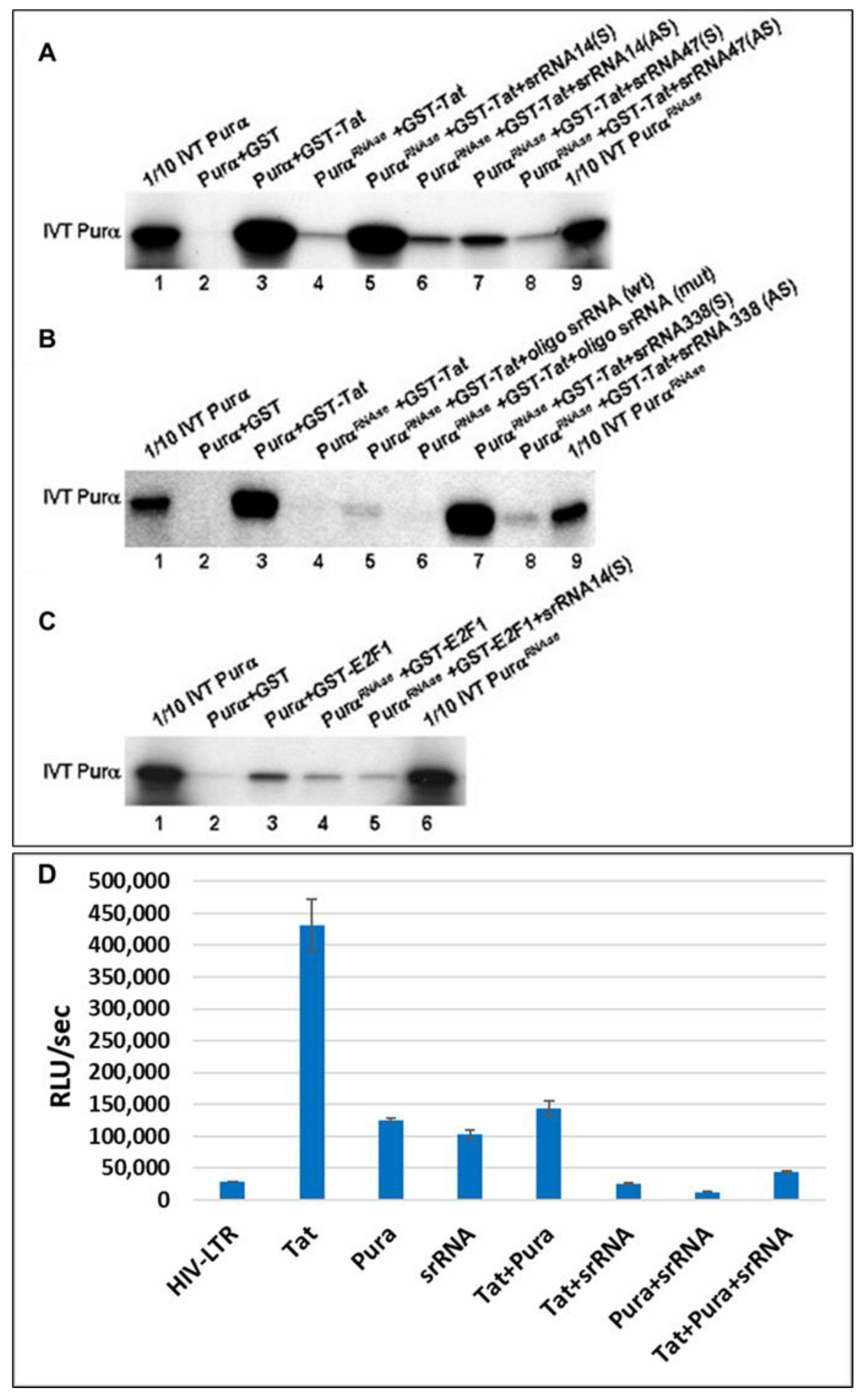
2.6. 18S Homologous srRNA Regulates Cell–Virus Association
2.7. Potential Targets of srRNA
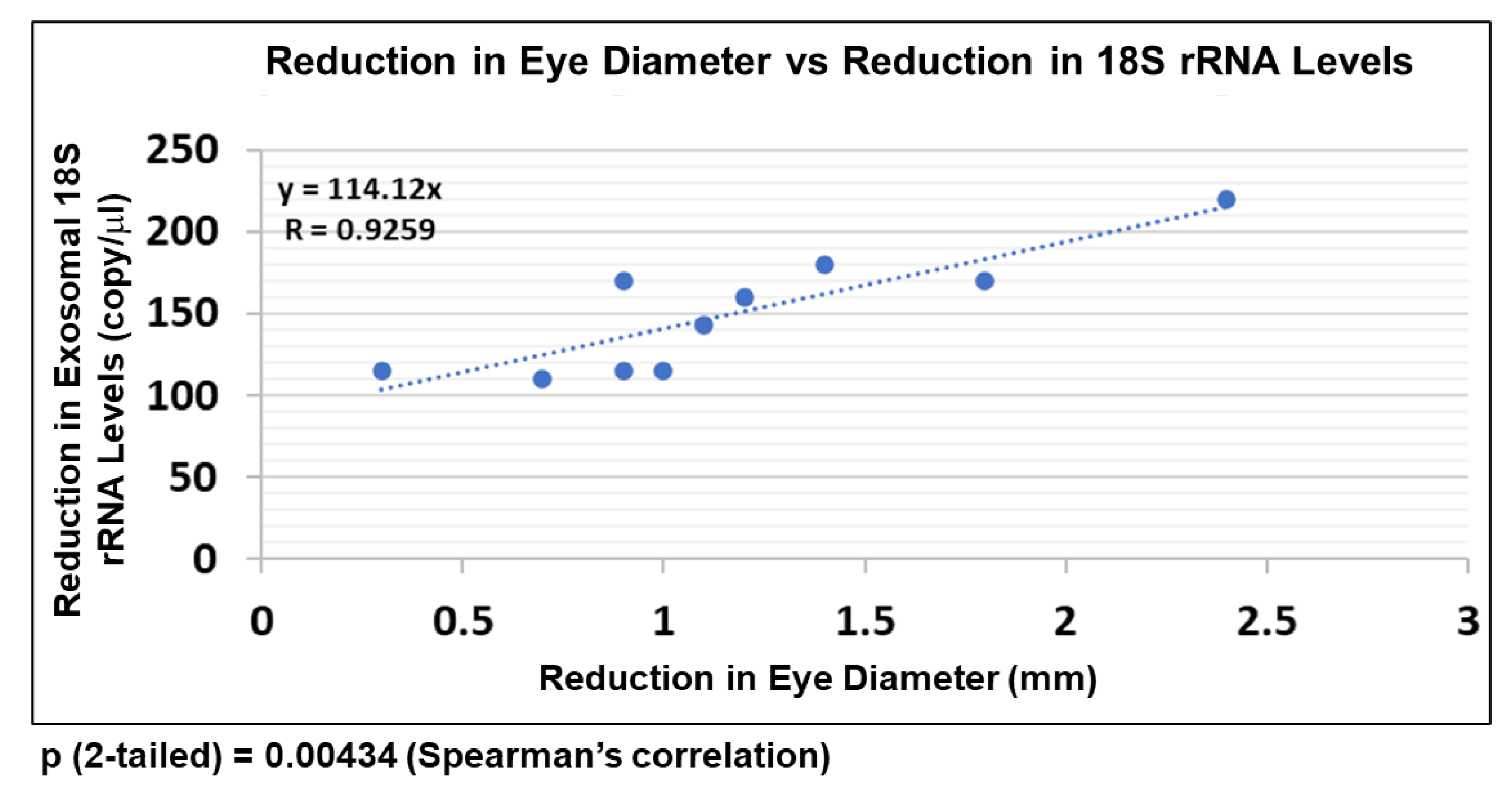
2.8. EtOH-Induced Reduction in FB-E srRNA Content Correlates with Reduction in Human Fetal Eye Size
3. Discussion
3.1. Small Ribosomal RNAs as a New Indicator of Alcohol-Associated Fetal Pathology
3.2. Potential Molecular Biomarkers, Using Novel Non-Invasive Tools
3.3. Formation of srRNAs
4. Methods
4.1. Clinical Samples
- Eligibility Criteria: The blood and tissue samples were obtained according to NIH Guidelines through a trained study coordinator. Samples were collected regardless of sex, ethnic background, and race.
- Treatment Plan: Each patient was asked to sign a separate consent form for research on blood and tissue samples. Blood obtained was processed for collection of serum and plasma. No invasive procedures were performed on the mother, other than those used in her routine medical care. Fetal tissues were processed for RNA or protein isolation.
- Risk and Benefits: There are very small risks of loss of privacy, as with any research study where protected health information is viewed. The samples were depersonalized before they were sent to the lab for analysis. There were no additional risks of blood sampling as this was only performed in patients with clinically indicated venous access. There was little anticipated risk from obtaining approximately 2–3 cc of blood, but a well-trained study coordinator collected all samples.
- d.
- Informed Consent: Consent forms were maintained by the study coordinator and were not sent to the investigator with the samples. The deidentified log sheets and IRB protocol were sent by the study coordinator to the principal investigator with each blood and tissue sample. This sheet contains an assigned accession number, the age, sex, ethnicity, and race of the patient. Except for an assigned accession number, no identification was kept on the blood and tissue samples.
4.2. Cell Culture
4.3. RNA Preparation and Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR)
4.4. Sex Determination Using Human Fetal Genomic DNA
4.5. Droplet Digital PCR (ddPCR)
4.6. Primers (IDT Inc., Coralville, IA, USA)
4.7. Isolation of Fetal Brain-Derived Exosomes (FB-Es) from Maternal Plasma
4.8. Ribosome Analysis
4.9. Electrophoretic Mobility Shift Assay (EMSA)
4.10. DNA Helix-Destabilizing Activity of Purα
4.11. Unwinding Assay
4.12. Nuclease Assay
4.13. GST Pull-Down Assays
4.14. Overexpression and Purification of Recombinant Proteins
4.15. In Vitro Transcription and RNA Binding Assays
4.16. DNA Transfections and Preparation of Cell Extracts
4.17. Quantification of Neuronal Cell Injury: Caspase-GLO 3/7 Activity Assay
4.18. Luciferase Reactions
4.19. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Control and Prevention (CDC). Alcohol use and binge drinking among women of childbearing age—United States, 2006–2010. MMWR Morb. Mortal. Wkly. Rep. 2012, 61, 534–538. [Google Scholar]
- Mutch, R.C.; Jones, H.M.; Bower, C.; Watkins, R.E. Fetal Alcohol Spectrum Disorders: Using Knowledge, Attitudes and Practice of Justice Professionals to Support their Educational Needs. J. Popul. Ther. Clin. Pharmacol. 2016, 23, e77–e89. Available online: https://pubmed.ncbi.nlm.nih.gov/27132254/ (accessed on 22 March 2022).
- Hoyme, H.E.; Kalberg, W.O.; Elliott, A.J.; Blankenship, J.; Buckley, D.; Marais, A.-S.; Manning, M.A.; Robinson, L.K.; Adam, M.P.; Abdul-Rahman, O.; et al. Updated Clinical Guidelines for Diagnosing Fetal Alcohol Spectrum Disorders. Pediatrics 2016, 138, e20154256. [Google Scholar] [CrossRef] [PubMed]
- Sampson, P.D.; Streissguth, A.P.; Bookstein, F.L.; Little, R.E.; Clarren, S.K.; Dehaene, P.; Hanson, J.W.; Graham, J.M., Jr. Incidence of fetal alcohol syndrome and prevalence of alcohol-related neurodevelopmental disorder. Teratology 1997, 56, 317–326. [Google Scholar] [CrossRef]
- Andersen, A.-M.N.; Andersen, P.K.; Olsen, J.; Gronbaek, M.; Strandberg-Larsen, K. Moderate alcohol intake during pregnancy and risk of fetal death. Int. J. Epidemiol. 2012, 41, 405–413. [Google Scholar] [CrossRef]
- Popova, S.; Lange, S.; Probst, C.; Gmel, G.; Rehm, J. Estimation of national, regional, and global prevalence of alcohol use during pregnancy and fetal alcohol syndrome: A systematic review and meta-analysis. Lancet Glob. Health 2017, 5, e290–e299. [Google Scholar] [CrossRef] [PubMed]
- May, P.A.; Tabachnick, B.; Hasken, J.M.; Marais, A.-S.; de Vries, M.M.; Barnard, R.; Joubert, B.; Cloete, M.; Botha, I.; Kalberg, W.; et al. Who is most affected by prenatal alcohol exposure: Boys or girls? Drug Alcohol Depend. 2017, 177, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Abbott, C.W.; Kozanian, O.O.; Kanaan, J.; Wendel, K.M.; Huffman, K.J. The Impact of Prenatal Ethanol Exposure on Neuroanatomical and Behavioral Development in Mice. Alcohol. Clin. Exp. Res. 2016, 40, 122–133. [Google Scholar] [CrossRef]
- Donald, K.A.; Eastman, E.; Howells, F.M.; Adnams, C.; Riley, E.P.; Woods, R.P.; Narr, K.L.; Stein, D.J. Neuroimaging effects of prenatal alcohol exposure on the developing human brain: A magnetic resonance imaging review. Acta Neuropsychiatr. 2015, 27, 251–269. [Google Scholar] [CrossRef]
- West, J.R. Recent findings on the mechanisms by which alcohol damages the developing nervous system. Alcohol Alcohol. 1994, 2, 395–399. Available online: https://pubmed.ncbi.nlm.nih.gov/8974361/ (accessed on 23 March 2022).
- Ikonomidou, C.; Bittigau, P.; Ishimaru, M.J.; Wozniak, D.F.; Koch, C.; Genz, K.; Price, M.T.; Stefovska, V.; Hörster, F.; Tenkova, T.; et al. Ethanol-induced apoptotic neurodegeneration and fetal alcohol syndrome. Science 2000, 287, 1056–1060. [Google Scholar] [CrossRef]
- Goodlett, C.R.; Horn, K.H. Mechanisms of alcohol-induced damage to the developing nervous system. Alcohol Res. Health J. Natl. Inst. Alcohol Abus. Alcohol. 2001, 25, 175–184. [Google Scholar]
- Smith, S.M.; Garic, A.; Flentke, G.R.; Berres, M.E. Neural crest development in fetal alcohol syndrome. Birth Defects Res. C Embryo Today 2014, 102, 210–220. [Google Scholar] [CrossRef]
- Green, M.L.; Singh, A.V.; Zhang, Y.; Nemeth, K.A.; Sulik, K.K.; Knudsen, T.B. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev. Dyn. 2017, 236, 613–631. [Google Scholar] [CrossRef] [PubMed]
- Downing, C.; Balderrama-Durbin, C.; Kimball, A.; Biers, J.; Wright, H.; Gilliam, D.; Johnson, T.E. Quantitative trait locus mapping for ethanol teratogenesis in BXD recombinant inbred mice. Alcohol Clin. Exp. Res. 2012, 36, 1340–1354. [Google Scholar] [CrossRef]
- Garic, A.; Berres, M.E.; Smith, S.M. High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to Fetal Alcohol Spectrum Disorders. Alcohol Clin. Exp. Res. 2014, 38, 1874–1882. [Google Scholar] [CrossRef] [PubMed]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef]
- Trainor, P.A. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am. J. Med. Genet. Part A 2010, 152A, 2984–2994. [Google Scholar] [CrossRef] [PubMed]
- Hetman, M.; Slomnicki, L.P. Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies. J. Neurochem. 2019, 148, 325–347. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef]
- Burman, L.G.; Mauro, V.P. Analysis of rRNA processing and translation in mammalian cells using a synthetic 18S rRNA expression system. Nucleic Acids Res. 2012, 40, 8085–8098. [Google Scholar] [CrossRef]
- Fuentes-Beals, C.; Olivares-Costa, M.; Andrés, M.E.; Haeger, P.A.; Riadi, G.; Oliva, C.; Faunes, F. Bioinformatic analysis predicts that ethanol exposure during early development causes alternative splicing alterations of genes involved in RNA post-transcriptional regulation. PLoS ONE 2023, 18, e0284357. [Google Scholar] [CrossRef] [PubMed]
- Cherlin, T.; Magee, R.; Jing, Y.; Pliatsika, V.; Loher, P.; Rigoutsos, I. Ribosomal RNA fragmentation into short RNAs (rRFs) is modulated in a sex- and population of origin-specific manner. BMC Biol. 2020, 18, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Sun, Q.; Yao, Y. Characterization of Small RNAs Derived from tRNAs, rRNAs and snoRNAs and Their Response to Heat Stress in Wheat Seedlings. PLoS ONE 2016, 11, e0150933. [Google Scholar] [CrossRef]
- Lambert, M.; Benmoussa, A.; Provost, P. Small Non-Coding RNAs Derived from Eukaryotic Ribosomal RNA. Non-coding RNA 2019, 5, 16. [Google Scholar] [CrossRef]
- Hashimoto, J.G.; Beadles-Bohling, A.S.; Wiren, K.M. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. Biotechniques 2004, 36, 54–60. [Google Scholar] [CrossRef]
- Yeung, M.L.; Bennasser, Y.; Le, S.Y.; Jeang, K.T. siRNA, miRNA and HIV: Promises and challenges. Cell Res. 2005, 15, 935–946. [Google Scholar] [CrossRef]
- Gallia, G.L.; Darbinian, N.; Jaffe, N.; Khalili, K. Single-stranded nucleic acid-binding protein, Pur alpha, interacts with RNA homologous to 18S ribosomal RNA and inhibits translation in vitro. J. Cell. Biochem. 2001, 83, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Gallia, G.L.; Darbinian, N.; Tretiakova, A.; Ansari, S.A.; Rappaport, J.; Brady, J.; Wortman, M.J.; Johnson, E.M.; Khalili, K. Association of HIV-1 Tat with the cellular protein, Puralpha, is mediated by RNA. Proc. Natl. Acad. Sci. USA 1999, 96, 11572–11577. [Google Scholar] [CrossRef]
- Amini, S.; Merabova, N.; Khalili, K.; Darbinian, N. p38SJ, a novel DINGG protein protects neuronal cells from alcohol induced injury and death. J. Cell. Physiol. 2009, 221, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Darbinyan, A.; Merabova, N.; Bajwa, A.; Tatevosian, G.; Martirosyan, D.; Zhao, H.; Selzer, M.E.; Goetzl, L. Ethanol-mediated alterations in oligodendrocyte differentiation in the developing brain. Neurobiol. Dis. 2021, 148, 105181. [Google Scholar] [CrossRef]
- Darbinian, N.; Darbinyan, A.; Sinard, J.; Tatevosian, G.; Merabova, N.; D’Amico, F.; Khader, T.; Bajwa, A.; Martirosyan, D.; Gawlinski, A.K.; et al. Molecular Markers in Maternal Blood Exosomes Allow Early Detection of Fetal Alcohol Spectrum Disorders. Int. J. Mol. Sci. 2023, 24, 135. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Sparks, E.S.; Darbinyan, A.; Merabova, N.; Tatevosian, G.; Vadachkoria, E.; Zhao, H.; Amini, S.; Goetzl, L.; Selzer, M.E. Maternal Blood Lipid Biomarkers of Oligodendrocyte Pathology to Predict Fetal Alcohol Spectrum Disorders. Obstet. Gynecol. Res. 2023, 6, 127–138. [Google Scholar] [CrossRef]
- Darbinian, N.; Darbinyan, A.; Merabova, N.; Kassem, M.; Tatevosian, G.; Amini, S.; Goetzl, G.; Selzer, M.E. In Utero Ethanol Exposure Induces Mitochondrial DNA Damage and Inhibits mtDNA Repair in Developing Brain. Front. Neurosci. 2023, 17, 1214958. [Google Scholar] [CrossRef]
- Hayashi, R. Gene Expression and the Impact of an Antioxidant Supplement in the Cataractous Lens. In Handbook of Nutrition, Diet, and the Eye, 2nd ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 551–568. ISBN 9780128152454. [Google Scholar] [CrossRef]
- Hayashi, R.; Hayashi, S.; Arai, K.; Chikuda, M.; Obara, Y. Effects of antioxidant supplementation on mRNA expression of glucose-6-phosphate dehydrogenase, β-actin and 18S rRNA in the anterior capsule of the lens in cataract patients. Exp. Eye Res. 2012, 96, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Gallia, G.L.; Darbinian, N.; Johnson, E.M.; Khalili, K. Self-association of Purα is mediated by RNA. J. Cell Biochem. 1999, 74, 334–348. [Google Scholar] [CrossRef]
- Chu, E.; Copur, S.M.; Ju, J.; Chen, T.M.; Khleif, S.; Voeller, D.M.; Mizunuma, N.; Patel, M.; Maley, G.F.; Maley, F.; et al. Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol. Cell Biol. 1999, 19, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Gallia, G.L.; Kundu, M.; Shcherbik, N.; Tretiakova, A.; Giordano, A.; Khalili, K. Association of Purα and E2F-1 suppresses transcriptional activity of E2F-1. Oncogene 1999, 18, 6398–6402. [Google Scholar] [CrossRef]
- Darbinian, N.; White, M.K.; Gallia, G.; Amini Sh Rappaport, J.; Khalili, K. Interaction between the Purα and E2F-1 transcription factors. Anticancer. Res. 2004, 24, 2585–2594. [Google Scholar]
- Darbinian, N.; White, M.K.; Khalili, K. Regulation of the Purα promoter by E2F-1. J. Cell Biochem. 2006, 99, 1052–1063. [Google Scholar] [CrossRef]
- Darbinian, N.; Gallia, G.L.; Khalili, K. Helix-destabilizing properties of the human single-stranded DNA- and RNA-binding protein Purα. J. Cell Biochem. 2001, 80, 589–595. [Google Scholar] [CrossRef]
- Darbinian, N.; Cui, J.; Basile, A.; Del Valle, L.; Miklossy, J.; Sawaya, B.E.; Amini, S.; Khalili, K.; Gordon, J. Negative regulation of AbPP gene expression by Purα. J. Alzheimer’s Dis. 2008, 15, 71–82. [Google Scholar] [CrossRef]
- Ramsey, P.H. Critical Values for Spearman’s Rank Order Correlation. J. Educ. Stat. 1989, 14, 245–253. [Google Scholar]
- Fowler, J.; Cohen, L.; Jarvis, P. Practical Statistics for Field Biology; John Wiley & Sons: Hoboken, NJ, USA, 2009; p. 132. [Google Scholar]
- Rosett, H.L. A Clinical Perspective of the Fetal Alcohol Syndrome. Alcohol. Clin. Exp. Res. 1980, 4, 119–122. [Google Scholar] [CrossRef]
- Abdelrahman, A.; Conn, R. Eye abnormalities in fetal alcohol syndrome. Ulst. Med. J. 2009, 78, 164–165. [Google Scholar]
- Strömland, K. Visual impairment and ocular abnormalities in children with fetal alcohol syndrome. Addict. Biol. 2004, 9, 153–157. [Google Scholar] [CrossRef]
- Rice, D.; Barone, S. Critical periods of vulnerability for the developing nervous system: Evidence from humans and animal models. Environ. Health Perspect. 2000, 108, 511–533. [Google Scholar]
- Bayer, S.A.; Altman, J. Directions in neurogenetic gradients and patterns of anatomical connections in the telencephalon. Prog. Neurobiol. 1987, 29, 57–106. [Google Scholar] [CrossRef]
- Gallia, G.L.; Johnson, E.M.; Khalili, K. Puralpha: A multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000, 28, 3197–3205. [Google Scholar] [CrossRef]
- Haas, S.; Steplewski, A.; Siracusa, L.D.; Amini, S.; Khalili, K. Identification of a sequence-specific single-stranded DNA binding protein that suppresses transcription of the mouse myelin basic protein gene. J. Biol Chem. 1995, 270, 12503–12510. [Google Scholar] [CrossRef]
- Haas, S.; Thatikunta, P.; Steplewski, A.; Johnson, E.M.; Khalili, K.; Amini, S. A 39-kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J. Cell Biol. 1995, 130, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Tretiakova, A.; Steplewski, A.; Johnson, E.M.; Khalili, K.; Amini, S. Regulation of myelin basic protein gene transcription by Sp1 and Puralpha: Evidence for association of Sp1 and Puralpha in brain. J. Cell Physiol. 1999, 181, 160–168. [Google Scholar] [CrossRef]
- Goetzl, L.; Darbinian, N.; Goetzl, E.J. Novel window on early human neurodevelopment via fetal exosomes in maternal blood. Ann. Clin. Transl. Neurol. 2016, 3, 381–385. [Google Scholar] [CrossRef]
- Goetzl, L.; Darbinian, N.; Merabova, N. Noninvasive assessment of fetal central nervous system insult: Potential application to prenatal diagnosis. Prenat. Diagn. 2019, 39, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Selzer, M.E. Oligodendrocyte pathology in fetal alcohol spectrum disorders. Neural Regen. Res. 2022, 17, 497–502. [Google Scholar] [CrossRef]
- Tretiakova, A.; Otte, J.; Croul, S.E.; Kim, J.H.; Johnson, E.M.; Amini, S.; Khalili, K. Association of JC virus large T antigen with myelin basic protein transcription factor (MEF-1/Puralpha) in hypomyelinated brains of mice transgenically expressing T antigen. J. Virol. 1999, 73, 6076–6084. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, S.; Koike, K.; Omori, A.; Ichinose, S.; Ohara, S.; Kobayashi, S.; Sato, T.A.; Anzai, K. Identification of mRNA/protein (mRNP) complexes containing Puralpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J. Biol. Chem. 2002, 277, 37804–37810. [Google Scholar] [CrossRef]
- Tretiakova, A.; Gallia, G.L.; Shcherbik, N.; Jameson, B.; Johnson, E.M.; Amini, S.; Khalili, K. Association of Puralpha with RNAs homologous to 7 SL determines its binding ability to the myelin basic protein promoter DNA sequence. J. Biol. Chem. 1998, 273, 22241–22247. [Google Scholar] [CrossRef]
- Goetzl, L.; Thompson-Felix, T.; Darbinian, N.; Merabova, N.; Merali, S.; Merali, C.; Sanserino, K.; Tatevosian, T.; Fant, B.; Wimmer, M.E. Novel biomarkers to assess in utero effects of maternal opioid use: First steps toward understanding short- and long-term neurodevelopmental sequelae. Genes Brain Behav. 2019, 18, e12583. [Google Scholar] [CrossRef]
- Dukes, K.A.; Burd, L.; Elliott, A.J.; Fifer, W.P.; Folkerth, R.D.; Hankins, G.D.V.; Hereld, D.; Hoffman, H.J.; Myers, M.M.; Odendaal, H.J.; et al. The Safe Passage Study: Design, Methods, Recruitment, and Follow-Up Approach. Paediatr. Perinat. Epidemiol. 2014, 28, 455–465. [Google Scholar] [CrossRef]
- Stewart, S.H.; Koch, D.G.; Burgess, D.M.; Willner, I.R.; Reuben, A. Sensitivity and specificity of urinary ethyl glucuronide and ethyl sulfate in liver disease patients. Alcohol Clin. Exp. Res. 2013, 37, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Spong, C.Y.; Mercer, B.M.; D’Alton, M.; Kilpatrick, S.; Blackwell, S.; Saade, G. Timing of indicated late-preterm and early-term birth. Obstet Gynecol. 2011, 118, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Darbinyan, A.; Kaminski, R.; White, M.K.; Pozniak, P.D.; Darbinian, N.; Khalili, K. Isolation and Propagation of Primary Human and Rodent Embryonic Neural Progenitor Cells and Cortical Neurons. Methods Mol. Biol. 2021, 2311, 51–61. [Google Scholar] [CrossRef]
- Darbinian, N.; Darbinyan, A.; Khalili, K.; Amini, S. Fetal Brain Injury Models of Fetal Alcohol Syndrome: Examination of Neuronal Morphologic Condition Using Sholl Assay. Methods Mol. Biol. 2021, 2311, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Khalili, K.; Amini, S. Neuroprotective activity of pDING in response to HIV-1 Tat. J. Cell Physiol. 2014, 229, 153–161. [Google Scholar] [CrossRef]
- Darbinyan, A.; Kaminski, R.; White, M.K.; Darbinian-Sarkissian, N.; Khalili, K. Polyomavirus JC infection inhibits differentiation of oligodendrocyte progenitor cells. J. Neurosci. Res. 2013, 91, 116–127. [Google Scholar] [CrossRef]
- De Simone, F.I.; Darbinian, N.; Amini, S.; Muniswamy, M.; White, M.K.; Elrod, J.W.; Datta, P.K.; Langford, D.; Khalili, K. HIV-1 Tat and Cocaine Impair Survival of Cultured Primary Neuronal Cells via a Mitochondrial Pathway. J. Neuroimmune Pharmacol. 2016, 11, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Darbinian, N.; Darbinyan, A.; Merabova, N.; Selzer, M.E.; Amini, S. HIV-1 and HIV-1-Tat Induce Mitochondrial DNA Damage in Human Neurons. J. HIV AIDS 2020, 6, 176. [Google Scholar] [CrossRef]
- Darbinian-Sarkissian, N.; Darbinyan, A.; Otte, J.; Radhakrishnan, S.; Sawaya, B.E.; Arzumanyan, A.; Chipitsyna, G.; Popov, Y.; Rappaport, J.; Amini, S.; et al. p27(SJ), a novel protein in St John’s Wort, that suppresses expression of HIV-1 genome. Gene Ther. 2006, 13, 288–295. [Google Scholar] [CrossRef]
- Yano, H.; Wang, B.E.; Ahmad, I.; Zhang, J.; Abo, T.; Nakayama, J.; Krempen, K.; Kohwi, Y. Identification of (CAG)(n) and (CGG)(n) repeat-binding proteins, CAGERs expressed in mature neurons of the mouse brain. Exp. Cell Res. 1999, 251, 388–400. [Google Scholar] [CrossRef]
- Stahl, F.W. Roles of double-strand breaks in generalized genetic recombination. Prog. Nucleic Acid Res. Mol. Biol. 1986, 33, 169–194. [Google Scholar] [PubMed]
- Rosner, B. Fundamentals of Biostatistics, 7th ed.; Brooks/Cole: Boston, MA, USA, 2011. [Google Scholar]
- Venter, J.C.; Adams, M.D.; Myers, E.W.; Li, P.W.; Mural, R.J.; Sutton, G.G.; Smith, H.O.; Yandell, M.; Evans, C.A.; Holt, R.A.; et al. The sequence of the human genome. Science 2001, 291, 1304–1351, Erratum in Science 2001, 292, 1838. [Google Scholar] [CrossRef]
- Chan, E.Y. Advances in sequencing technology. Mutat Res. 2005, 573, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Pospísilová, S.; Tichý, B.; Mayer, J. Human genome sequencing—Next generation technology or will the routine sequencing of human genome be possible? Cas. Lek. Cesk. 2009, 148, 296–302. [Google Scholar] [PubMed]
- Hutchison, C.A., 3rd. DNA sequencing: Bench to bedside and beyond. Nucleic Acids Res. 2007, 35, 6227–6237. [Google Scholar] [CrossRef]
- Roth, B.L.; Lopez, E.; Beischel, S.; Westkaemper, R.B.; Evans, J.M. Screening the receptorome to discover the molecular targets for plant-derived psychoactive compounds: A novel approach for CNS drug discovery. Pharmacol. Ther. 2004, 102, 99–110. [Google Scholar] [CrossRef]
- Altshuler, D.; Pollara, V.J.; Cowles, C.R.; Van Etten, W.J.; Baldwin, J.; Linton, L.; Lander, E.S. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 2000, 407, 513–516. [Google Scholar] [CrossRef]
- Aparicio, S.A. How to count…human genes. Nat. Genet. 2000, 25, 129–130. [Google Scholar] [CrossRef]
- Galtier, N.; Piganeau, G.; Mouchiroud, D.; Duret, L. GC-content evolution in mammalian genomes: The biased gene conversion hypothesis. Genetics 2001, 159, 907–911. [Google Scholar] [CrossRef]
- Gerhard, D.S.; Wagner, L.; Feingold, E.A.; Shenmen, C.M.; Grouse, L.H.; Schuler, G.; Klein, S.L.; Old, S.; Rasooly, R.; Good, P.; et al. The status, quality, and expansion of the NIH full-length cDNA project: The Mammalian Gene Collection (MGC). Genome Res. 2004, 14, 2121–2127, Erratum in Genome Res. 2006, 16, 804. [Google Scholar]
- Guigó, R.; Reese, M.G. EGASP: Collaboration through competition to find human genes. Nat. Methods 2005, 2, 575–577. [Google Scholar] [CrossRef]
- Guigó, R.; Flicek, P.; Abril, J.F.; Reymond, A.; Lagarde, J.; Denoeud, F.; Antonarakis, S.; Ashburner, M.; Bajic, V.B.; Birney, E.; et al. EGASP: The human ENCODE Genome Annotation Assessment Project. Genome Biol. 2006, 7, S2.1–S2.31. [Google Scholar] [CrossRef]
- Guigó, R.; Valcárcel, J. Unweaving the meanings of messenger RNA sequences. Mol. Cell. 2006, 23, 150–151. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921, Erratum in Nature 2001, 412, 565. Nature 2001, 411, 720. [Google Scholar] [PubMed]
- Liu, Y.; Shaw, S. The human genome: An immuno-centric view of evolutionary strategies. Trends Immunol. 2001, 22, 227–229. [Google Scholar] [CrossRef]
- López-Bigas, N.; Audit, B.; Ouzounis, C.; Parra, G.; Guigó, R. Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 2005, 579, 1900–1903. [Google Scholar] [CrossRef]
- Sachidanandam, R.; Weissman, D.; Schmidt, S.C.; Kakol, J.M.; Stein, L.D.; Marth, G.; Sherry, S.; Mullikin, J.C.; Mortimore, B.J.; Willey, D.L.; et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 2001, 409, 928–933. [Google Scholar] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Bikandi, J.; San Millán, R.; Rementeria, A.; Garaizar, J. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR, and endonuclease restriction. Bioinformatics 2004, 20, 798–799. [Google Scholar] [CrossRef]
- Jensen, L.J.; Gupta, R.; Blom, N.; Devos, D.; Tamames, J.; Kesmir, C.; Nielsen, H.; Staerfeldt, H.H.; Rapacki, K.; Workman, C.; et al. Prediction of human orphan protein function from post-translational modifications and localization features. J. Mol. Biol. 2002, 319, 1257–1265. [Google Scholar] [CrossRef]
- Jensen, L.J.; Gupta, R.; Staerfeldt, H.H.; Brunak, S. Prediction of human protein function according to Gene Ontology categories. Bioinformatics 2003, 19, 635–642. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Wang, X.; Carlson, V.C.C.; Studholme, C.; Newman, N.; Ford, M.M.; Grant, K.A.; Kroenke, C.D. In utero MRI identifies consequences of early-gestation alcohol drinking on fetal brain development in rhesus macaques. Proc. Natl. Acad. Sci. USA 2020, 117, 10035–10044. [Google Scholar] [CrossRef] [PubMed]
- Mooney, S.M.; Miller, M.W. Time-specific effects of ethanol exposure on cranial nerve nuclei: Gastrulation and neuronogenesis. Exp. Neurol. 2007, 205, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Chiappelli, F.; Taylor, A.N.; Espinosa de los Monteros, A.; de Vellis, J. Fetal alcohol delays the developmental expression of myelin basic protein and transferrin in rat primary oligodendrocyte cultures. Int. J. Dev. Neurosci. Off. J. Int. Soc. Dev. Neurosci. 1991, 9, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Zoeller, R.T.; Butnariu, O.V.; Fletcher, D.L.; Riley, E.P. Limited postnatal ethanol exposure permanently alters the expression of mRNAS encoding myelin basic protein and myelin-associated glycoprotein in cerebellum. Alcohol. Clin. Exp. Res. 1994, 18, 909–916. [Google Scholar] [CrossRef]
- Bichenkov, E.; Ellingson, J.S. Ethanol alters the expressions of c-Fos and myelin basic protein in differentiating oligodendrocytes. Alcohol 2009, 43, 627–634. [Google Scholar] [CrossRef]



| EtOH Consumed Women (n = 20) | Controls (No EtOH Users, n = 20) | |
|---|---|---|
| Maternal Age (years ± SD) | 28.0 ± 2.7 | 24.15 ± 2.3 |
| Gestational Age (weeks ± SD) | 15.22 ± 1.6 | 14.92 ± 1.58 |
| Race: White (%) | 50 | 50 |
| Race: Black (%) | 50 | 50 |
| Fetal Sex, Male (%) | 50 | 50 |
| Fetal Sex, Female (%) | 50 | 50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darbinian, N.; Gallia, G.L.; Darbinyan, A.; Vadachkoria, E.; Merabova, N.; Moore, A.; Goetzl, L.; Amini, S.; Selzer, M.E. Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder. Int. J. Mol. Sci. 2023, 24, 13714. https://doi.org/10.3390/ijms241813714
Darbinian N, Gallia GL, Darbinyan A, Vadachkoria E, Merabova N, Moore A, Goetzl L, Amini S, Selzer ME. Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder. International Journal of Molecular Sciences. 2023; 24(18):13714. https://doi.org/10.3390/ijms241813714
Chicago/Turabian StyleDarbinian, Nune, Gary L. Gallia, Armine Darbinyan, Ekaterina Vadachkoria, Nana Merabova, Amos Moore, Laura Goetzl, Shohreh Amini, and Michael E. Selzer. 2023. "Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder" International Journal of Molecular Sciences 24, no. 18: 13714. https://doi.org/10.3390/ijms241813714
APA StyleDarbinian, N., Gallia, G. L., Darbinyan, A., Vadachkoria, E., Merabova, N., Moore, A., Goetzl, L., Amini, S., & Selzer, M. E. (2023). Effects of In Utero EtOH Exposure on 18S Ribosomal RNA Processing: Contribution to Fetal Alcohol Spectrum Disorder. International Journal of Molecular Sciences, 24(18), 13714. https://doi.org/10.3390/ijms241813714






