Regional Gene Expression in the Retina, Optic Nerve Head, and Optic Nerve of Mice with Optic Nerve Crush and Experimental Glaucoma
Abstract
:1. Introduction
2. Results
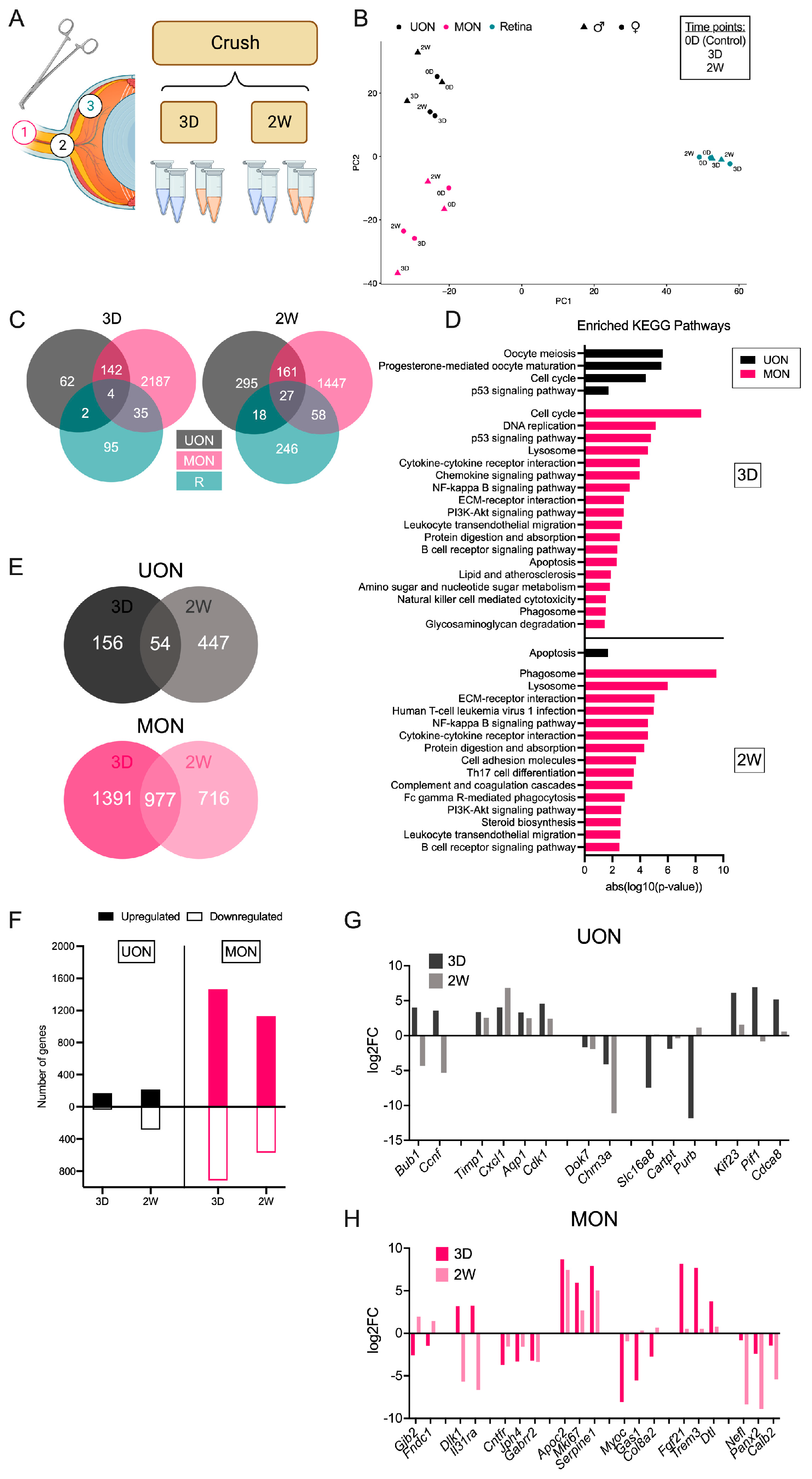
2.1. Distinct Expression Patterns in Naïve UON, MON, and Retinal Tissues
2.2. Differential Gene Expression after ON Crush Injury
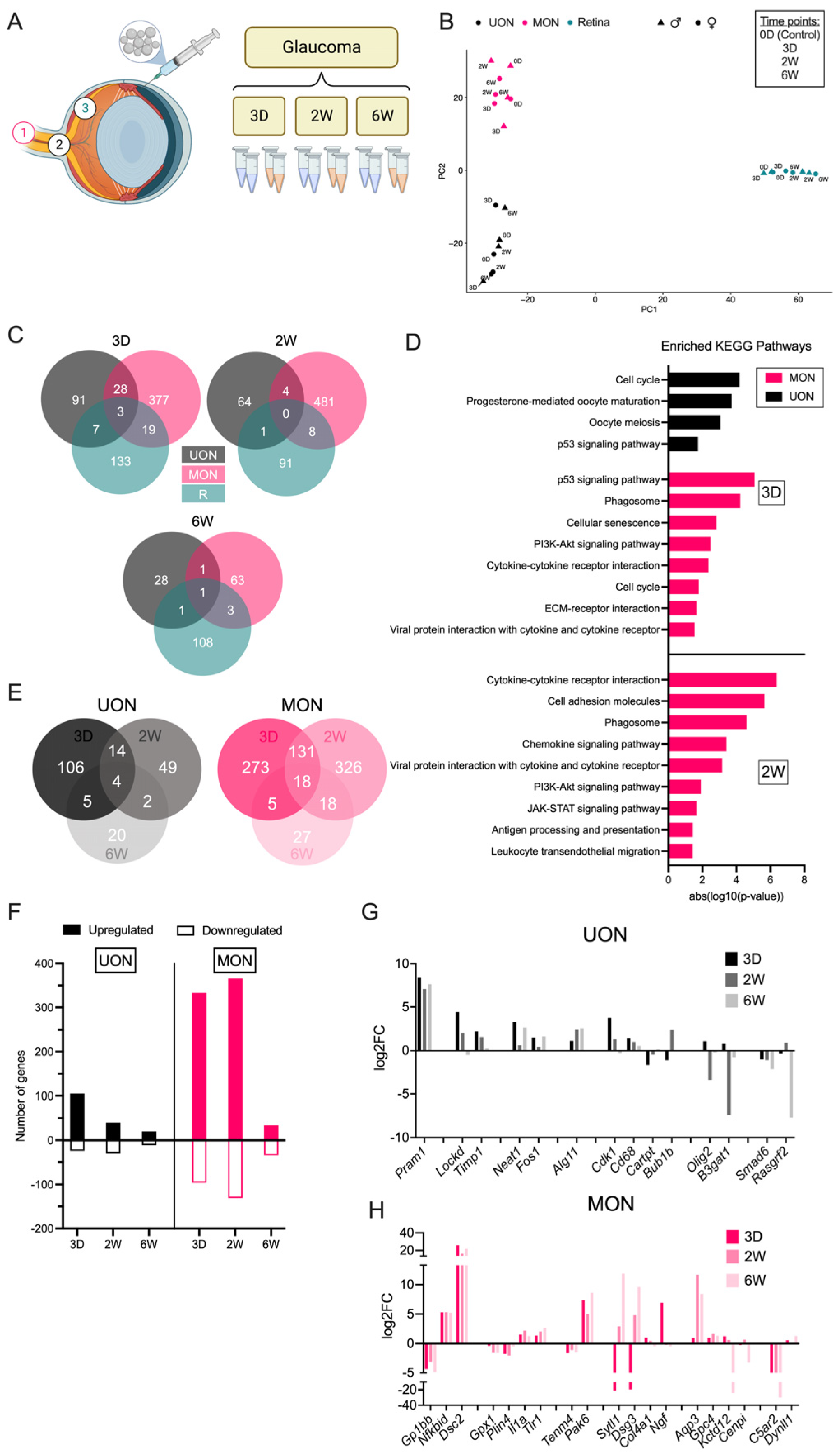
2.3. Differential Gene Expression in the Glaucoma Model
2.4. Injury-Specific Responses in ON Tissue Regions
2.5. Astrocyte Responses to ON Injury
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Anesthesia
4.3. IOP Measurements
4.4. Microbead Injections (Ocular Hypertension/Glaucoma Model)
4.5. ON Crush
4.6. Tissue Collection
4.7. Total RNA Extraction
4.8. RNA-Seq Library Preparation and Sequencing
4.9. RNA-Seq Analysis
4.10. Quantitative Polymerase Chain Reaction (qPCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boland, M.V.; Quigley, H.A. Risk factors and open-angle glaucoma: Classification and application. J. Glaucoma 2007, 16, 406–418. [Google Scholar] [CrossRef]
- Morrison, J.C.; Johnson, E.C.; Cepurna, W.; Jia, L. Understanding mechanisms of pressure-induced optic nerve damage. Prog. Retin. Eye Res. 2005, 24, 217–240. [Google Scholar] [CrossRef]
- Howell, G.R.; Libby, R.T.; Jakobs, T.C.; Smith, R.S.; Phalan, F.C.; Barter, J.W.; Barbay, J.M.; Marchant, J.K.; Mahesh, N.; Porciatti, V.; et al. Axons of retinal ganglion cells are insulted in the optic nerve early in DBA/2J glaucoma. J. Cell Biol. 2007, 179, 1523–1537. [Google Scholar] [CrossRef]
- Quigley, H.A.; Addicks, E.M.; Green, W.R.; Maumenee, A.E. Optic nerve damage in human glaucoma. II. The site of injury and susceptibility to damage. Arch. Ophthalmol. 1981, 99, 635–649. [Google Scholar] [CrossRef]
- Sun, D.; Lye-Barthel, M.; Masland, R.H.; Jakobs, T.C. The morphology and spatial arrangement of astrocytes in the optic nerve head of the mouse. J. Comp. Neurol. 2009, 516, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Downs, J.C.; Roberts, M.D.; Burgoyne, C.F. Mechanical environment of the optic nerve head in glaucoma. Optom. Vis. Sci. 2008, 85, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Vidal, I.; Pena, J.D.; Hernandez, M.R. Expression of neural cell adhesion molecule (NCAM) characterizes a subpopulation of type 1 astrocytes in human optic nerve head. Glia 1997, 20, 262–273. [Google Scholar] [CrossRef]
- Quillen, S.; Schaub, J.; Quigley, H.; Pease, M.; Korneva, A.; Kimball, E. Astrocyte responses to experimental glaucoma in mouse optic nerve head. PLoS ONE 2020, 15, e0238104. [Google Scholar] [CrossRef] [PubMed]
- Morrison, J.C. Integrins in the optic nerve head: Potential roles in glaucomatous optic neuropathy (an American Ophthalmological Society thesis). Trans. Am. Ophthalmol. Soc. 2006, 104, 453–477. [Google Scholar] [PubMed]
- Forrest, S.L.; Kim, J.H.; Crockford, D.R.; Huynh, K.; Cheong, R.; Knott, S.; Kane, M.A.; Ittner, L.M.; Halliday, G.M.; Kril, J.J. Distribution Patterns of Astrocyte Populations in the Human Cortex. Neurochem. Res. 2022, 48, 1222–1232. [Google Scholar] [CrossRef]
- Mazumder, A.G.; Julé, A.M.; Cullen, P.F.; Sun, D. Astrocyte heterogeneity within white matter tracts and a unique subpopulation of optic nerve head astrocytes. iScience 2022, 25, 105568. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.; Brown, K.M.; Stephan, D.A.; Morrison, J.C.; Johnson, E.C.; Tomarev, S.I. Microarray analysis of changes in mRNA levels in the rat retina after experimental elevation of intraocular pressure. Investig. Ophthalmol. Vis. Sci. 2004, 45, 1247–1258. [Google Scholar] [CrossRef]
- Qu, J.; Jakobs, T.C. The Time Course of Gene Expression during Reactive Gliosis in the Optic Nerve. PLoS ONE 2013, 8, e67094. [Google Scholar] [CrossRef]
- Johnson, E.C.; Doser, T.A.; Cepurna, W.O.; Dyck, J.A.; Jia, L.; Guo, Y.; Lambert, W.S.; Morrison, J.C. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Investig. Ophthalmol. Vis. Sci. 2011, 52, 504–518. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.C.; Jia, L.; Cepurna, W.O.; Doser, T.A.; Morrison, J.C. Global changes in optic nerve head gene expression after exposure to elevated intraocular pressure in a rat glaucoma model. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3161–3177. [Google Scholar] [CrossRef] [PubMed]
- Guttenplan, K.A.; Stafford, B.K.; El-Danaf, R.N.; Adler, D.I.; Münch, A.E.; Weigel, M.K.; Huberman, A.D.; Liddelow, S.A. Neurotoxic Reactive Astrocytes Drive Neuronal Death after Retinal Injury. Cell Rep. 2020, 31, 107776. [Google Scholar] [CrossRef]
- Tezel, G.; Yang, X.; Luo, C.; Cai, J.; Powell, D.W. An astrocyte-specific proteomic approach to inflammatory responses in experimental rat glaucoma. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4220–4233. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, G.-L.; Xu, M.-X.; Zhou, H.; Li, F.; Miao, Y.; Lei, B.; Yang, X.-L.; Wang, Z. Interplay between Muller cells and microglia aggravates retinal inflammatory response in experimental glaucoma. J. Neuroinflamm. 2021, 18, 303. [Google Scholar] [CrossRef]
- Sun, D.; Qu, J.; Jakobs, T.C. Reversible reactivity by optic nerve astrocytes. Glia 2013, 61, 1218–1235. [Google Scholar] [CrossRef]
- Karasawa, K.; Tanaka, A.; Jung, K.; Matsuda, A.; Okamoto, N.; Oida, K.; Ohmori, K.; Matsuda, H. Patterns of aquaporin expression in the canine eye. Vet. J. 2011, 190, e72–e77. [Google Scholar] [CrossRef]
- Yang, P.; Agapova, O.; Parker, A.; Shannon, W.; Pecen, P.; Duncan, J.; Salvador-Silva, M.; Hernandez, M.R. DNA microarray analysis of gene expression in human optic nerve head astrocytes in response to hydrostatic pressure. Physiol. Genom. 2004, 17, 157–169. [Google Scholar] [CrossRef]
- Salvador-Silva, M.; Aoi, S.; Parker, A.; Yang, P.; Pecen, P.; Hernandez, M.R. Responses and signaling pathways in human optic nerve head astrocytes exposed to hydrostatic pressure in vitro. Glia 2004, 45, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Cone-Kimball, E.; Nguyen, C.; Oglesby, E.N.; Pease, M.E.; Steinhart, M.R.; Quigley, H.A. Scleral structural alterations associated with chronic experimental intraocular pressure elevation in mice. Mol. Vis. 2013, 19, 2023–2039. [Google Scholar]
- Kimball, E.; Schaub, J.; Quillen, S.; Keuthan, C.; Pease, M.E.; Korneva, A.; Quigley, H. The role of aquaporin-4 in optic nerve head astrocytes in experimental glaucoma. PLoS ONE 2021, 16, e0244123. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.T.T.; Pease, M.E.; Jefferys, J.L.; Kimball, E.C.; Quigley, H.A.; Nguyen, T.D. Pressure-Induced Changes in Astrocyte GFAP, Actin, and Nuclear Morphology in Mouse Optic Nerve. Investig. Ophthalmol. Vis. Sci. 2020, 61, 14. [Google Scholar] [CrossRef] [PubMed]
- Paisley, C.E.; Kay, J.N. Seeing stars: Development and function of retinal astrocytes. Dev. Biol. 2021, 478, 144–154. [Google Scholar] [CrossRef]
- Kimball, E.C.; Quillen, S.; Pease, M.E.; Keuthan, C.; Nagalingam, A.; Zack, D.J.; Johnson, T.V.; Quigley, H.A. Aquaporin 4 is not present in normal porcine and human lamina cribrosa. PLoS ONE 2022, 17, e0268541. [Google Scholar] [CrossRef] [PubMed]
- Khakh, B.S.; Sofroniew, M.V. Diversity of astrocyte functions and phenotypes in neural circuits. Nat. Neurosci. 2015, 18, 942–952. [Google Scholar] [CrossRef]
- Westergard, T.; Rothstein, J.D. Astrocyte Diversity: Current Insights and Future Directions. Neurochem. Res. 2020, 45, 1298–1305. [Google Scholar] [CrossRef] [PubMed]
- Endo, F.; Kasai, A.; Soto, J.S.; Yu, X.; Qu, Z.; Hashimoto, H.; Gradinaru, V.; Kawaguchi, R.; Khakh, B.S. Molecular basis of astrocyte diversity and morphology across the CNS in health and disease. Science 2022, 378, eadc9020. [Google Scholar] [CrossRef] [PubMed]
- Chumakov, P.M. Function of the p53 gene: Choice between life and death. Biochemistry 2000, 65, 28–40. [Google Scholar]
- Hernandez Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef]
- Stracquadanio, G.; Wang, X.; Wallace, M.D.; Grawenda, A.M.; Zhang, P.; Hewitt, J.; Zeron-Medina, J.; Castro-Giner, F.; Tomlinson, I.P.; Goding, C.R.; et al. The importance of p53 pathway genetics in inherited and somatic cancer genomes. Nat. Rev. Cancer 2016, 16, 251–265. [Google Scholar] [CrossRef]
- Wiggs, J.L.; Hewitt, A.W.; Fan, B.J.; Wang, D.Y.; Sena, D.R.F.; O’brien, C.; Realini, A.; Craig, J.E.; Dimasi, D.P.; Mackey, D.A.; et al. The p53 codon 72 PRO/PRO genotype may be associated with initial central visual field defects in caucasians with primary open angle glaucoma. PLoS ONE 2012, 7, e45613. [Google Scholar] [CrossRef]
- Asefa, N.G.; Kamali, Z.; Pereira, S.; Vaez, A.; Jansonius, N.; Bergen, A.A.; Snieder, H. Bioinformatic Prioritization and Functional Annotation of GWAS-Based Candidate Genes for Primary Open-Angle Glaucoma. Genes 2022, 13, 1055. [Google Scholar] [CrossRef] [PubMed]
- Son, J.L.; Soto, I.; Oglesby, E.; Lopez-Roca, T.; Pease, M.E.; Quigley, H.A.; Marsh-Armstrong, N. Glaucomatous optic nerve injury involves early astrocyte reactivity and late oligodendrocyte loss. Glia 2010, 58, 780–789. [Google Scholar] [CrossRef]
- Paralkar, V.R.; Taborda, C.C.; Huang, P.; Yao, Y.; Kossenkov, A.V.; Prasad, R.; Luan, J.; Davies, J.O.; Hughes, J.R.; Hardison, R.C.; et al. Unlinking an lncRNA from Its Associated cis Element. Mol. Cell 2016, 62, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Howell, G.R.; Macalinao, D.G.; Sousa, G.L.; Walden, M.; Soto, I.; Kneeland, S.C.; Barbay, J.M.; King, B.L.; Marchant, J.K.; Hibbs, M.; et al. Molecular clustering identifies complement and endothelin induction as early events in a mouse model of glaucoma. J. Clin. Investig. 2011, 121, 1429–1444. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Moore, S.; Jakobs, T.C. Optic nerve astrocyte reactivity protects function in experimental glaucoma and other nerve injuries. J. Exp. Med. 2017, 214, 1411–1430. [Google Scholar] [CrossRef]
- Okada, S.; Nakamura, M.; Katoh, H.; Miyao, T.; Shimazaki, T.; Ishii, K.; Yamane, J.; Yoshimura, A.; Iwamoto, Y.; Toyama, Y.; et al. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat. Med. 2006, 12, 829–834. [Google Scholar] [CrossRef]
- Liddelow, S.A.; Guttenplan, K.A.; Clarke, L.E.; Bennett, F.C.; Bohlen, C.J.; Schirmer, L.; Bennett, M.L.; Münch, A.E.; Chung, W.-S.; Peterson, T.C.; et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017, 541, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Smith, M.D.; Kersbergen, C.J.; Kam, T.-I.; Viswanathan, M.; Martin, K.; Dawson, T.M.; Dawson, V.L.; Zack, D.J.; Whartenby, K.; et al. Glial pathology and retinal neurotoxicity in the anterior visual pathway in experimental autoimmune encephalomyelitis. Acta Neuropathol. Commun. 2019, 7, 125. [Google Scholar] [CrossRef]
- Lanjakornsiripan, D.; Pior, B.-J.; Kawaguchi, D.; Furutachi, S.; Tahara, T.; Katsuyama, Y.; Suzuki, Y.; Fukazawa, Y.; Gotoh, Y. Layer-specific morphological and molecular differences in neocortical astrocytes and their dependence on neuronal layers. Nat. Commun. 2018, 9, 1623. [Google Scholar] [CrossRef] [PubMed]
- Khakh, B.S.; Deneen, B. The Emerging Nature of Astrocyte Diversity. Annu. Rev. Neurosci. 2019, 42, 187–207. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.; Diaz-Castro, B.; Shigetomi, E.; Monte, E.; Octeau, J.C.; Yu, X.; Cohn, W.; Rajendran, P.S.; Vondriska, T.M.; Whitelegge, J.P.; et al. Neural Circuit-Specialized Astrocytes: Transcriptomic, Proteomic, Morphological, and Functional Evidence. Neuron 2017, 95, 531–549.e9. [Google Scholar] [CrossRef]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of region-specific astrocyte subtypes at single cell resolution. Nat. Commun. 2020, 11, 1220. [Google Scholar] [CrossRef]
- Hinkle, J.T.; Dawson, V.L.; Dawson, T.M. The A1 astrocyte paradigm: New avenues for pharmacological intervention in neurodegeneration. Mov. Disord. 2019, 34, 959–969. [Google Scholar] [CrossRef]
- Zamanian, J.L.; Xu, L.; Foo, L.C.; Nouri, N.; Zhou, L.; Giffard, R.G.; Barres, B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012, 32, 6391–6410. [Google Scholar] [CrossRef]
- Cuenca, N.; Fernández-Sánchez, L.; Campello, L.; Maneu, V.; De la Villa, P.; Lax, P.; Pinilla, I. Cellular responses following retinal injuries and therapeutic approaches for neurodegenerative diseases. Prog. Retin. Eye Res. 2014, 43, 17–75. [Google Scholar] [CrossRef]
- Cui, Q.N.; Stein, L.M.; Fortin, S.M.; Hayes, M.R. The role of glia in the physiology and pharmacology of glucagon-like peptide-1: Implications for obesity, diabetes, neurodegeneration and glaucoma. Br. J. Pharmacol. 2022, 179, 715–726. [Google Scholar] [CrossRef]
- Gharagozloo, M.; Smith, M.D.; Jin, J.; Garton, T.; Taylor, M.; Chao, A.; Meyers, K.; Kornberg, M.D.; Zack, D.J.; Ohayon, J.; et al. Complement component 3 from astrocytes mediates retinal ganglion cell loss during neuroinflammation. Acta Neuropathol. 2021, 142, 899–915. [Google Scholar] [CrossRef]
- Guttenplan, K.A.; Weigel, M.K.; Prakash, P.; Wijewardhane, P.R.; Hasel, P.; Rufen-Blanchette, U.; Münch, A.E.; Blum, J.A.; Fine, J.; Neal, M.C.; et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature 2021, 599, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Hasel, P.; Rose, I.V.L.; Sadick, J.S.; Kim, R.D.; Liddelow, S.A. Neuroinflammatory astrocyte subtypes in the mouse brain. Nat. Neurosci. 2021, 24, 1475–1487. [Google Scholar] [CrossRef]
- Neufeld, A.H. Microglia in the optic nerve head and the region of parapapillary chorioretinal atrophy in glaucoma. Arch. Ophthalmol. 1999, 117, 1050–1056. [Google Scholar] [CrossRef]
- Bordone, M.P.; Fleitas, M.F.G.; Pasquini, L.A.; Bosco, A.; Sande, P.H.; Rosenstein, R.E.; Dorfman, D. Involvement of microglia in early axoglial alterations of the optic nerve induced by experimental glaucoma. J. Neurochem. 2017, 142, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Hilla, A.M.; Diekmann, H.; Fischer, D. Microglia Are Irrelevant for Neuronal Degeneration and Axon Regeneration after Acute Injury. J. Neurosci. 2017, 37, 6113–6124. [Google Scholar] [CrossRef]
- Yang, X.; Zeng, Q.; Baris, M.; Tezel, G. Transgenic inhibition of astroglial NF-kappaB restrains the neuroinflammatory and neurodegenerative outcomes of experimental mouse glaucoma. J. Neuroinflamm. 2020, 17, 252. [Google Scholar] [CrossRef]
- Ferreira-Silva, J.; Aires, I.D.; Boia, R.; Ambrósio, A.F.; Santiago, A.R. Activation of Adenosine A(3) Receptor Inhibits Microglia Reactivity Elicited by Elevated Pressure. Int. J. Mol. Sci. 2020, 21, 7218. [Google Scholar] [CrossRef]
- Ohnishi, M.; Urasaki, T.; Ochiai, H.; Matsuoka, K.; Takeo, S.; Harada, T.; Ohsugi, Y.; Inoue, A. Selective enhancement of wnt4 expression by cyclic AMP-associated cooperation between rat central astrocytes and microglia. Biochem. Biophys. Res. Commun. 2015, 467, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, B.D.; Grashoff, C.; Schwartz, M.A. Dynamic molecular processes mediate cellular mechanotransduction. Nature 2011, 475, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Martino, F.; Perestrelo, A.R.; Vinarsky, V.; Pagliari, S.; Forte, G. Cellular Mechanotransduction: From Tension to Function. Front. Physiol. 2018, 9, 824. [Google Scholar] [CrossRef]
- KKorneva, A.; Schaub, J.; Jefferys, J.; Kimball, E.; Pease, M.E.; Nawathe, M.; Johnson, T.V.; Pitha, I.; Quigley, H. A method to quantify regional axonal transport blockade at the optic nerve head after short term intraocular pressure elevation in mice. Exp. Eye Res. 2020, 196, 108035. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- de Sena Brandine, G.; Smith, A.D. Falco: High-speed FastQC emulation for quality control of sequencing data. F1000Res 2019, 8, 1874. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keuthan, C.J.; Schaub, J.A.; Wei, M.; Fang, W.; Quillen, S.; Kimball, E.; Johnson, T.V.; Ji, H.; Zack, D.J.; Quigley, H.A. Regional Gene Expression in the Retina, Optic Nerve Head, and Optic Nerve of Mice with Optic Nerve Crush and Experimental Glaucoma. Int. J. Mol. Sci. 2023, 24, 13719. https://doi.org/10.3390/ijms241813719
Keuthan CJ, Schaub JA, Wei M, Fang W, Quillen S, Kimball E, Johnson TV, Ji H, Zack DJ, Quigley HA. Regional Gene Expression in the Retina, Optic Nerve Head, and Optic Nerve of Mice with Optic Nerve Crush and Experimental Glaucoma. International Journal of Molecular Sciences. 2023; 24(18):13719. https://doi.org/10.3390/ijms241813719
Chicago/Turabian StyleKeuthan, Casey J., Julie A. Schaub, Meihan Wei, Weixiang Fang, Sarah Quillen, Elizabeth Kimball, Thomas V. Johnson, Hongkai Ji, Donald J. Zack, and Harry A. Quigley. 2023. "Regional Gene Expression in the Retina, Optic Nerve Head, and Optic Nerve of Mice with Optic Nerve Crush and Experimental Glaucoma" International Journal of Molecular Sciences 24, no. 18: 13719. https://doi.org/10.3390/ijms241813719
APA StyleKeuthan, C. J., Schaub, J. A., Wei, M., Fang, W., Quillen, S., Kimball, E., Johnson, T. V., Ji, H., Zack, D. J., & Quigley, H. A. (2023). Regional Gene Expression in the Retina, Optic Nerve Head, and Optic Nerve of Mice with Optic Nerve Crush and Experimental Glaucoma. International Journal of Molecular Sciences, 24(18), 13719. https://doi.org/10.3390/ijms241813719





