rs71327024 Associated with COVID-19 Hospitalization Reduces CXCR6 Promoter Activity in Human CD4+ T Cells via Disruption of c-Myb Binding
Abstract
1. Introduction
2. Results
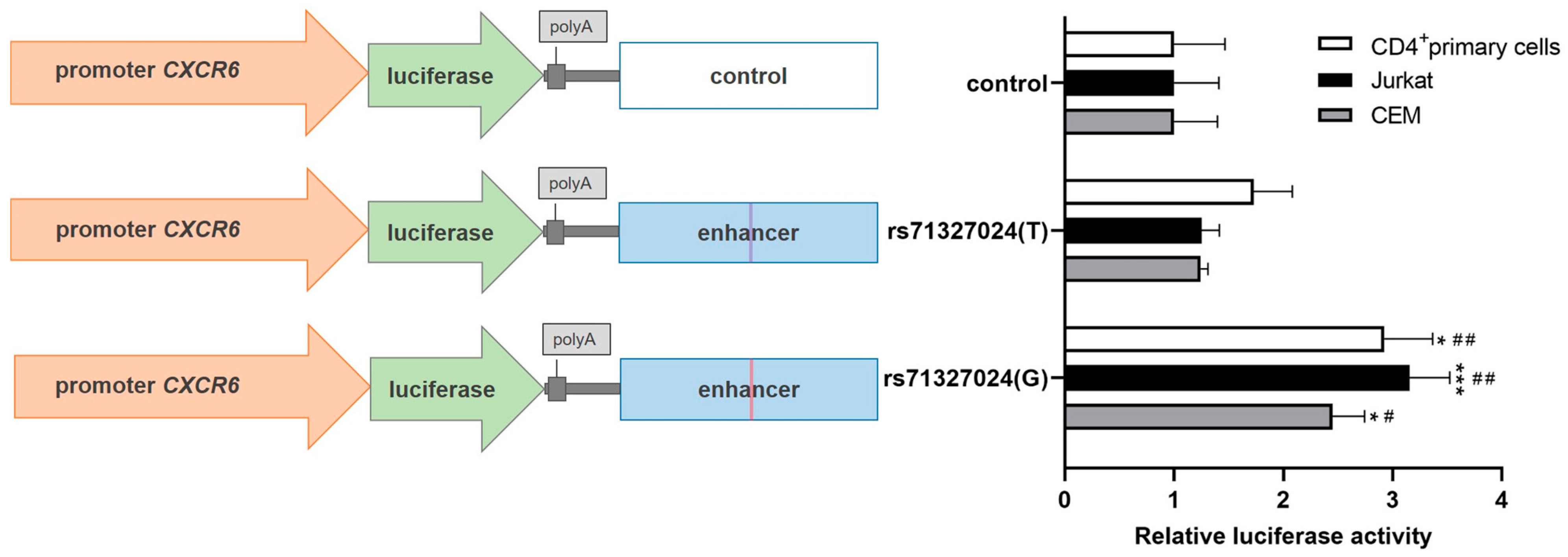
2.1. The Risk Allele (T) of rs71327024 Decreases the CXCR6 Promoter Activity in Jurkat, CEM and CD4+ Primary Cells
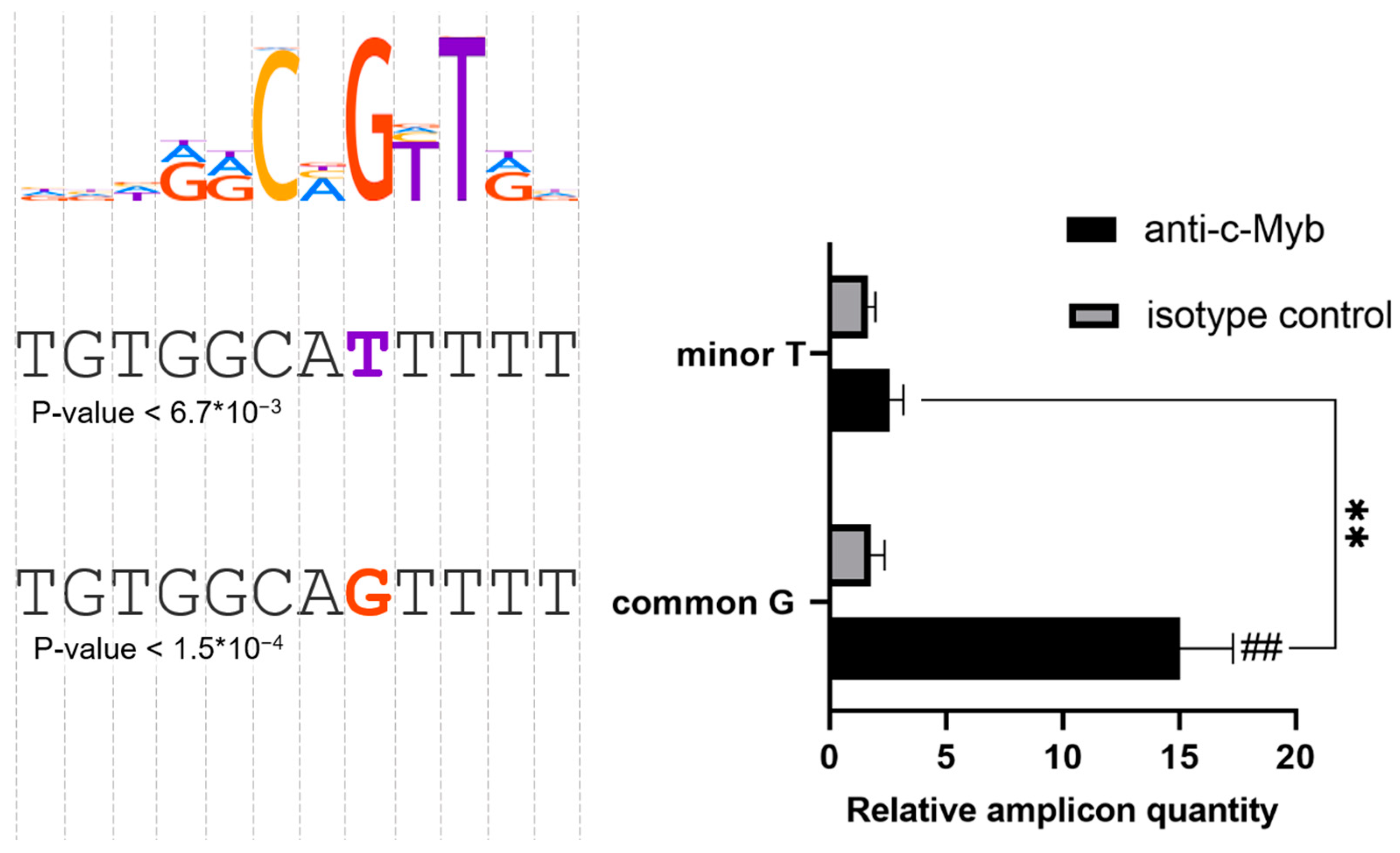
2.2. The Common rs71327024(G) Makes a c-Myb Binding Site in PMA-Treated Jurkat Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Reporter Plasmids
4.3. Cell Transfection and Luciferase Reporter Assay
4.4. siRNA-Mediated c-Myb Knockdown
4.5. DNA Pull-Down Assay
4.6. RNA Extraction, cDNA Synthesis and Real-Time-PCR
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- COVID-19 Host Genetics Initiative. A First Update on Mapping the Human Genetic Architecture of COVID-19. Nature 2022, 608, E1–E10. [Google Scholar] [CrossRef] [PubMed]
- The Severe COVID-19 GWAS Group. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Roberts, G.H.L.; Partha, R.; Rhead, B.; Knight, S.C.; Park, D.S.; Coignet, M.V.; Zhang, M.; Berkowitz, N.; Turrisini, D.A.; Gaddis, M.; et al. Expanded COVID-19 Phenotype Definitions Reveal Distinct Patterns of Genetic Association and Protective Effects. Nat. Genet. 2022, 54, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Velavan, T.P.; Pallerla, S.R.; Rüter, J.; Augustin, Y.; Kremsner, P.G.; Krishna, S.; Meyer, C.G. Host Genetic Factors Determining COVID-19 Susceptibility and Severity. eBioMedicine 2021, 72, 103629. [Google Scholar] [CrossRef]
- Nakanishi, T.; Pigazzini, S.; Degenhardt, F.; Cordioli, M.; Butler-Laporte, G.; Maya-Miles, D.; Bujanda, L.; Bouysran, Y.; Niemi, M.E.K.; Palom, A.; et al. Age-Dependent Impact of the Major Common Genetic Risk Factor for COVID-19 on Severity and Mortality. J. Clin. Investig. 2021, 131, 49. [Google Scholar] [CrossRef] [PubMed]
- Shelton, J.F.; Shastri, A.J.; Ye, C.; Weldon, C.H.; Filshtein-Sonmez, T.; Coker, D.; Symons, A.; Esparza-Gordillo, J.; Chubb, A.; Fitch, A.; et al. Trans-Ancestry Analysis Reveals Genetic and Nongenetic Associations with COVID-19 Susceptibility and Severity. Nat. Genet. 2021, 53, 801–808. [Google Scholar] [CrossRef]
- Kasela, S.; Daniloski, Z.; Bollepalli, S.; Jordan, T.X.; tenOever, B.R.; Sanjana, N.E.; Lappalainen, T. Integrative Approach Identifies SLC6A20 and CXCR6 as Putative Causal Genes for the COVID-19 GWAS Signal in the 3p21.31 Locus. Genome Biol. 2021, 22, 242. [Google Scholar] [CrossRef]
- Yao, Y.; Ye, F.; Li, K.; Xu, P.; Tan, W.; Feng, Q.; Rao, S. Genome and Epigenome Editing Identify CCR9 and SLC6A20 as Target Genes at the 3p21.31 Locus Associated with Severe COVID-19. Signal Transduct. Target. Ther. 2021, 6, 85. [Google Scholar] [CrossRef]
- Fricke-Galindo, I.; Falfán-Valencia, R. Genetics Insight for COVID-19 Susceptibility and Severity: A Review. Front. Immunol. 2021, 12, 622176. [Google Scholar] [CrossRef]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and Chemokine Receptors during COVID-19 Infection. Comput. Struct. Biotechnol. J. 2021, 19, 976. [Google Scholar] [CrossRef]
- Korbecki, J.; Bajdak-Rusinek, K.; Kupnicka, P.; Kapczuk, P.; Simińska, D.; Chlubek, D.; Baranowska-Bosiacka, I. The Role of CXCL16 in the Pathogenesis of Cancer and Other Diseases. Int. J. Mol. Sci. 2021, 22, 3490. [Google Scholar] [CrossRef]
- Wehr, A.; Baeck, C.; Heymann, F.; Niemietz, P.M.; Hammerich, L.; Martin, C.; Zimmermann, H.W.; Pack, O.; Gassler, N.; Hittatiya, K.; et al. Chemokine Receptor CXCR6-Dependent Hepatic NK T Cell Accumulation Promotes Inflammation and Liver Fibrosis. J. Immunol. 2013, 190, 5226–5236. [Google Scholar] [CrossRef]
- Butcher, M.J.; Wu, C.I.; Waseem, T.; Galkina, E.V. CXCR6 Regulates the Recruitment of Pro-Inflammatory IL-17A-Producing T Cells into Atherosclerotic Aortas. Int. Immunol. 2016, 28, 255. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.N.; Ronan, E.O.; de Lara, C.; Franken, K.L.M.C.; Ottenhoff, T.H.M.; Tchilian, E.Z.; Beverley, P.C.L. CXCR6 Is a Marker for Protective Antigen-Specific Cells in the Lungs after Intranasal Immunization against Mycobacterium Tuberculosis. Infect. Immun. 2011, 79, 3328. [Google Scholar] [CrossRef]
- Ashhurst, A.S.; Flórido, M.; Lin, L.C.W.; Quan, D.; Armitage, E.; Stifter, S.A.; Stambas, J.; Britton, W.J. CXCR6-Deficiency Improves the Control of Pulmonary Mycobacterium Tuberculosis and Influenza Infection Independent of T-Lymphocyte Recruitment to the Lungs. Front. Immunol. 2019, 10, 339. [Google Scholar] [CrossRef] [PubMed]
- The GTEx Consortium. The GTEx Consortium Atlas of Genetic Regulatory Effects across Human Tissues. Science 2020, 369, 1318–1330. [Google Scholar] [CrossRef]
- Payne, D.J.; Dalal, S.; Leach, R.; Parker, R.; Griffin, S.; McKimmie, C.S.; Cook, G.P.; Richards, S.J.; Hillmen, P.; Munir, T.; et al. The CXCR6/CXCL16 Axis Links Inflamm-Aging to Disease Severity in COVID-19 Patients. bioRxiv 2021. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Wang, J.; Jeong, H.-H.; Chen, W.; Jia, P.; Zhao, Z. Association of CXCR6 with COVID-19 Severity: Delineating the Host Genetic Factors in Transcriptomic Regulation. Hum. Genet. 2021, 140, 1313–1328. [Google Scholar] [CrossRef]
- Melms, J.C.; Biermann, J.; Huang, H.; Wang, Y.; Nair, A.; Tagore, S.; Katsyv, I.; Rendeiro, A.F.; Amin, A.D.; Schapiro, D.; et al. A Molecular Single-Cell Lung Atlas of Lethal COVID-19. Nature 2021, 595, 114–119. [Google Scholar] [CrossRef]
- Li, S.; Jiang, L.; Li, X.; Lin, F.; Wang, Y.; Li, B.; Jiang, T.; An, W.; Liu, S.; Liu, H.; et al. Clinical and Pathological Investigation of Patients with Severe COVID-19. JCI Insight 2020, 5, e138070. [Google Scholar] [CrossRef]
- Odak, I.; Barros-Martins, J.; Bošnjak, B.; Stahl, K.; David, S.; Wiesner, O.; Busch, M.; Hoeper, M.M.; Pink, I.; Welte, T.; et al. Reappearance of Effector T Cells Is Associated with Recovery from COVID-19. EBioMedicine 2020, 57, 102885. [Google Scholar] [CrossRef]
- Mathew, D.; Giles, J.R.; Baxter, A.E.; Oldridge, D.A.; Greenplate, A.R.; Wu, J.E.; Alanio, C.; Kuri-Cervantes, L.; Pampena, M.B.; D’Andrea, K.; et al. Deep Immune Profiling of COVID-19 Patients Reveals Distinct Immunotypes with Therapeutic Implications. Science 2020, 369, eabc8511. [Google Scholar] [CrossRef]
- Wen, X.-S.; Jiang, D.; Gao, L.; Zhou, J.-Z.; Xiao, J.; Cheng, X.-C.; He, B.; Chen, Y.; Lei, P.; Tan, X.-W.; et al. Clinical Characteristics and Predictive Value of Lower CD4+T Cell Level in Patients with Moderate and Severe COVID-19: A Multicenter Retrospective Study. BMC Infect. Dis. 2021, 21, 57. [Google Scholar] [CrossRef]
- Chen, Z.; John Wherry, E. T Cell Responses in Patients with COVID-19. Nat. Rev. Immunol. 2020, 20, 529–536. [Google Scholar] [CrossRef]
- COVID-19 Host Genetics Initiative. Mapping the Human Genetic Architecture of COVID-19. Nature 2021, 600, 472–477. [Google Scholar] [CrossRef]
- Mora, A.; Sandve, G.K.; Gabrielsen, O.S.; Eskeland, R. In the Loop: Promoter–Enhancer Interactions and Bioinformatics. Brief. Bioinform. 2016, 17, 980. [Google Scholar] [CrossRef] [PubMed]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Roadmap Epigenomics Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative Analysis of 111 Reference Human Epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef]
- Ahnadi, C.E.; Giguère, P.; Gravel, S.; Gagné, D.; Goulet, A.; Fülöp, T.; Payet, M.D.; Dupuis, G. Chronic PMA Treatment of Jurkat T Lymphocytes Results in Decreased Protein Tyrosine Phosphorylation and Inhibition of CD3- but Not Ti-dependent Antibody-triggered Ca 2+ Signaling. J. Leukoc. Biol. 2000, 68, 293–300. [Google Scholar] [CrossRef]
- Smeets, R.L.; Fleuren, W.W.M.; He, X.; Vink, P.M.; Wijnands, F.; Gorecka, M.; Klop, H.; Bauerschmidt, S.; Garritsen, A.; Koenen, H.J.P.M.; et al. Molecular Pathway Profiling of T Lymphocyte Signal Transduction Pathways; Th1 and Th2 Genomic Fingerprints Are Defined by TCR and CD28-Mediated Signaling. BMC Immunol. 2012, 13, 12. [Google Scholar] [CrossRef]
- Perbellini, O.; Cavallini, C.; Chignola, R.; Galasso, M.; Scupoli, M.T. Phospho-Specific Flow Cytometry Reveals Signaling Heterogeneity in T-Cell Acute Lymphoblastic Leukemia Cell Lines. Cells 2022, 11, 2072. [Google Scholar] [CrossRef]
- Forrest, A.R.R.; Kawaji, H.; Rehli, M.; Baillie, J.K.; De Hoon, M.J.L.; Haberle, V.; Lassmann, T.; Kulakovskiy, I.V.; Lizio, M.; Itoh, M.; et al. A Promoter-Level Mammalian Expression Atlas. Nature 2014, 507, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Abramov, S.; Boytsov, A.; Bykova, D.; Penzar, D.D.; Yevshin, I.; Kolmykov, S.K.; Fridman, M.V.; Favorov, A.V.; Vorontsov, I.E.; Baulin, E.; et al. Landscape of Allele-Specific Transcription Factor Binding in the Human Genome. Nat. Commun. 2021, 12, 2751. [Google Scholar] [CrossRef]
- Boytsov, A.; Abramov, S.; Aiusheeva, A.Z.; Kasianova, A.M.; Baulin, E.; Kuznetsov, I.A.; Aulchenko, Y.S.; Kolmykov, S.; Yevshin, I.; Kolpakov, F.; et al. ANANASTRA: Annotation and Enrichment Analysis of Allele-Specific Transcription Factor Binding at SNPs. Nucleic Acids Res. 2022, 50, W51–W56. [Google Scholar] [CrossRef]
- Cicirò, Y.; Sala, A. MYB Oncoproteins: Emerging Players and Potential Therapeutic Targets in Human Cancer. Oncogenesis 2021, 10, 19. [Google Scholar] [CrossRef]
- Nakata, Y.; Brignier, A.C.; Jin, S.; Shen, Y.; Rudnick, S.I.; Sugita, M.; Gewirtz, A.M. C-Myb, Menin, GATA-3, and MLL Form a Dynamic Transcription Complex That Plays a Pivotal Role in Human T Helper Type 2 Cell Development. Blood 2010, 116, 1280–1290. [Google Scholar] [CrossRef]
- Zeng, Z.; Yang, H.; Huang, Z.Z.; Chen, C.; Wang, J.; Lu, S.C. The Role of C-Myb in the up-Regulation of Methionine Adenosyltransferase 2A Expression in Activated Jurkat Cells. Biochem. J. 2001, 353, 163–168. [Google Scholar] [CrossRef]
- Jagoda, E.; Marnetto, D.; Senevirathne, G.; Gonzalez, V.; Baid, K.; Montinaro, F.; Richard, D.; Falzarano, D.; Leblanc, E.V.; Colpitts, C.C.; et al. Regulatory Dissection of the Severe COVID-19 Risk Locus Introgressed by Neanderthals. eLife 2023, 12, e71235. [Google Scholar] [CrossRef]
- Stikker, B.S.; Stik, G.; van Ouwerkerk, A.F.; Trap, L.; Spicuglia, S.; Hendriks, R.W.; Stadhouders, R. Severe COVID-19-Associated Variants Linked to Chemokine Receptor Gene Control in Monocytes and Macrophages. Genome Biol. 2022, 23, 96. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.J.; Guillen, C.; Symon, F.A.; Huynh, T.T.; Berry, M.A.; Entwisle, J.J.; Briskin, M.; Pavord, I.D.; Wardlaw, A.J. Expression of CXCR6 and Its Ligand CXCL16 in the Lung in Health and Disease. Clin. Exp. Allergy 2005, 35, 1572–1580. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2021, 591, 92–98. [Google Scholar] [CrossRef]
- Smieszek, S.P.; Polymeropoulos, V.M.; Polymeropoulos, C.M.; Przychodzen, B.P.; Birznieks, G.; Polymeropoulos, M.H. Elevated Plasma Levels of CXCL16 in Severe COVID-19 Patients. Cytokine 2022, 152, 155810. [Google Scholar] [CrossRef]
- Ustiugova, A.S.; Korneev, K.V.; Kuprash, D.V.; Afanasyeva, M.A. Functional SNPs in the Human Autoimmunity-Associated Locus 17q12-21. Genes 2019, 10, 77. [Google Scholar] [CrossRef]
- Lorenzo, P.I.; Brendeford, E.M.; Gilfillan, S.; Gavrilov, A.A.; Leedsak, M.; Razin, S.V.; Eskeland, R.; Sæther, T.; Gabrielsen, O.S. Identification of C-Myb Target Genes in K562 Cells Reveals a Role for c-Myb as a Master Regulator. Genes Cancer 2011, 2, 805–817. [Google Scholar] [CrossRef][Green Version]
- Korneev, K.V.; Kondakova, A.N.; Sviriaeva, E.N.; Mitkin, N.A.; Palmigiano, A.; Kruglov, A.A.; Telegin, G.B.; Drutskaya, M.S.; Sturiale, L.; Garozzo, D.; et al. Hypoacylated LPS from Foodborne Pathogen Campylobacter Jejuni Induces Moderate TLR4-Mediated Inflammatory Response in Murine Macrophages. Front. Cell. Infect. Microbiol. 2018, 8, 58. [Google Scholar] [CrossRef]
- Mitkin, N.A.; Korneev, K.V.; Gorbacheva, A.M.; Kuprash, D.V. Relative Efficiency of Transcription Factor Binding to Allelic Variants of Regulatory Regions of Human Genes in Immunoprecipitation and Real-Time PCR. Mol. Biol. 2019, 53, 346–353. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uvarova, A.N.; Stasevich, E.M.; Ustiugova, A.S.; Mitkin, N.A.; Zheremyan, E.A.; Sheetikov, S.A.; Zornikova, K.V.; Bogolyubova, A.V.; Rubtsov, M.A.; Kulakovskiy, I.V.; et al. rs71327024 Associated with COVID-19 Hospitalization Reduces CXCR6 Promoter Activity in Human CD4+ T Cells via Disruption of c-Myb Binding. Int. J. Mol. Sci. 2023, 24, 13790. https://doi.org/10.3390/ijms241813790
Uvarova AN, Stasevich EM, Ustiugova AS, Mitkin NA, Zheremyan EA, Sheetikov SA, Zornikova KV, Bogolyubova AV, Rubtsov MA, Kulakovskiy IV, et al. rs71327024 Associated with COVID-19 Hospitalization Reduces CXCR6 Promoter Activity in Human CD4+ T Cells via Disruption of c-Myb Binding. International Journal of Molecular Sciences. 2023; 24(18):13790. https://doi.org/10.3390/ijms241813790
Chicago/Turabian StyleUvarova, Aksinya N., Ekaterina M. Stasevich, Alina S. Ustiugova, Nikita A. Mitkin, Elina A. Zheremyan, Savely A. Sheetikov, Ksenia V. Zornikova, Apollinariya V. Bogolyubova, Mikhail A. Rubtsov, Ivan V. Kulakovskiy, and et al. 2023. "rs71327024 Associated with COVID-19 Hospitalization Reduces CXCR6 Promoter Activity in Human CD4+ T Cells via Disruption of c-Myb Binding" International Journal of Molecular Sciences 24, no. 18: 13790. https://doi.org/10.3390/ijms241813790
APA StyleUvarova, A. N., Stasevich, E. M., Ustiugova, A. S., Mitkin, N. A., Zheremyan, E. A., Sheetikov, S. A., Zornikova, K. V., Bogolyubova, A. V., Rubtsov, M. A., Kulakovskiy, I. V., Kuprash, D. V., Korneev, K. V., & Schwartz, A. M. (2023). rs71327024 Associated with COVID-19 Hospitalization Reduces CXCR6 Promoter Activity in Human CD4+ T Cells via Disruption of c-Myb Binding. International Journal of Molecular Sciences, 24(18), 13790. https://doi.org/10.3390/ijms241813790






