In Silico Neuroprotective Effects of Specific Rheum palmatum Metabolites on Parkinson’s Disease Targets
Abstract
:1. Introduction
2. Results and Discussion
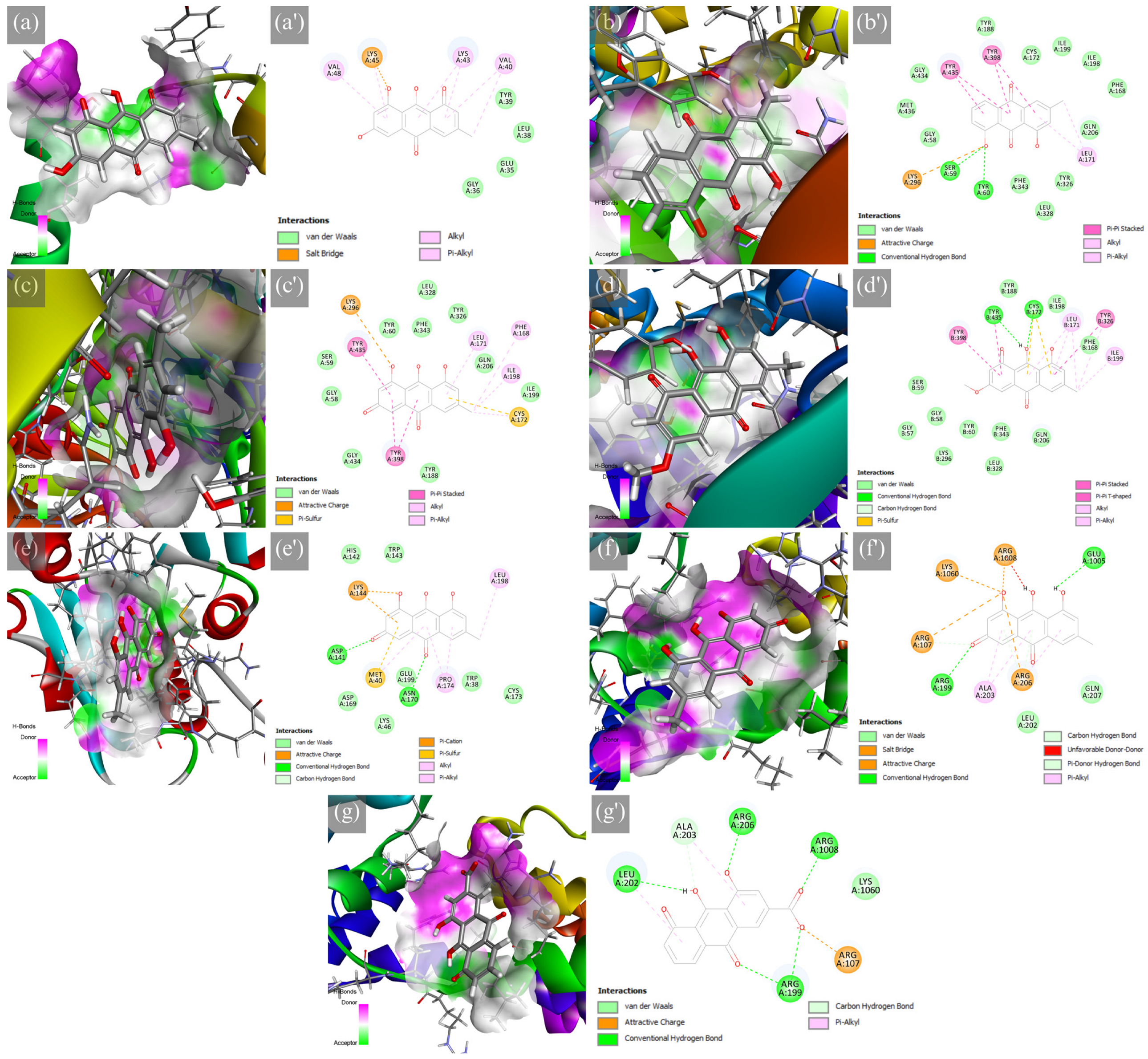
2.1. Molecular Docking
2.2. Density Functional Theory (DFT) Analysis
2.3. Predicted Inhibitory Activity from 3D QSAR Models
2.4. ADMET Properties
3. Materials and Methods
3.1. Preparation of Ligands
3.2. Preparation of Proteins
3.3. Molecular Docking
3.4. 3D QSAR Modeling and Activity Prediction
3.5. Density Functional Theory (DFT) Analysis
3.6. ADMET Properties Prediction
4. Conclusions
5. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chopade, P.; Chopade, N.; Zhao, Z.; Mitragotri, S.; Liao, R.; Chandran Suja, V. Alzheimer’s and Parkinson’s Disease Therapies in the Clinic. Bioeng. Transl. Med. 2023, 8, e10367. [Google Scholar] [CrossRef]
- Marino, B.L.B.; de Souza, L.R.; Sousa, K.P.A.; Ferreira, J.V.; Padilha, E.C.; da Silva, C.H.T.P.; Taft, C.A.; Hage-Melim, L.I.S. Parkinson’s Disease: A Review from Pathophysiology to Treatment. Mini-Rev. Med. Chem. 2020, 20, 754–767. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Hamilton, J.L.; Kopil, C.; Beck, J.C.; Tanner, C.M.; Albin, R.L.; Ray Dorsey, E.; Dahodwala, N.; Cintina, I.; Hogan, P.; et al. Current and Projected Future Economic Burden of Parkinson’s Disease in the U.S. NPJ Park. Dis. 2020, 6, 15. [Google Scholar] [CrossRef]
- Bhat, S.; Acharya, U.R.; Hagiwara, Y.; Dadmehr, N.; Adeli, H. Parkinson’s Disease: Cause Factors, Measurable Indicators, and Early Diagnosis. Comput. Biol. Med. 2018, 102, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Lang, A.E. Parkinson’s Disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Kim, S.; Seo, J.-H.; Suh, Y.-H. α-Synuclein, Parkinson’s Disease, and Alzheimer’s Disease. Parkinsonism Relat. Disord. 2004, 10, S9–S13. [Google Scholar] [CrossRef]
- Dauer, W.; Przedborski, S. Parkinson’s Disease: Mechanisms and Models. Neuron 2003, 39, 889–909. [Google Scholar] [CrossRef] [PubMed]
- Gnanaraj, C.; Sekar, M.; Fuloria, S.; Swain, S.S.; Gan, S.H.; Chidambaram, K.; Rani, N.N.I.M.; Balan, T.; Stephenie, S.; Lum, P.T.; et al. In Silico Molecular Docking Analysis of Karanjin against Alzheimer’s and Parkinson’s Diseases as a Potential Natural Lead Molecule for New Drug Design, Development and Therapy. Molecules 2022, 27, 2834. [Google Scholar] [CrossRef] [PubMed]
- Latif, S.; Jahangeer, M.; Maknoon Razia, D.; Ashiq, M.; Ghaffar, A.; Akram, M.; El Allam, A.; Bouyahya, A.; Garipova, L.; Ali Shariati, M.; et al. Dopamine in Parkinson’s Disease. Clin. Chim. Acta 2021, 522, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Singh, S.; Shukla, S. Physiological and Functional Basis of Dopamine Receptors and Their Role in Neurogenesis: Possible Implication for Parkinson’s Disease. J. Exp. Neurosci. 2018, 12, 117906951877982. [Google Scholar] [CrossRef]
- Takakusaki, K. Functional Neuroanatomy for Posture and Gait Control. J. Mov. Disord. 2017, 10, 1–17. [Google Scholar] [CrossRef]
- Rubin, J.E.; McIntyre, C.C.; Turner, R.S.; Wichmann, T. Basal Ganglia Activity Patterns in Parkinsonism and Computational Modeling of Their Downstream Effects. Eur. J. Neurosci. 2012, 36, 2213–2228. [Google Scholar] [CrossRef]
- Galvan, A.; Devergnas, A.; Wichmann, T. Alterations in Neuronal Activity in Basal Ganglia-Thalamocortical Circuits in the Parkinsonian State. Front. Neuroanat. 2015, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-C.; Chen, Y.; Xu Yang ShenTu, C.-Y.; Peng, L.-H. Signaling Pathways in Parkinson’s Disease: Molecular Mechanisms and Therapeutic Interventions. Signal Transduct. Target. Ther. 2023, 8, 73. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, Y.; Przedborski, S. Oxidative Stress in Parkinson’s Disease. Ann. N. Y. Acad. Sci. 2008, 1147, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Reichmann, H.; Jost, W. Parkinson’s Disease: A Never Ending Story. J. Neural. Transm. 2023, 130, 735–736. [Google Scholar] [CrossRef]
- Tanaka, M.; Szabó, Á.; Spekker, E.; Polyák, H.; Tóth, F.; Vécsei, L. Mitochondrial Impairment: A Common Motif in Neuropsychiatric Presentation? The Link to the Tryptophan–Kynurenine Metabolic System. Cells 2022, 11, 2607. [Google Scholar] [CrossRef]
- González-Rodríguez, P.; Zampese, E.; Stout, K.A.; Guzman, J.N.; Ilijic, E.; Yang, B.; Tkatch, T.; Stavarache, M.A.; Wokosin, D.L.; Gao, L.; et al. Disruption of Mitochondrial Complex I Induces Progressive Parkinsonism. Nature 2021, 599, 650–656. [Google Scholar] [CrossRef]
- Funayama, M.; Ohe, K.; Amo, T.; Furuya, N.; Yamaguchi, J.; Saiki, S.; Li, Y.; Ogaki, K.; Ando, M.; Yoshino, H.; et al. CHCHD2 Mutations in Autosomal Dominant Late-Onset Parkinson’s Disease: A Genome-Wide Linkage and Sequencing Study. Lancet Neurol. 2015, 14, 274–282. [Google Scholar] [CrossRef]
- Battaglia, M.R.; Di Fazio, C.; Battaglia, S. Activated Tryptophan-Kynurenine Metabolic System in the Human Brain Is Associated with Learned Fear. Front. Mol. Neurosci. 2023, 16, 1217090. [Google Scholar] [CrossRef]
- Behl, T.; Kaur, I.; Sehgal, A.; Singh, S.; Bhatia, S.; Al-Harrasi, A.; Zengin, G.; Bumbu, A.G.; Andronie-Cioara, F.L.; Nechifor, A.C.; et al. The Footprint of Kynurenine Pathway in Neurodegeneration: Janus-Faced Role in Parkinson’s Disorder and Therapeutic Implications. Int. J. Mol. Sci. 2021, 22, 6737. [Google Scholar] [CrossRef]
- Bohár, Z.; Toldi, J.; Fülöp, F.; Vécsei, L. Changing the Face of Kynurenines and Neurotoxicity: Therapeutic Considerations. Int. J. Mol. Sci. 2015, 16, 9772–9793. [Google Scholar] [CrossRef]
- Arias-Vera, J. Abnormalities in Blood Pressure Regulation in a Patient with Parkinson’s Disease. Am. J. Hypertens. 2003, 16, 612–613. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, H.; Noda, K.; Mitsui, R.; Hashitani, H. Role of Enteric Dopaminergic Neurons in Regulating Peristalsis of Rat Proximal Colon. Neurogastroenterol. Motil. 2021, 33, e14127. [Google Scholar] [CrossRef]
- Bogdanov, V.; Kim, A.; Nodel, M.; Pavlenko, T.; Pavlova, E.; Blokhin, V.; Chesnokova, N.; Ugrumov, M. A Pilot Study of Changes in the Level of Catecholamines and the Activity of α-2-Macroglobulin in the Tear Fluid of Patients with Parkinson’s Disease and Parkinsonian Mice. Int. J. Mol. Sci. 2021, 22, 4736. [Google Scholar] [CrossRef]
- Stefanis, L. α-Synuclein in Parkinson’s Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef]
- Finberg, J.P.M. Inhibitors of MAO-B and COMT: Their Effects on Brain Dopamine Levels and Uses in Parkinson’s Disease. J. Neural Transm. 2019, 126, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Pinna, A. Adenosine A2A Receptor Antagonists in Parkinson’s Disease: Progress in Clinical Trials from the Newly Approved Istradefylline to Drugs in Early Development and Those Already Discontinued. CNS Drugs 2014, 28, 455–474. [Google Scholar] [CrossRef]
- Kulisevsky, J.; Poyurovsky, M. Adenosine A2A-Receptor Antagonism and Pathophysiology of Parkinson’s Disease and Drug-Induced Movement Disorders. Eur. Neurol. 2012, 67, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Pillay, V.; Choonara, Y.E. Advances in the Treatment of Parkinson’s Disease. Prog. Neurobiol. 2007, 81, 29–44. [Google Scholar] [CrossRef]
- Sengupta, T.; Vinayagam, J.; Singh, R.; Jaisankar, P.; Mohanakumar, K.P. Plant-Derived Natural Products for Parkinson’s Disease Therapy. In Advances in Neurobiology; Springer: New York, NY, USA, 2016; Volume 12, pp. 415–496. [Google Scholar]
- Rahman, M.M.; Wang, X.; Islam, M.R.; Akash, S.; Supti, F.A.; Mitu, M.I.; Harun-Or-Rashid, M.; Aktar, M.N.; Khatun Kali, M.S.; Jahan, F.I.; et al. Multifunctional Role of Natural Products for the Treatment of Parkinson’s Disease: At a Glance. Front. Pharmacol. 2022, 13, 976385. [Google Scholar] [CrossRef]
- Khattak, A.K.; Hassan, S.M.; Mughal, S.S. General Overview of Phytochemistry and Pharmacological Potential of Rheum Palmatum (Chinese Rhubarb). Innovare J. Ayurvedic Sci. 2020, 8, 5–9. [Google Scholar] [CrossRef]
- Tsai, P.-W.; Hsieh, C.-Y.; Ting, J.U.; Rogio, K.G.G.; Lee, C.-J.; De Castro-Cruz, K.A.; Ciou, Y.-R.; Lien, T.-K.; Yang, L.-L.; Hsueh, C.-C.; et al. Unraveling the Bioenergy Production and Electron Transport Characteristics of Processed Rheum palmatum L. for Antiviral Drug Development. Ind. Crops Prod. 2023, 195, 116488. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Lin, Y.-H.; Wu, Y.-C.; Hsueh, C.-C. Deciphering Electron-Shuttling Characteristics of Neurotransmitters to Stimulate Bioelectricity-Generating Capabilities in Microbial Fuel Cells. Appl. Biochem. Biotechnol. 2020, 191, 59–73. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Wu, Y.-C.; Lin, Y.-H.; Tayo, L.L.; Tacas, A.C.; Hsueh, C.-C. Deciphering Electron-Shuttling Characteristics of Parkinson’s Disease Medicines via Bioenergy Extraction in Microbial Fuel Cells. Ind. Eng. Chem. Res. 2020, 59, 17124–17136. [Google Scholar] [CrossRef]
- Mitra, S.; Anjum, J.; Muni, M.; Das, R.; Rauf, A.; Islam, F.; Bin Emran, T.; Semwal, P.; Hemeg, H.A.; Alhumaydhi, F.A.; et al. Exploring the Journey of Emodin as a Potential Neuroprotective Agent: Novel Therapeutic Insights with Molecular Mechanism of Action. Biomed. Pharmacother. 2022, 149, 112877. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Meng, J.; Jiang, K.; Lan, H.; Wang, Z.; Lin, M.; Li, W.; Guo, H.; Wei, Y.; Mu, Y. Improving Protein–Ligand Docking and Screening Accuracies by Incorporating a Scoring Function Correction Term. Brief. Bioinform. 2022, 23, bbac051. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, D.; Caballero, J. Is It Reliable to Take the Molecular Docking Top Scoring Position as the Best Solution without Considering Available Structural Data? Molecules 2018, 23, 1038. [Google Scholar] [CrossRef]
- Mor, D.E.; Ischiropoulos, H. The Convergence of Dopamine and α-Synuclein: Implications for Parkinson’s Disease. J. Exp. Neurosci. 2018, 12, 117906951876136. [Google Scholar] [CrossRef]
- Sousa, V.L.; Bellani, S.; Giannandrea, M.; Yousuf, M.; Valtorta, F.; Meldolesi, J.; Chieregatti, E. α-Synuclein and Its A30P Mutant Affect Actin Cytoskeletal Structure and Dynamics. Mol. Biol. Cell 2009, 20, 3725–3739. [Google Scholar] [CrossRef]
- Olanow, C.W.; Brundin, P. Parkinson’s Disease and Alpha Synuclein: Is Parkinson’s Disease a Prion-Like Disorder? Mov. Disord. 2013, 28, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, R.; Schapira, A.H.V. Parkinson Disease. Eur. J. Neurol. 2020, 27, 27–42. [Google Scholar] [CrossRef]
- Riederer, P.; Müller, T. Monoamine Oxidase-B Inhibitors in the Treatment of Parkinson’s Disease: Clinical–Pharmacological Aspects. J. Neural Transm. 2018, 125, 1751–1757. [Google Scholar] [CrossRef] [PubMed]
- Ferreira de Freitas, R.; Schapira, M. A Systematic Analysis of Atomic Protein–Ligand Interactions in the PDB. Med. Chem. Commun. 2017, 8, 1970–1981. [Google Scholar] [CrossRef]
- Pearson, R.G. Absolute Electronegativity and Hardness: Application to Inorganic Chemistry. Inorg. Chem. 1988, 27, 734–740. [Google Scholar] [CrossRef]
- Koopmans, T. Über Die Zuordnung von Wellenfunktionen Und Eigenwerten Zu Den Einzelnen Elektronen Eines Atoms. Physica 1934, 1, 104–113. [Google Scholar] [CrossRef]
- Janak, J.F. Proof That ∂E/∂ni = ε in Density-Functional Theory. Phys. Rev. B 1978, 18, 7165–7168. [Google Scholar] [CrossRef]
- Chandrakumar, K.; Pal, S. The Concept of Density Functional Theory Based Descriptors and Its Relation with the Reactivity of Molecular Systems: A Semi-Quantitative Study. Int. J. Mol. Sci. 2002, 3, 324–337. [Google Scholar] [CrossRef]
- Choudhary, V.; Bhatt, A.; Dash, D.; Sharma, N. DFT Calculations on Molecular Structures, HOMO–LUMO Study, Reactivity Descriptors and Spectral Analyses of Newly Synthesized Diorganotin(IV) 2-chloridophenylacetohydroxamate Complexes. J. Comput. Chem. 2019, 40, 2354–2363. [Google Scholar] [CrossRef]
- Gazquez, J.L.; Mendez, F. The Hard and Soft Acids and Bases Principle: An Atoms in Molecules Viewpoint. J. Phys. Chem. 1994, 98, 4591–4593. [Google Scholar] [CrossRef]
- Oliveri, V. Toward the Discovery and Development of Effective Modulators of α-Synuclein Amyloid Aggregation. Eur. J. Med. Chem. 2019, 167, 10–36. [Google Scholar] [CrossRef] [PubMed]
- Park, H.R.; Kim, J.; Kim, T.; Jo, S.; Yeom, M.; Moon, B.; Choo, I.H.; Lee, J.; Lim, E.J.; Park, K.D.; et al. Oxazolopyridines and Thiazolopyridines as Monoamine Oxidase B Inhibitors for the Treatment of Parkinson’s Disease. Bioorg. Med. Chem. 2013, 21, 5480–5487. [Google Scholar] [CrossRef] [PubMed]
- von Kleist, L.; Michaelis, S.; Bartho, K.; Graebner, O.; Schlief, M.; Dreger, M.; Schrey, A.K.; Sefkow, M.; Kroll, F.; Koester, H.; et al. Identification of Potential Off-Target Toxicity Liabilities of Catechol-O-Methyltransferase Inhibitors by Differential Competition Capture Compound Mass Spectrometry. J. Med. Chem. 2016, 59, 4664–4675. [Google Scholar] [CrossRef] [PubMed]
- Jörg, M.; Shonberg, J.; Mak, F.S.; Miller, N.D.; Yuriev, E.; Scammells, P.J.; Capuano, B. Novel Adenosine A2A Receptor Ligands: A Synthetic, Functional and Computational Investigation of Selected Literature Adenosine A2A Receptor Antagonists for Extending into Extracellular Space. Bioorg. Med. Chem. Lett. 2013, 23, 3427–3433. [Google Scholar] [CrossRef]
- Kong, L.D.; Cheng, C.H.K.; Tan, R.X. Inhibition of MAO A and B by Some Plant-Derived Alkaloids, Phenols and Anthraquinones. J. Ethnopharmacol. 2004, 91, 351–355. [Google Scholar] [CrossRef]
- Buchwald, P. Soft Drugs: Design Principles, Success Stories, and Future Perspectives. Expert Opin. Drug Metab. Toxicol. 2020, 16, 645–650. [Google Scholar] [CrossRef]
- Tian, H.; Ketkar, R.; Tao, P. ADMETboost: A Web Server for Accurate ADMET Prediction. J. Mol. Model. 2022, 28, 408. [Google Scholar] [CrossRef]
- Sakaeda, T.; Okamura, N.; Nagata, S.; Yagami, T.; Horinouichi, M.; Okumura, K.; Yamashita, F.; Hashida, M. Molecular and Pharmacokinetic Properties of 222 Commercially Available Oral Drugs in Humans. Biol. Pharm. Bull. 2001, 24, 935–940. [Google Scholar] [CrossRef]
- Gomtsyan, A. Heterocycles in Drugs and Drug Discovery. Chem. Heterocycl. Compd. 2012, 48, 7–10. [Google Scholar] [CrossRef]
- Keen, P. Effect of Binding to Plasma Proteins on the Distribution, Activity and Elimination of Drugs. In Concepts in Biochemical Pharmacology; Springer: Berlin/Heidelberg, Germany, 1971; pp. 213–233. [Google Scholar]
- Di, L. An Update on the Importance of Plasma Protein Binding in Drug Discovery and Development. Expert Opin. Drug Discov. 2021, 16, 1453–1465. [Google Scholar] [CrossRef]
- Chico, L.K.; Behanna, H.A.; Hu, W.; Zhong, G.; Roy, S.M.; Watterson, D.M. Molecular Properties and CYP2D6 Substrates: Central Nervous System Therapeutics Case Study and Pattern Analysis of a Substrate Database. Drug Metab. Dispos. 2009, 37, 2204–2211. [Google Scholar] [CrossRef]
- Dong, X.; Fu, J.; Yin, X.; Cao, S.; Li, X.; Lin, L.; Ni, J. Emodin: A Review of Its Pharmacology, Toxicity and Pharmacokinetics. Phytother. Res. 2016, 30, 1207–1218. [Google Scholar] [CrossRef]
- Ulmer, T.S.; Bax, A.; Cole, N.B.; Nussbaum, R.L. Structure and Dynamics of Micelle-Bound Human α-Synuclein. J. Biol. Chem. 2005, 280, 9595–9603. [Google Scholar] [CrossRef]
- Hubálek, F.; Binda, C.; Li, M.; Herzig, Y.; Sterling, J.; Youdim, M.B.H.; Mattevi, A.; Edmondson, D.E. Inactivation of Purified Human Recombinant Monoamine Oxidases A and B by Rasagiline and Its Analogues. J. Med. Chem. 2004, 47, 1760–1766. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Hubálek, F.; Li, M.; Herzig, Y.; Sterling, J.; Edmondson, D.E.; Mattevi, A. Binding of Rasagiline-Related Inhibitors to Human Monoamine Oxidases: A Kinetic and Crystallographic Analysis. J. Med. Chem. 2005, 48, 8148–8154. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Le Trong, I.; Stenkamp, R.E.; Parson, W.W. Crystal Structures of Human 108V and 108M Catechol O-Methyltransferase. J. Mol. Biol. 2008, 380, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, V.-P.; Griffith, M.T.; Hanson, M.A.; Cherezov, V.; Chien, E.Y.T.; Lane, J.R.; IJzerman, A.P.; Stevens, R.C. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science 2008, 322, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.V.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting Small-Molecule Pharmacokinetic and Toxicity Properties Using Graph-Based Signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
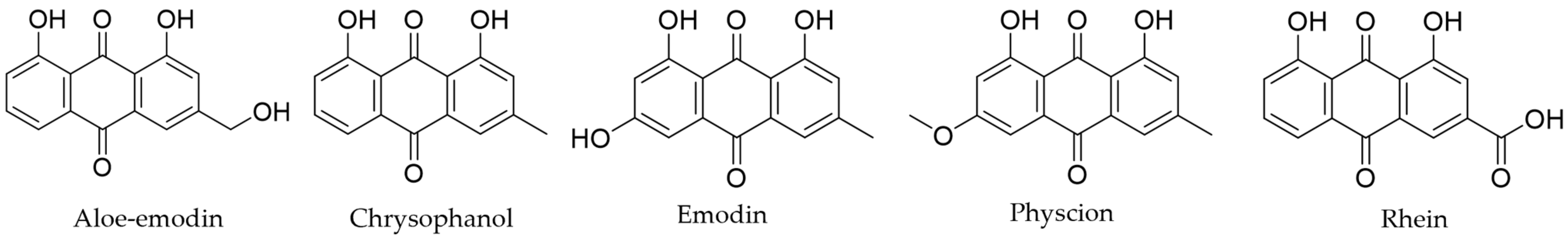
| Ligand | Form No. | ASN (PDB: 1XQ8) | MAOB (PDB: 2C65) | COMT (PDB: 3BWM) | A2AAR (PDB: 3EML) |
|---|---|---|---|---|---|
| Aloe-emodin | 1 | 83.2529 | 121.666 | 102.147 | 103.938 |
| 2 | 88.0637 | 125.89 | 122.628 | 119.698 | |
| 3 | 86.1153 | 122.002 | 108.446 | 106.68 | |
| 4 | 81.5402 | 121.551 | 101.605 | 103.695 | |
| 5 | 85.6552 | 121.304 | 115.675 | 107.195 | |
| 6 | 81.7519 | 122.844 | 101.408 | 104.847 | |
| Chrysophanol | 1 | 78.9452 | 114.483 | 92.5595 | 98.8463 |
| 2 | 79.5134 | 115.194 | 102.129 | 101.463 | |
| 3 | 77.4547 | 114.957 | 95.9588 | 100.238 | |
| 4 | 72.7176 | 115.208 | 93.4339 | 96.1831 | |
| 5 | 72.0571 | 114.407 | 93.139 | 98.3591 | |
| 6 | 64.9091 | 115.13 | 93.61 | 96.686 | |
| Emodin | 1 | 81.1783 | 120.25 | 96.7097 | 97.0337 |
| 2 | 84.4642 | 119.622 | 101.456 | 100.972 | |
| 3 | 83.4505 | 121.64 | 104.623 | 101.864 | |
| 4 | 82.0401 | 119.959 | 89.9023 | 95.9239 | |
| 5 | 83.5858 | 119.78 | 101.984 | 99.3968 | |
| Physcion | 1 | 84.0608 | 124.603 | 98.9597 | 102.614 |
| 2 | 88.5065 | 128.092 | 116.342 | 114.673 | |
| 3 | 86.15 | 128.415 | 118.163 | 105.413 | |
| 4 | 83.3815 | 125.267 | 97.6397 | 100.39 | |
| 5 | 88.3428 | 124.502 | 113.259 | 118.168 | |
| 6 | 83.8676 | 125.486 | 89.3911 | 99.4669 | |
| Rhein | 1 | 86.5327 | 123.458 | 106.972 | 106.412 |
| 2 | 84.0283 | 124.371 | 119.125 | 108.289 | |
| 3 | 87.7195 | 125.19 | 107.756 | 109.683 | |
| 4 | 84.7186 | 123.958 | 94.2238 | 106.914 | |
| 5 | 86.7861 | 124.038 | 98.3459 | 108.707 | |
| 6 | 82.2617 | 124.203 | 96.3977 | 106.131 | |
| Dopamine 1 | 65.7182 | 85.4606 | 89.5598 | 77.5231 | |
| Levodopa 1 | 75.4623 | 103.043 | 110.127 | 91.2493 |
| Ligand | ASN (PDB: 1XQ8) | MAOB (PDB: 2C65) | COMT (PDB: 3BWM) | A2AAR (PDB: 3EML) |
|---|---|---|---|---|
| Aloe-emodin | −10.9777 | −3.33321 | −12.2531 | — |
| Chrysophanol | −9.65713 | −39.7554 | −10.606 | −15.9783 |
| Emodin | −34.4961 | −44.9155 | −32.431 | −57.5427 |
| Physcion | −10.5583 | −30.3292 | −15.7917 | — |
| Rhein | −29.9042 | −22.7501 | −14.9567 | −39.4557 |
| Dopamine 1 | −31.0394 | −30.2512 | −36.1948 | −39.3845 |
| Levodopa 1 | −49.7879 | −53.7193 | −46.8662 | −61.8193 |
| Protein | Ligand | εHOMO | εLUMO | Δε | η | µ | ω |
|---|---|---|---|---|---|---|---|
| ASN | Emodin | −5.2390 | −2.7854 | 2.4536 | 1.2268 | −4.0122 | 6.5608 |
| MAOB | Chrysophanol | −5.0907 | −2.4882 | 2.6025 | 1.3013 | −3.7895 | 5.5178 |
| Emodin | −5.1769 | −2.8436 | 2.3333 | 1.1667 | −4.0102 | 6.8924 | |
| Physcion | −6.3604 | −3.3253 | 3.0351 | 1.5175 | −4.8429 | 7.7275 | |
| COMT | Emodin | −5.1784 | −2.8472 | 2.3313 | 1.1656 | −4.0128 | 6.9073 |
| A2AAR | Emodin | −5.1753 | −2.8598 | 2.3155 | 1.1578 | −4.0175 | 6.9706 |
| Rhein | −6.0684 | −3.3462 | 2.7222 | 1.3611 | −4.7073 | 8.1400 | |
| Dopamine 1 | −5.1946 | −2.7709 | 2.4237 | 1.2119 | −3.9828 | 6.5447 | |
| Levodopa 1 | −4.5353 | −1.8210 | 2.7143 | 1.3571 | −3.1782 | 3.7213 |
| Parameter | ASN | MAOB | COMT | A2AAR |
|---|---|---|---|---|
| N | 36 | 112 | 84 | 96 |
| pIC50 range | 3.897–6.721 | 4.311–9.511 | 3.886–8.700 | 4.26–11.721 |
| IC50 range (µM) | 0.190–126.77 | 0.0003–48.87 | 0.002–130.02 | 1.9 × 10−6–75.86 |
| Internal validation | ||||
| r | 0.971 | 0.974 | 0.975 | 0.969 |
| r2 | 0.942 | 0.948 | 0.951 | 0.939 |
| r2adj | 0.941 | 0.946 | 0.950 | 0.938 |
| RMS residual error | 0.183 | 0.249 | 0.243 | 0.245 |
| Cross-validation | ||||
| q2 | 0.225 | 0.488 | 0.303 | 0.400 |
| RMS residual error | 0.674 | 0.781 | 0.927 | 0.772 |
| External validation | ||||
| q2 | 0.387 | 0.458 | 0.433 | 0.175 |
| RMS error | 0.925 | 1.073 | 0.787 | 0.917 |
| Mean absolute error | 0.821 | 0.901 | 0.620 | 0.805 |
| Ligand | ASN | MAOB | COMT | A2AAR |
|---|---|---|---|---|
| Aloe-emodin | 26.1921 | 0.2404 | 0.3297 | 1.5034 |
| Chrysophanol | 21.4724 | 0.0880 | 0.7712 | 0.8162 |
| Emodin | 15.9566 | 0.1277 | 0.7888 | 2.8790 |
| Physcion | 20.8521 | 1.2842 | 0.8107 | 23.0096 |
| Rhein | 18.4570 | 0.4140 | 0.7221 | 4.1237 |
| ALO | CHR | EMO | PHY | RHE | DOP | LDP | |
|---|---|---|---|---|---|---|---|
| Physicochemical Properties | |||||||
| Molecular weight | 270.24 | 254.24 | 270.24 | 284.27 | 284.22 | 153.18 | 197.19 |
| Hydrogen bond acceptors | 5 | 4 | 5 | 5 | 5 | 3 | 4 |
| Hydrogen bond donors | 3 | 2 | 3 | 2 | 3 | 3 | 4 |
| Rotational bonds | 1 | 0 | 0 | 1 | 1 | 2 | 3 |
| TPSA (Å2) | 94.83 | 74.6 | 94.83 | 83.83 | 111.9 | 66.48 | 103.78 |
| log KO/W | 1.37 | 2.18 | 1.89 | 2.19 | 1.57 | 0.60 | 0.05 |
| Lipinski rule | (+) | (+) | (+) | (+) | (+) | (+) | (+) |
| Pfizer rule | (+) | (−) | (+) | (+) | (+) | (+) | (+) |
| GSK rule | (+) | (−) | (+) | (−) | (+) | (+) | (+) |
| Golden triangle | (+) | (+) | (+) | (+) | (+) | (−) | (−) |
| Absorption | |||||||
| C2P (log cm/s) | −5.30 | −5.06 | −5.25 | −5.11 | −5.37 | −5.33 | −5.34 |
| HIA (%) | 73.81 | 73.93 | 73.93 | 73.93 | 73.81 | 73.58 | 73.99 |
| log D7.4 | 1.77 | 1.93 | 1.86 | 1.98 | 1.89 | 1.51 | 1.43 |
| log S (log mol/L) | −4.79 | −5.00 | −4.79 | −5.09 | −4.72 | −4.24 | −4.35 |
| Oral bioavailability (%) | 42.60 | 42.67 | 41.87 | 45.12 | 43.18 | 41.13 | 48.56 |
| Distribution | |||||||
| BBB penetration (%) | 29.99 | 33.86 | 29.47 | 31.65 | 33.29 | 29.16 | 30.49 |
| PPBR (%) | 42.67 | 48.22 | 45.28 | 45.46 | 54.58 | 39.48 | 50.56 |
| Metabolism | |||||||
| CYP2C9 inhibitor (%) | 56.55 | 58.07 | 63.77 | 58.02 | 54.56 | 47.99 | 47.19 |
| CYP2C9 substrate * (%) | 35.14 | 34.41 | 34.33 | 36.50 | 37.90 | 28.59 | 36.27 |
| CYP2D6 inhibitor (%) | 83.88 | 92.34 | 91.24 | 88.36 | 86.22 | 89.63 | 91.19 |
| CYP2D6 substrate * (%) | 56.44 | 56.88 | 57.39 | 58.14 | 55.49 | 51.70 | 59.00 |
| CYP3A4 inhibitor (%) | 31.48 | 32.59 | 33.51 | 40.29 | 30.01 | 33.23 | 34.31 |
| CYP3A4 substrate * (%) | 34.94 | 33.79 | 36.14 | 37.18 | 35.42 | 40.46 | 38.97 |
| Excretion | |||||||
| Half-life * (h) | 68.11 | 67.24 | 67.41 | 67.56 | 67.78 | 39.82 | 54.73 |
| HPC * (uL/min/106 cells) | 36.10 | 32.75 | 36.66 | 36.18 | 35.07 | 45.91 | 48.32 |
| MSC * (mL/min/g−1) | 36.05 | 34.16 | 35.58 | 41.62 | 35.93 | 30.30 | 27.97 |
| Toxicity | |||||||
| hERG blockers (%) | 35.28 | 35.97 | 37.25 | 37.52 | 36.65 | 32.27 | 32.43 |
| AMES toxicity (%) | 43.30 | 44.46 | 43.93 | 42.63 | 42.30 | 41.60 | 40.60 |
| DILI (%) | 46.46 | 42.09 | 51.68 | 46.05 | 50.41 | 43.00 | 40.09 |
| ROALD50 (mmol/kg) | 2.82 | 2.34 | 6.92 | 5.75 | 4.90 | 29.51 | 15.14 |
| hMTD (mg/kg/day) | 1.23 | 1.80 | 0.70 | 1.80 | 0.19 | 0.19 | 0.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, P.J.B.; Huang, S.K.-H.; De Castro-Cruz, K.A.; Leron, R.B.; Tsai, P.-W. In Silico Neuroprotective Effects of Specific Rheum palmatum Metabolites on Parkinson’s Disease Targets. Int. J. Mol. Sci. 2023, 24, 13929. https://doi.org/10.3390/ijms241813929
Garcia PJB, Huang SK-H, De Castro-Cruz KA, Leron RB, Tsai P-W. In Silico Neuroprotective Effects of Specific Rheum palmatum Metabolites on Parkinson’s Disease Targets. International Journal of Molecular Sciences. 2023; 24(18):13929. https://doi.org/10.3390/ijms241813929
Chicago/Turabian StyleGarcia, Patrick Jay B., Steven Kuan-Hua Huang, Kathlia A. De Castro-Cruz, Rhoda B. Leron, and Po-Wei Tsai. 2023. "In Silico Neuroprotective Effects of Specific Rheum palmatum Metabolites on Parkinson’s Disease Targets" International Journal of Molecular Sciences 24, no. 18: 13929. https://doi.org/10.3390/ijms241813929
APA StyleGarcia, P. J. B., Huang, S. K.-H., De Castro-Cruz, K. A., Leron, R. B., & Tsai, P.-W. (2023). In Silico Neuroprotective Effects of Specific Rheum palmatum Metabolites on Parkinson’s Disease Targets. International Journal of Molecular Sciences, 24(18), 13929. https://doi.org/10.3390/ijms241813929







