What about Current Diversity of Mycolactone-Producing Mycobacteria? Implication for the Diagnosis and Treatment of Buruli Ulcer
Abstract
1. Introduction
2. History of BU Diagnosis and M. ulcerans DNA Testing
2.1. Clinical and Microbial Diagnosis
2.2. First Molecular Diagnosis
2.3. Discovery and Implementation of IS2404 and IS2606 PCR Assays
2.4. Implementation of qPCR Assays
3. On the Origin of Mycolactone-Producing Mycobacteria (MPM) Species Assignment
4. Lack of Specificity of (q)PCR-Based Tests for M. ulcerans Variant Detection
4.1. Lack of Specificity of IS2404, IS2606 and KR-B
4.2. Lack of Reliability of IS2404 and IS2606 CT-Values
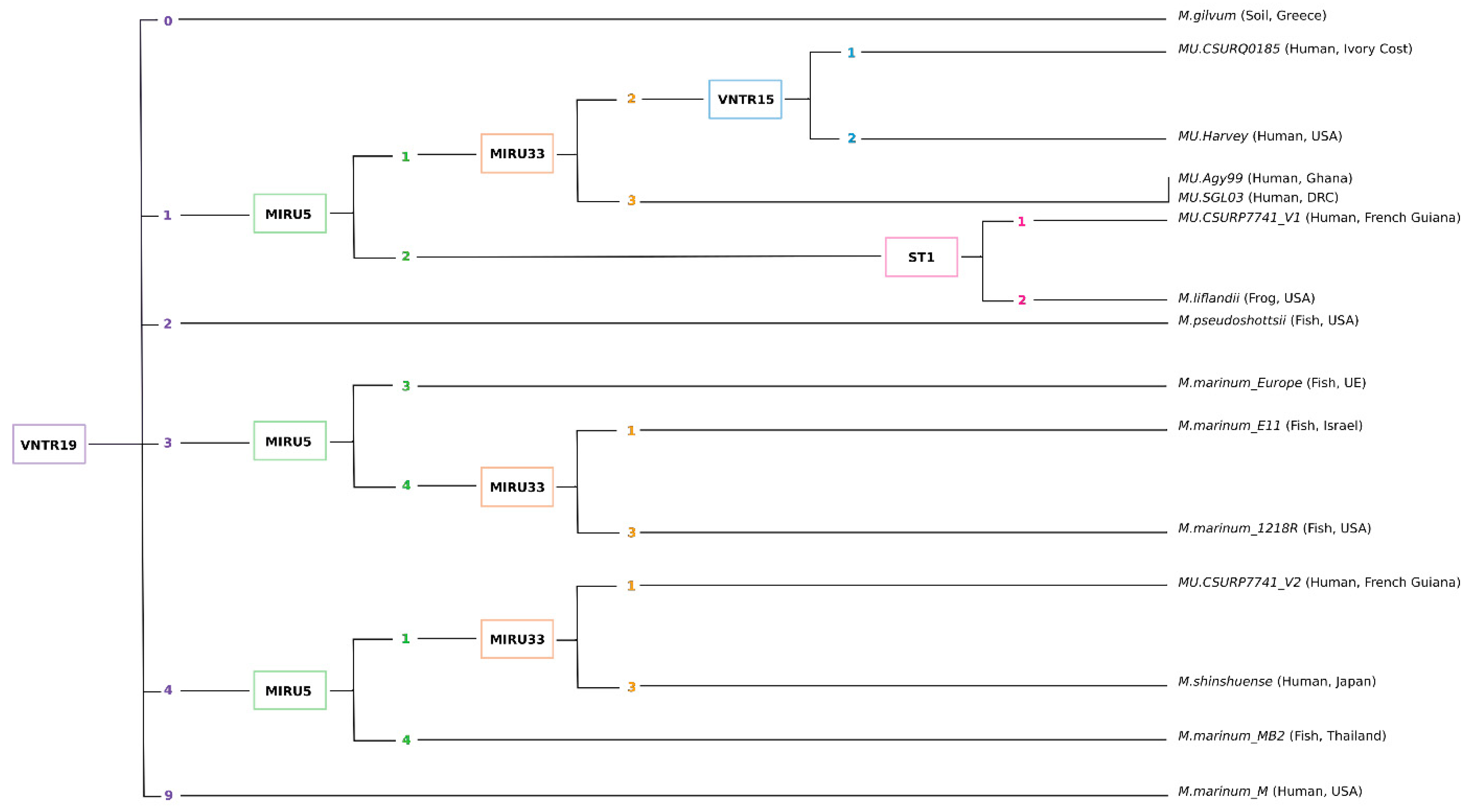
5. Implementation of MIRU-VNTR to Assess Genotypic Diversity
5.1. Implementation of MIRU-VNTR Typing
5.2. Main Limitations and Bias
5.3. The Use of a Universal Identification Key
6. Implication for the Diagnosis and Treatment of BU
6.1. Implication for BU Diagnosis
6.2. Implication for M. ulcerans Genetic Diversity
6.3. What about the Pathogenicity of Other Mycobacterium Species
6.4. Implication for BU Treatment
7. Conclusions
8. Material and Methods
8.1. Literature Search
8.2. In Silico Evaluation of M. ulcerans Detection Primers
8.3. VNTR-Based Identification Key
8.4. Copy Number Variation of IS2404, IS2606 and KR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, K.; Patel, N.; Levy, M.; Storeygard, A.; Balk, D.; Gittleman, J.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Dong, J.; Olano, J.P.; McBride, J.W.; Walker, D.H. Emerging pathogens: Challenges and successes of molecular diagnostics. J. Mol. Diagn. 2008, 10, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Le Turnier, P.; Epelboin, L. Mise au point sur la leptospirose. Rev. Médecine Interne 2019, 40, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Guillemi, E.C.; Tomassone, L.; Farber, M.D. Tick-borne Rickettsiales: Molecular tools for the study of an emergent group of pathogens. J. Microbiol. Methods 2015, 119, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Radomski, N.; Kreitmann, L.; Mcintosh, F.; Behr, M.A. The Critical Role of DNA Extraction for Detection of Mycobacteria in Tissues. PLoS ONE 2013, 8, e78749. [Google Scholar] [CrossRef][Green Version]
- Rezazadeh, E.; Moazeni, M.; Sabokbar, A. Use of cost effective and rapid molecular tools for identification of Candida species, opportunistic pathogens. Curr. Med. Mycol. 2016, 2, 1–4. [Google Scholar] [CrossRef][Green Version]
- Wei, E.; Bou-Nader, C.; Perry, M.L.; Fattah, R.; Zhang, J.; Leppla, S.H.; Bothra, A. S9.6 Antibody-Enzyme Conjugates for the Detection of DNA-RNA Hybrids. Bioconjugate Chem. 2023, 34, 834–844. [Google Scholar] [CrossRef]
- Williamson, H.R.; Benbow, M.E.; Nguyen, K.D.; Beachboard, D.C.; Kimbirauskas, R.K.; McIntosh, M.D.; Quaye, C.; Ampadu, E.O.; Boakye, D.; Merritt, R.W.; et al. Distribution of Mycobacterium ulcerans in Buruli ulcer endemic and non-endemic aquatic sites in Ghana. PLoS Negl. Trop. Dis. 2008, 2, e205. [Google Scholar] [CrossRef]
- Combe, M.; Velvin, C.J.; Morris, A.; Garchitorena, A.; Carolan, K.; Sanhueza, D.; Roche, B.; Couppié, P.; Guégan, J.-F.; Gozlan, R.E. Global and local environmental changes as drivers of Buruli ulcer emergence. Emerg. Microbes Infect. 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Portaels, F.; Aguiar, J.; Fissette, K.; Fonteyne, P.A.; de Beenhouwer, H.; de Rijk, P.; Guédénon, A.; Lemans, R.; Steunou, C.; Zinsou, C.; et al. Direct detection and identification of Mycobacterium ulcerans in clinical speciemns by PCR and oligonucleotide-specific capture plate hybridization. J. Clin. Microbiol. 1997, 35, 1097–1100. [Google Scholar] [CrossRef]
- Merritt, R.W.; Walker, E.D.; Small, P.L.C.; Wallace, J.R.; Johnson, P.D.R.; Benbow, M.E.; Boakye, D.A. Ecology and Transmission of Buruli Ulcer Disease: A Systematic Review. PLoS Negl. Trop. Dis. 2010, 4, e911. [Google Scholar] [CrossRef]
- Giles-Vernick, T.; Owona-Ntsama, J.; Landier, J.; Eyangoh, S. The puzzle of Buruli ulcer transmission, ethno-ecological history and the end of “love” in the Akonolinga district, Cameroon. Soc. Sci. Med. 2015, 129, 20–27. [Google Scholar] [CrossRef] [PubMed]
- MacCULLUM, P. A new mycobacterial infection in man; clinical aspects. J. Pathol. Bacteriol. 1948, 60, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Portaels, F.; Fonteyne, P.A.; De Beenhouwer, H.; De Rijk, P.; Guédénon, A.; Hayman, J.; Meyers, M.W. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J. Clin. Microbiol. 1996, 34, 962–965. [Google Scholar] [CrossRef] [PubMed]
- Ross, B.C.; Marino, L.; Oppedisano, F.; Edwards, R.; Robins-Browne, R.M.; Johnson, P.D.R. Development of a PCR assay for rapid diagnosis of Mycobacterium ulcerans infection. J. Clin. Microbiol. 1997, 35, 1696–1700. [Google Scholar] [CrossRef]
- Yip, M.J.; Porter, J.L.; Fyfe, J.A.M.; Lavender, C.J.; Portaels, F.; Rhodes, M.; Kator, H.; Colorni, A.; Jenkin, G.A.; Stinear, T. Evolution of Mycobacterium ulcerans and other mycolactone-producing mycobacteria from a common Mycobacterium marinum progenitor. J. Bacteriol. 2007, 189, 2021–2029. [Google Scholar] [CrossRef]
- Tobias, N.J.; Doig, K.D.; Medema, M.H.; Chen, H.; Haring, V.; Moore, R.; Seemann, T.; Stinear, T.P. Complete genome sequence of the frog pathogen Mycobacterium ulcerans ecovar Liflandii. J. Bacteriol. 2013, 195, 556–564. [Google Scholar] [CrossRef]
- Demangel, C.; Stinear, T.P.; Cole, S.T. Buruli ulcer: Reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 2009, 7, 50–60. [Google Scholar] [CrossRef]
- Pidot, S.J.; Asiedu, K.; Käser, M.; Fyfe, J.A.M.; Stinear, T.P. Mycobacterium ulcerans and other Mycolactone-producing mycobacteria should be considered a single species. PLoS Negl. Trop. Dis. 2010, 4, e663. [Google Scholar] [CrossRef]
- Doig, K.D.; Holt, K.E.; Fyfe, J.A.M.; Lavender, C.J.; Eddyani, M.; Portaels, F.; Yeboah-Manu, D.; Pluschke, G.; Seemann, T.; Stinear, T.P. On the origin of Mycobacterium ulcerans, the causative agent of Buruli ulcer. BMC Genom. 2012, 13, 258. [Google Scholar] [CrossRef]
- Nakanaga, K.; Ishii, N.; Suzuki, K.; Tanigawa, K.; Goto, M.; Okabe, T.; Imada, H.; Kodama, A.; Iwamoto, T.; Takahashi, H.; et al. “Mycobacteriumulcerans subsp. shinshuense” isolated from a skin ulcer lesion: Identification based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2007, 45, 3840–3843. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M.; Kikuchi, N.; Yamamoto, T.; Suzutani, T.; Nakanaga, K.; Suzuki, K.; Ishii, N. Buruli ulcer caused by Mycobacterium ulcerans subsp shinshuense. JAMA Dermatol. 2014, 150, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Nguetta, A.; Coulibaly, N.D.; Kouamé-Elogne, N.C.; Acquah, K.J.R.; Amon, A.C.; Kouamé, K.; Konan, N.; Koffi, A.; Kadio, M.C.; Yao, A.; et al. Phenotypic and Genotypic Characterization of Mycobacteria Isolates from Buruli Ulcer Suspected Patients Reveals the Involvement of Several Mycobacteria in Chronic Skin Lesions. Am. J. Microbiol. Res. 2018, 6, 79–87. [Google Scholar] [CrossRef]
- Saad, J.; Combe, M.; Hammoudi, N.; Couppié, P.; Blaizot, R.; Jedir, F.; Gozlan, R.E.; Drancourt, M.; Bouam, A. Whole-genome sequence of Mycobacterium ulcerans CSURP7741, a French Guianan clinical isolate. Microbiol. Resour. Announc. 2019, 8, e00215-19. [Google Scholar] [CrossRef]
- Combe, M.; Couppié, P.; Blaizot, R.; Valentini, A.; Gozlan, R.E. Are all Buruli ulcers caused by Mycobacterium ulcerans? Br. J. Dermatol. 2020, 183, 968–970. [Google Scholar] [CrossRef]
- Diaz, D.; Döbeli, H.; Yeboah-Manu, D.; Mensah-Quaino, E.; Friedlein, A.; Soder, N.; Rondini, S.; Bodmer, T.; Pluschke, G. Use of the immunodominant 18-kiloDalton small heat shock protein as a serological marker for exposure to Mycobacterium ulcerans. Clin. Vaccine Immunol. 2006, 13, 1314–11321. [Google Scholar] [CrossRef]
- Stinear, T.; Davies, J.K.; Jenkin, G.A.; Portaels, F.; Ross, B.C.; Oppedisano, F.; Purcell, M.; Hayman, J.A.; Johnson, P.D.R. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 2000, 38, 1482–1487. [Google Scholar] [CrossRef]
- Hayman, J. Postulated epidemiology of Mycobacterium ulcerans infection. Int. J. Epidemiol. 1991, 20, 1093–1098. [Google Scholar] [CrossRef]
- Marston, B.J.; Diallo, M.O.; Horsburgh, C.R.; Diomande, I.; Saki, M.Z.; Kanga, J.M.; Patrice, G.; Lipman, H.B.; Ostroff, S.M.; Good, R.C. Emergence of Buruli ulcer disease in the Daola region of Côte d’Ivoire. Am. J. Trop. Med. Hyg. 1995, 52, 219–224. [Google Scholar] [CrossRef]
- Meyers, W.M. Mycobacterial infections of the skin. In Tropical Pathology; Seifert, G., Ed.; Springer: Heidelberg, Germany, 1994. [Google Scholar]
- Ross, B.C.; Johnson, P.D.R.; Oppedisano, F.; Marino, L.; Sievers, A.; Stinear, T.; Hayman, J.A.; Veitch, M.G.; Robins-Browne, R.M. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl. Environ. Microbiol. 1997, 63, 4135–4138. [Google Scholar] [CrossRef]
- Hofer, M.; Hirschel, B.; Kirschner, P.; Beghetti, M.; Kaelin, A.; Siegrist, C.-A.; Suter, S.; Teske, A.; Bottger, E.C. Brief report: Dissaminated osteomyelitis from Mycobacterium ulcerans after snakebite. N. Engl. J. Med. 1993, 328, 1007–1009. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.; Edwards, R.; Leslie, D.E.; Hayman, J. Molecular method for typing Mycobacterium ulcerans. J. Clin. Microbiol. 1995, 33, 2250–2253. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, T.M.H.; Nomaguchi, H.; Anderson, D.C.; Young, R.A.; Gillis, T.P.; Britton, W.J.; Ivanyi, J.; Kolk, A.H.; Closs, O.; Bloom, B.R. Characterization of antibofy-reactive epitopes on the 65-kilodalton protein of Mycobacterium leprae. Infect. Immun. 1987, 55, 1000–1003. [Google Scholar] [CrossRef] [PubMed]
- Hance, A.J.; Grandchamp, B.; Lévy-Frébault, V.; Lecossier, D.; Rauzier, J.; Bocart, D.; Gicquel, B. Detection and identification of mycobacteria by amplification pf mycobacterial DNA. Mol. Microbiol. 1989, 3, 843–849. [Google Scholar] [CrossRef]
- Stinear, T.; Ross, B.C.; Davies, J.K.; Marino, L.; Robins-Browne, R.M.; Oppedisano, F.; Sievers, A.; Johnson, P.D.R. Identification and characterization of IS2404 and IS2606: Two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 1999, 37, 1018–1023. [Google Scholar] [CrossRef]
- Guerrero, C.; Bernasconi, C.; Burki, D.; Bodmer, T.; Talenti, A. A novel insertion element from Mycobacteriul avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 1995, 33, 304–307. [Google Scholar] [CrossRef]
- Picardeau, M.; Bull, T.J.; Vincent, V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol. Lett. 1997, 154, 95–102. [Google Scholar] [CrossRef][Green Version]
- Shinoda, N.; Nakamura, H.; Watanabe, M. Detection of Mycobacterium ulcerans by real-time PCR with improved primers. Trop. Med. Health 2016, 44, 28. [Google Scholar] [CrossRef]
- Rondini, S.; Mensah-Quainoo, E.; Troll, H.; Bodmer, T.; Pluschke, G. Development and application of real-time PCR assay for quantification of Mycobacterium ulcerans DNA. J. Clin. Microbiol. 2003, 41, 4231–4237. [Google Scholar] [CrossRef]
- Fyfe, J.A.M.; Lavender, C.J.; Johnson, P.D.R.; Globan, M.; Sievers, A.; Azuolas, J.; Stinear, T.P. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl. Environ. Microbiol. 2007, 73, 4733–4740. [Google Scholar] [CrossRef]
- George, K.M. Mycolactone: A polyketide toxin from Mycobacterium ulcerans required for virulence. Science 1999, 283, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Mve-Obiang, A.; Lee, R.E.; Umstot, E.S.; Trott, K.A.; Grammer, T.C.; Parker, J.M.; Ranger, B.S.; Grainger, R.; Mahrous, E.A.; Small, P.L.C. A newly discovered mycobacterial pathogen isolated from laboratory colonies of Xenopus species with lethal infections produces a novel form of mycolactone, the Mycobacterium ulcerans macrolide toxin. Infect. Immun. 2005, 73, 3307–3312. [Google Scholar] [CrossRef] [PubMed]
- Stinear, T.P.; Pryor, M.J.; Porter, J.L.; Cole, S.T. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology 2005, 151, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Trott, K.A.; Stacy, B.A.; Lifland, B.D.; Diggs, H.E.; Harland, R.M.; Khokha, M.K.; Grammer, T.C.; Parker, J.M. Characterization of a Mycobacterium ulcerans-like infection in a colony of African tropical clawed frogs (Xenopus tropicalis). Comp. Med. 2004, 54, 309–317. [Google Scholar]
- Ranger, B.S.; Mahrous, E.A.; Mosi, L.; Adusumilli, S.; Lee, R.E.; Colorni, A.; Rhodes, M.; Small, P.L.C. Globally distributed mycobacterial fish pathogens produce a novel plasmid-encoded toxic macrolide, mycolactone F. Infect. Immun. 2006, 74, 6037–6045. [Google Scholar] [CrossRef]
- Rhodes, M.W.; Kator, H.; McNabb, A.; Deshayes, C.; Reyrat, J.M.; Brown-Elliott, B.A.; Wallace, R.; Trott, K.A.; Parker, J.M.; Lifland, B.; et al. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 2005, 55, 1139–1147. [Google Scholar] [CrossRef]
- Ucko, M.; Colorni, A.; Kvitt, H.; Diamant, A.; Zlotkin, A.; Knibb, W.R. Strain variation in Mycobacterium marinum fish isolates. Appl. Environ. Microbiol. 2002, 68, 5281–5287. [Google Scholar] [CrossRef]
- Tsukamura, M.; Mikoshiba, H. A New Mycobacterium which Caused Skin Infection. Microbiol. Immunol. 1982, 26, 951–955. [Google Scholar] [CrossRef]
- Tsukamura, M.; Kaneda, K.; Imaeda, T.; Mikoshiba, H. A taxonomic study on mycobacterium which caused a skin ulcer in a Japanese girl and resembled Mycobacterium ulcerans. Kekkaku 1989, 64, 691–697. [Google Scholar]
- Kim, H.; Kim, S.H.; Shim, T.S.; Kim, M.N.; Bai, G.H.; Park, Y.G.; Lee, S.-H.; Chae, G.-T.; Cha, C.-Y.; Kook, Y.-H.; et al. Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 2005, 55, 1649–1656. [Google Scholar] [CrossRef]
- Stinear, T.P.; Seemann, T.; Pidot, S.; Frigui, W.; Reysset, G.; Garnier, T.; Meurice, G.; Simon, D.; Bouchier, C.; Ma, L.; et al. Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer. Genome Res. 2007, 17, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Käzer, M.; Rondini, S.; Naegeli, M.; Stinear, T.; Portaels, F.; Certa, U.; Pluschke, G. Evolution of two distinct phylogenetic lineages of the emerging human pathogen Mycobacterium ulcerans. BMC Evol. Biol. 2007, 7, 177. [Google Scholar] [CrossRef] [PubMed]
- Stragier, P.; Ablordey, A.; Durnez, L.; Portaels, F. VNTR analysis differentiates Mycobacterium ulcerans and IS2404 positive mycobacteria. Syst. Appl. Microbiol. 2007, 30, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Chemlal, K.; Huys, G.; Fonteyne, P.-A.; Vincent, V.; Lopez, G.; Rigouts, L.; Swings, J.; Meyers, W.M.; Portaels, F. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphisms and amplified fragment length polymorphisms fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 2001, 39, 3272–3278. [Google Scholar] [CrossRef]
- Chemlal, K.; De Ridder, K.; Fonteyne, P.-A.; Meyers, W.M.; Swings, J.; Portaels, F. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 2001, 64, 270–273. [Google Scholar] [CrossRef][Green Version]
- Stragier, P.; Ablordey, A.; Meyers, W.M.; Portaels, F. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by Using Mycobacterial Interspersed Repetitive Units. J. Bacteriol. 2005, 187, 1639–1647. [Google Scholar] [CrossRef]
- Ablordey, A.; Swings, J.; Hubans, C.; Chemlal, K.; Locht, C.; Portaels, F.; Supply, P. Multilocus Variable-Number Tandem Repeat Typing of Mycobacterium ulcerans. J. Clin. Microbiol. 2005, 43, 1546–1551. [Google Scholar] [CrossRef]
- Hilty, M.; Yeboah-Manu, D.; Boakye, D.; Mensah-Quainoo, E.; Rondini, S.; Schelling, E.; Ofori-Adjei, D.; Portaels, F.; Zinsstag, J.; Pluschke, G. Genetic Diversity in Mycobacterium ulcerans Isolates from Ghana Revealed by a Newly Identified Locus Containing a Variable Number of Tandem Repeats. J. Bacteriol. 2006, 188, 1462–1465. [Google Scholar] [CrossRef]
- Supply, P.; Magdalena, J.; Himpens, S.; Locht, C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 1997, 26, 991–1003. [Google Scholar] [CrossRef]
- Bull, T.J.; Sidi-Boumedine, K.; McMinn, E.J.; Stevenson, K.; Pickup, R.; Hermon-Taylor, J. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 2003, 17, 157–164. [Google Scholar] [CrossRef]
- Hilty, M.; Käser, M.; Zinsstag, J.; Stinear, T.; Pluschke, G. Analysis of the Mycobacterium ulcerans genome sequence reveals new loci for variable number tandem repeats (VNTR) typing. Microbiology 2007, 153, 1483–1487. [Google Scholar] [CrossRef] [PubMed]
- Ablordey, A.; Fonteyne, P.; Stragier, P.; Vandamme, P.; Portaels, F. Identification of a new variable number tandem repeat locus in Mycobacterium ulcerans for potential strain discrimination among African isolates. Clin. Microbiol. Infect. 2007, 13, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Williamson, H.; Phillips, R.; Sarfo, S.; Wansbrough-Jones, M.; Small, P. Genetic diversity of PCR-positive, culture-negative and culture-positive Mycobacterium ulcerans isolated from buruli ulcer patients in Ghana. PLoS ONE 2014, 9, e88007. [Google Scholar] [CrossRef] [PubMed]
- Benbow, M.E.; Kimbirauskas, R.; McIntosh, M.D.; Williamson, H.; Quaye, C.; Boakye, D.; Small, P.L.; Merritt, R.W. Aquatic Macroinvertebrate Assemblages of Ghana, West Africa: Understanding the Ecology of a Neglected Tropical Disease. EcoHealth 2014, 11, 168–183. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, M.; Williamson, H.; Benbow, E.; Kimbirauskas, R.; Quaye, C.; Boakye, D.; Small, P.; Merritt, R. Associations between Mycobacterium ulcerans and aquatic plant communities of West Africa: Implications for Buruli ulcer disease. EcoHealth 2014, 11, 184–196. [Google Scholar] [CrossRef]
- Narh, C.A.; Mosi, L.; Quaye, C.; Dassi, C.; Konan, D.O.; Tay, S.C.K.; de Souza, D.K.; Boakye, D.A.; Bonfoh, B. Source Tracking Mycobacterium ulcerans Infections in the Ashanti Region, Ghana. PLoS Negl. Trop. Dis. 2015, 9, e0003437. [Google Scholar] [CrossRef]
- Lavender, C.J.; Stinear, T.P.; Johnson, P.D.R.; Azuolas, J.; Benbow, M.E.; Wallace, J.R.; Fyfe, J.A. Evaluation of VNTR typing for the identification of Mycobacterium ulcerans in environmental samples from Victoria, Australia. FEMS Microbiol. Lett. 2008, 287, 250–255. [Google Scholar] [CrossRef]
- Reynaud, Y.; Millet, J.; Couvin, D.; Rastogi, N.; Brown, C. Heterogeneity among Mycobacterium ulcerans from French Guiana Revealed by Multilocus Variable Number Tandem Repeat Analysis (MLVA). PLoS ONE 2015, 10, e0118597. [Google Scholar] [CrossRef]
- Ahlstrom, C.; Barkeman, H.W.; Stevenson, K.; Zadoks, R.N.; Biek, R.; Kao, R.; Trewby, H.; Haupstein, D.; Kelton, D.F.; Fecteau, G.; et al. Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level. BMC Genom. 2015, 16, 161. [Google Scholar] [CrossRef]
- Röltgen, K.; Pluschke, G. Buruli ulcer: History and Disease Burden. In Buruli Ulcer; Pluschke, G., Röltgen, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; p. 5. [Google Scholar] [CrossRef]
- Bonamonte, D.; De Vito, D.; Vestita, M.; Delvecchio, S.; Ranieri, L.D.; Santantonio, M.; Angelini, G. Aquarium-borne Mycobacterium marinum skin infection. Report of 15 cases and review of the literature. Eur. J. Dermatol. 2013, 23, 510–516. [Google Scholar] [CrossRef]
- Nakanaga, K.; Hoshino, Y.; Yotsu, R.R.; Makino, M.; Ishii, N. Nineteen cases of Buruliulcer diagnosed in Japan from 1980 to 2010. J. Clin. Microbiol. 2011, 49, 3829–3836. [Google Scholar] [CrossRef] [PubMed]
- Dixit, P.; Kotra, L.P. Diseases caused by acid-fast bacteria. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–5. [Google Scholar]
- World Health Organization (WHO). 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/buruli-ulcer-(mycobacterium-ulcerans-infection) (accessed on 30 July 2023).
- Koichi, S.; Yuqian, L.; Yuji, M.; Chiaki, M.; Mariko, M.-S.; Rie Yotsu, R.; Ishii, N. Buruli Ulcer in Japan. In Buruli Ulcer; Pluschke, G., Röltgen, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; p. 87. [Google Scholar] [CrossRef]
- Couppié, P.; Blaizot, R.; Velvin, C.J.; Douine, M.; Combe, M.; Nacher, M.; Gozlan, R.E. Mycobacterium ulcerans infection in French Guiana; current state of knowledge. In Buruli Ulcer, Mycobacterium Ulcerans Disease; Springer: Berlin/Heidelberg, Germany, 2019; p. 77. [Google Scholar] [CrossRef]
- Honore, N.; Cole, S.T. Molecular basis of rifampin resistance in Mycobacterium leprae. Antimicrob. Agents Chemother. 1993, 37, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Narh, C.A.; Mosi, L.; Quaye, C.; Tay, S.C.K.; Bonfoh, B.; de Souza, D.K. Genotyping Tools for Mycobacterium ulcerans Drawbacks and Future Prospects. Mycobact. Dis. 2014, 4, 1000149. [Google Scholar] [CrossRef] [PubMed]
- Aubry, A.; Jarlier, V.; Escolano, S.; Truffot-Pernot, C.; Cambau, E. Antibiotic susceptibility pattern of Mycobacterium marinum. Antimicrob. Agents Chemother. 2000, 44, 3133–3136. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.; Brown-Elliott, B.A.; Catanzaro, A.; Daley, C.; Gordin, F.; Holland, S.M.; Horsburgh, R.; Huitt, G.; Iademarco, M.F.; et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–415. [Google Scholar] [CrossRef]
- Johnson, M.G.; Stout, J.E. Twenty-eight cases of Mycobacterium marinum infection: Retrospective case series and literature review. Infection 2015, 43, 655–662. [Google Scholar] [CrossRef]
- Matsumoto, M.; Machida, Y.; Kanemaru, M.; Yamamoto, M.; Sano, M. and Kato, G. Infection with Mycobacterium pseudoshottsii in cultured yellowtail Seriola quinqueradiata in Owase Bay, Japan. Fish Pathol. 2022, 57, 35–40. [Google Scholar] [CrossRef]
- Nigou, J.; Gilleron, M.; Puzo, G. Lipoarabinomannans: From structure to biosynthesis. Biochimie 2003, 85, 153–166. [Google Scholar] [CrossRef]
- Suykerbuyk, P.; Vleminckx, K.; Pasmans, F.; Stragier, P.; Ablordey, A.; Tran, H.T.; Hermans, K.; Fleetwood, M.; Meyers, W.M.; Portaels, F. Mycobacterium liflandii infection in European colony of Silurana tropicalis. Emerg. Infect. Dis. 2007, 13, 743–746. [Google Scholar] [CrossRef]
- Stanford, J.L.; Gunthorpe, W.J. A study of some fast-growing scotochromogenic mycobacteria including species descritpions of Mycobacterium gilvum (new species) and Mycobacterium duvalii (new species). Br. J. Exp. Pathol. 1971, 52, 627–673. [Google Scholar]
- Ablordey, A.; Amissah, D.A.; Aboagye, I.F.; Hatano, B.; Yamazaki, T.; Sata, T.; Ishikawa, K.; Katano, H. Detection of Mycobacterium ulcerans by the loop mediated isothermal amplification method. PLoS Neglect. Trop. Dis. 2012, 6, e1590. [Google Scholar] [CrossRef] [PubMed]
- Njiru, Z.K.; Yeboah-Manu, D.; Stinear, T.P.; Fyfe, J.A.M. Rapid and sensitive detection of Mycobacterium ulcerans by use of a loop-mediated isothermal amplification test. J. Clin. Microbiol. 2012, 50, 1737–1741. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, W71–W74. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]

| MPM | First Isolation | Origin | Host | Colony Coloration | Growth Rate | Growth Temperature | Mycolactone | Source |
|---|---|---|---|---|---|---|---|---|
| M. ulcerans | 1948 | Australia | Human | Greenish or brownish yellow | Slow growth (>4 weeks incubation) | 30 °C–33 °C | A/B | [13] |
| M. shinshuense (strain 753) | 1980 | Japan | Human | Yellowish, pigmentation in dark | Slow growth (>3 weeks incubation) | 28 °C | A/B | [49] |
| M. marinum (strain CC240299) | 2002 | Israel | Koi (Cyprinus carpio) | NA | NA | NA | F | [48] |
| M. marinum (strain DL240490) | 2002 | Red Sea | European sea bass (Dicentrarchus labrax) | NA | NA | NA | F | [48] |
| M. marinum (strain DL045) | 2002 | Mediterranean Sea | European sea bass (Dicentrarchus labrax) | NA | NA | NA | F | [48] |
| M. liflandii (strain 128FXT) | 2004 | US | African tropical clawed frogs (Xenopus tropicalis) | Buff-colored, non-pigmented | Slow growth (>4 weeks incubation) | 28 °C | E | [45] |
| M. pseudoshottsii (strain L15) | 2005 | US | Striped bass (Morone saxatilis) | Pale-yellow to gold, non-pigmented | Slow growth (>4 weeks incubation) | 23 °C | F | [47] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Combe, M.; Cherif, E.; Blaizot, R.; Breugnot, D.; Gozlan, R.E. What about Current Diversity of Mycolactone-Producing Mycobacteria? Implication for the Diagnosis and Treatment of Buruli Ulcer. Int. J. Mol. Sci. 2023, 24, 13727. https://doi.org/10.3390/ijms241813727
Combe M, Cherif E, Blaizot R, Breugnot D, Gozlan RE. What about Current Diversity of Mycolactone-Producing Mycobacteria? Implication for the Diagnosis and Treatment of Buruli Ulcer. International Journal of Molecular Sciences. 2023; 24(18):13727. https://doi.org/10.3390/ijms241813727
Chicago/Turabian StyleCombe, Marine, Emira Cherif, Romain Blaizot, Damien Breugnot, and Rodolphe Elie Gozlan. 2023. "What about Current Diversity of Mycolactone-Producing Mycobacteria? Implication for the Diagnosis and Treatment of Buruli Ulcer" International Journal of Molecular Sciences 24, no. 18: 13727. https://doi.org/10.3390/ijms241813727
APA StyleCombe, M., Cherif, E., Blaizot, R., Breugnot, D., & Gozlan, R. E. (2023). What about Current Diversity of Mycolactone-Producing Mycobacteria? Implication for the Diagnosis and Treatment of Buruli Ulcer. International Journal of Molecular Sciences, 24(18), 13727. https://doi.org/10.3390/ijms241813727






