Defining the Basal and Immunomodulatory Mediator-Induced Phosphoprotein Signature in Pediatric B Cell Acute Lymphoblastic Leukemia (B-ALL) Diagnostic Samples
Abstract
1. Introduction
2. Results
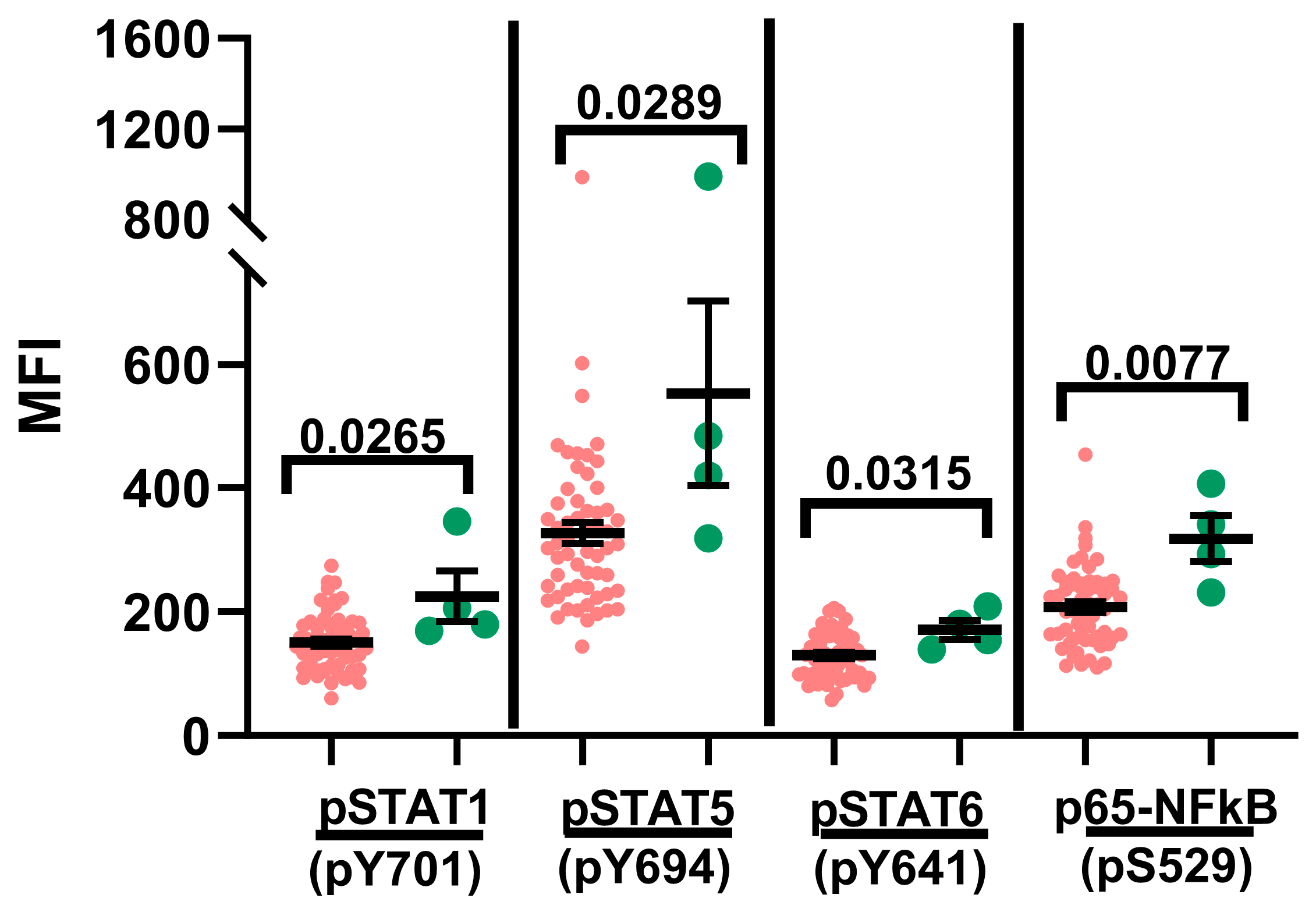
2.1. Basal Hyperphosphorylation in Leukemic Blasts
2.2. Phosphoprotein Signature of Pediatric B-ALL Blasts Treated with Immunomodulatory Mediators
2.3. Cytokine Receptor Expression Profile
2.3.1. IL-7Rα (CD127) and the Common γ Chain (CD132)
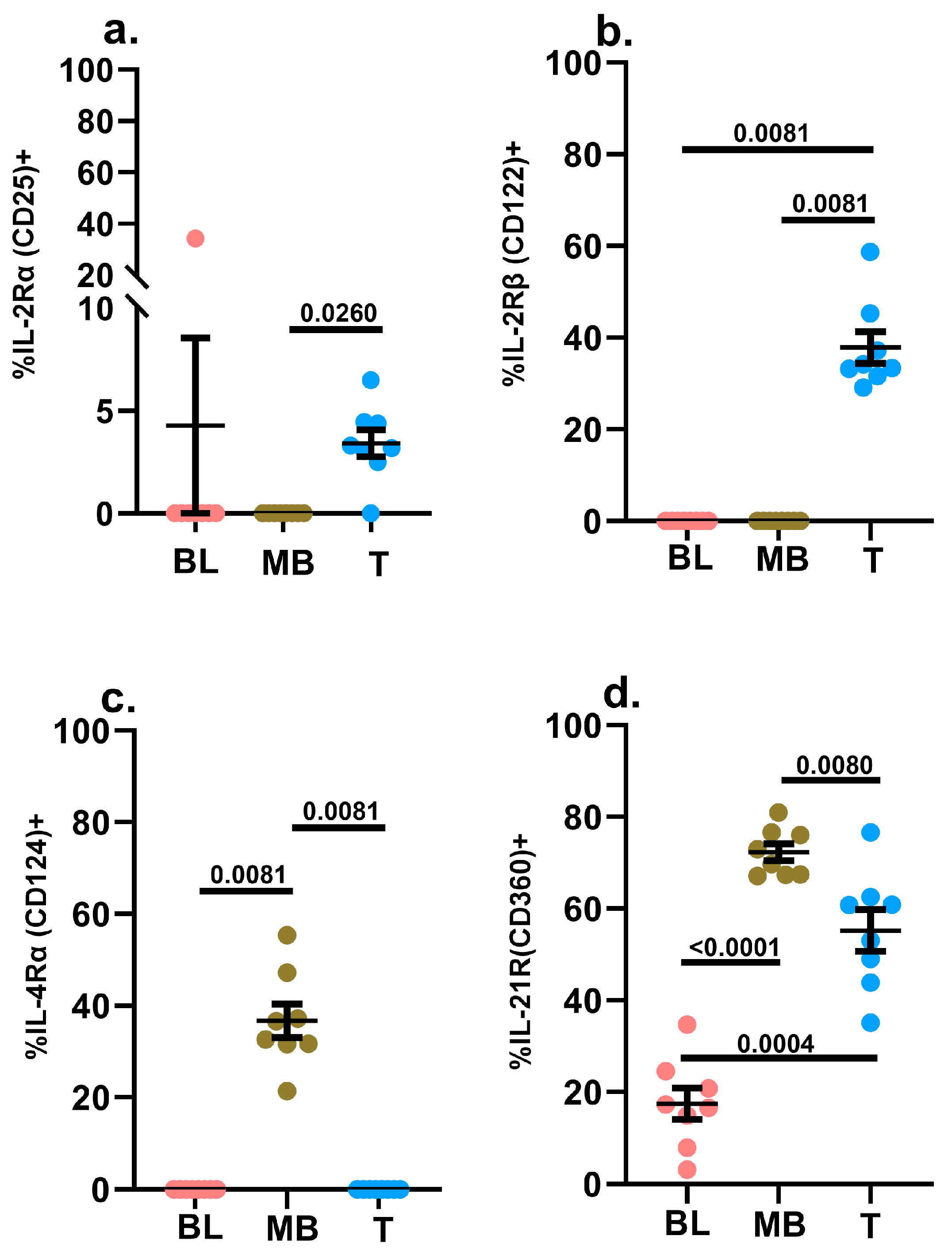
2.3.2. IL-2Rα (CD25) and IL-2Rβ (CD122)
2.3.3. IL-4Rα (CD124)
2.3.4. IL-21R (CD360)
2.3.5. IFNγR1 (CD119) and IFNγR2
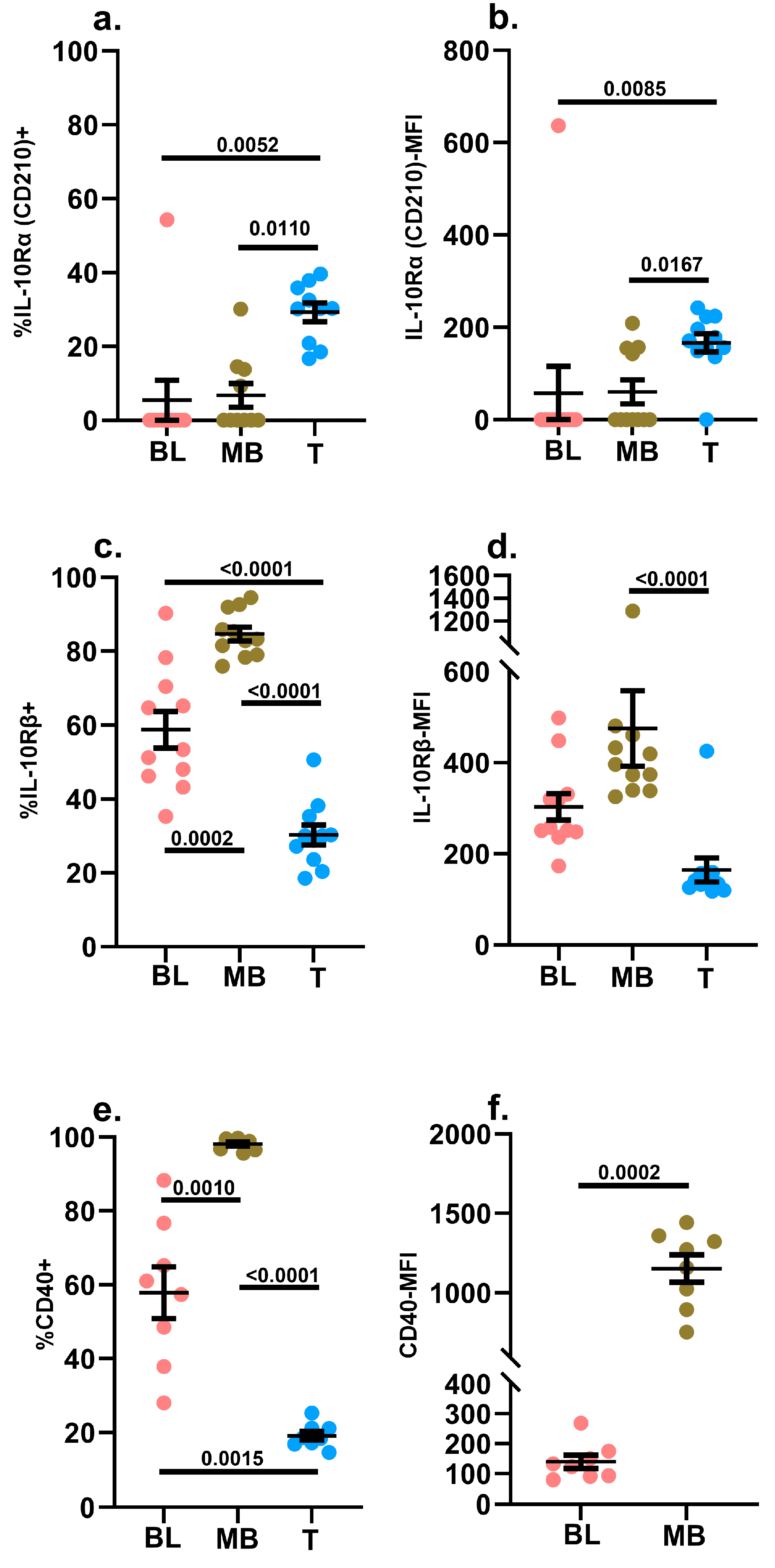
2.3.6. IL-10Rα (CD210) and IL-10Rβ
2.3.7. CD40
2.4. Phosphoprotein Signature and Overall Survival Analyses
3. Discussion
4. Materials and Methods
4.1. Study Subjects
4.2. Phosflow
4.3. Cell Surface Receptor Expression
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balkwill, F.; Mantovani, A. Inflammation and cancer: Back to Virchow? Lancet 2001, 357, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Shalapour, S.; Karin, M. Immunity, inflammation, and cancer: An eternal fight between good and evil. J. Clin. Investig. 2015, 125, 3347–3355. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Trinchieri, G. Cancer and inflammation: An old intuition with rapidly evolving new concepts. Annu. Rev. Immunol. 2012, 30, 677–706. [Google Scholar] [CrossRef]
- Karin, M. Nuclear factor-kappaB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef]
- Chavez, J.S.; Rabe, J.L.; Loeffler, D.; Higa, K.C.; Hernandez, G.; Mills, T.S.; Ahmed, N.; Gessner, R.L.; Ke, Z.; Idler, B.M.; et al. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J. Exp. Med. 2021, 218, e20201169. [Google Scholar] [CrossRef]
- Higa, K.C.; Goodspeed, A.; Chavez, J.S.; De Dominici, M.; Danis, E.; Zaberezhnyy, V.; Rabe, J.L.; Tenen, D.G.; Pietras, E.M.; DeGregori, J. Chronic interleukin-1 exposure triggers selection for Cebpa-knockout multipotent hematopoietic progenitors. J. Exp. Med. 2021, 218, e20200560. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Recher, C. Clinical Implications of Inflammation in Acute Myeloid Leukemia. Front. Oncol. 2021, 11, 623952. [Google Scholar] [CrossRef] [PubMed]
- Vilchis-Ordonez, A.; Contreras-Quiroz, A.; Vadillo, E.; Dorantes-Acosta, E.; Reyes-Lopez, A.; Quintela-Nunez del Prado, H.M.; Venegas-Vazquez, J.; Mayani, H.; Ortiz-Navarrete, V.; Lopez-Martinez, B.; et al. Bone Marrow Cells in Acute Lymphoblastic Leukemia Create a Proinflammatory Microenvironment Influencing Normal Hematopoietic Differentiation Fates. Biomed. Res. Int. 2015, 2015, 386165. [Google Scholar] [CrossRef]
- Rothwell, P.M.; Fowkes, F.G.; Belch, J.F.; Ogawa, H.; Warlow, C.P.; Meade, T.W. Effect of daily aspirin on long-term risk of death due to cancer: Analysis of individual patient data from randomised trials. Lancet 2011, 377, 31–41. [Google Scholar] [CrossRef]
- Rothwell, P.M.; Wilson, M.; Price, J.F.; Belch, J.F.; Meade, T.W.; Mehta, Z. Effect of daily aspirin on risk of cancer metastasis: A study of incident cancers during randomised controlled trials. Lancet 2012, 379, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; MacFadyen, J.G.; Thuren, T.; Everett, B.M.; Libby, P.; Glynn, R.J.; Group, C.T. Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: Exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 2017, 390, 1833–1842. [Google Scholar] [CrossRef]
- Mantovani, A.; Ponzetta, A.; Inforzato, A.; Jaillon, S. Innate immunity, inflammation and tumour progression: Double-edged swords. J. Intern. Med. 2019, 285, 524–532. [Google Scholar] [CrossRef]
- Andriani, G.A.; Almeida, V.P.; Faggioli, F.; Mauro, M.; Tsai, W.L.; Santambrogio, L.; Maslov, A.; Gadina, M.; Campisi, J.; Vijg, J.; et al. Whole Chromosome Instability induces senescence and promotes SASP. Sci. Rep. 2016, 6, 35218. [Google Scholar] [CrossRef]
- Komarova, E.A.; Krivokrysenko, V.; Wang, K.; Neznanov, N.; Chernov, M.V.; Komarov, P.G.; Brennan, M.L.; Golovkina, T.V.; Rokhlin, O.W.; Kuprash, D.V.; et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005, 19, 1030–1032. [Google Scholar] [CrossRef]
- Schwitalla, S.; Ziegler, P.K.; Horst, D.; Becker, V.; Kerle, I.; Begus-Nahrmann, Y.; Lechel, A.; Rudolph, K.L.; Langer, R.; Slotta-Huspenina, J.; et al. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 2013, 23, 93–106. [Google Scholar] [CrossRef]
- Liao, W.; Overman, M.J.; Boutin, A.T.; Shang, X.; Zhao, D.; Dey, P.; Li, J.; Wang, G.; Lan, Z.; Li, J.; et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell 2019, 35, 559–572.e7. [Google Scholar] [CrossRef] [PubMed]
- Davalos, A.R.; Coppe, J.P.; Campisi, J.; Desprez, P.Y. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010, 29, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Kortlever, R.M.; Sodir, N.M.; Wilson, C.H.; Burkhart, D.L.; Pellegrinet, L.; Brown Swigart, L.; Littlewood, T.D.; Evan, G.I. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell 2017, 171, 1301–1315.e14. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.P.; Mullighan, C.G. Redefining ALL classification: Toward detecting high-risk ALL and implementing precision medicine. Blood 2015, 125, 3977–3987. [Google Scholar] [CrossRef]
- Hauer, J.; Fischer, U.; Borkhardt, A. Toward prevention of childhood ALL by early-life immune training. Blood 2021, 138, 1412–1428. [Google Scholar] [CrossRef]
- Wiemels, J.L.; Cazzaniga, G.; Daniotti, M.; Eden, O.B.; Addison, G.M.; Masera, G.; Saha, V.; Biondi, A.; Greaves, M.F. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet 1999, 354, 1499–1503. [Google Scholar] [CrossRef]
- Greaves, M.F.; Wiemels, J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer 2003, 3, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Vrooman, L.M.; Blonquist, T.M.; Harris, M.H.; Stevenson, K.E.; Place, A.E.; Hunt, S.K.; O’Brien, J.E.; Asselin, B.L.; Athale, U.H.; Clavell, L.A.; et al. Refining risk classification in childhood B acute lymphoblastic leukemia: Results of DFCI ALL Consortium Protocol 05-001. Blood Adv. 2018, 2, 1449–1458. [Google Scholar] [CrossRef]
- Kinlen, L. Evidence for an infective cause of childhood leukaemia: Comparison of a Scottish new town with nuclear reprocessing sites in Britain. Lancet 1988, 2, 1323–1327. [Google Scholar] [CrossRef]
- Greaves, M.F. Speculations on the cause of childhood acute lymphoblastic leukemia. Leukemia 1988, 2, 120–125. [Google Scholar] [PubMed]
- Richardson, R.B. Promotional etiology for common childhood acute lymphoblastic leukemia: The infective lymphoid recovery hypothesis. Leuk. Res. 2011, 35, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.A.; Simon, R.; Strickler, H.D.; McQuillan, G.; Ries, L.A.; Linet, M.S. Evidence that childhood acute lymphoblastic leukemia is associated with an infectious agent linked to hygiene conditions. Cancer Causes Control 1998, 9, 285–298. [Google Scholar] [CrossRef] [PubMed]
- He, J.R.; Ramakrishnan, R.; Hirst, J.E.; Bonaventure, A.; Francis, S.S.; Paltiel, O.; Haberg, S.E.; Lemeshow, S.; Olsen, S.; Tikellis, G.; et al. Maternal Infection in Pregnancy and Childhood Leukemia: A Systematic Review and Meta-analysis. J. Pediatr. 2020, 217, 98–109.e8. [Google Scholar] [CrossRef] [PubMed]
- Schmiegelow, K.; Vestergaard, T.; Nielsen, S.M.; Hjalgrim, H. Etiology of common childhood acute lymphoblastic leukemia: The adrenal hypothesis. Leukemia 2008, 22, 2137–2141. [Google Scholar] [CrossRef]
- Chang, J.S.; Zhou, M.; Buffler, P.A.; Chokkalingam, A.P.; Metayer, C.; Wiemels, J.L. Profound deficit of IL10 at birth in children who develop childhood acute lymphoblastic leukemia. Cancer Epidemiol. Biomarkers Prev. 2011, 20, 1736–1740. [Google Scholar] [CrossRef]
- Ouyang, W.; O’Garra, A. IL-10 Family Cytokines IL-10 and IL-22: From Basic Science to Clinical Translation. Immunity 2019, 50, 871–891. [Google Scholar] [CrossRef]
- Soegaard, S.H.; Rostgaard, K.; Skogstrand, K.; Wiemels, J.L.; Schmiegelow, K.; Hjalgrim, H. Neonatal Inflammatory Markers Are Associated with Childhood B-cell Precursor Acute Lymphoblastic Leukemia. Cancer Res. 2018, 78, 5458–5463. [Google Scholar] [CrossRef]
- Cloppenborg, T.; Stanulla, M.; Zimmermann, M.; Schrappe, M.; Welte, K.; Klein, C. Immunosurveillance of childhood ALL: Polymorphic interferon-gamma alleles are associated with age at diagnosis and clinical risk groups. Leukemia 2005, 19, 44–48. [Google Scholar] [CrossRef]
- Wu, S.; Gessner, R.; von Stackelberg, A.; Kirchner, R.; Henze, G.; Seeger, K. Cytokine/cytokine receptor gene expression in childhood acute lymphoblastic leukemia: Correlation of expression and clinical outcome at first disease recurrence. Cancer 2005, 103, 1054–1063. [Google Scholar] [CrossRef]
- Belardelli, F.; Ferrantini, M. Cytokines as a link between innate and adaptive antitumor immunity. Trends Immunol. 2002, 23, 201–208. [Google Scholar] [CrossRef]
- Takatsu, K. Cytokines involved in B-cell differentiation and their sites of action. Proc. Soc. Exp. Biol. Med. 1997, 215, 121–133. [Google Scholar] [CrossRef]
- Acosta-Rodriguez, E.V.; Merino, M.C.; Montes, C.L.; Motran, C.C.; Gruppi, A. Cytokines and chemokines shaping the B-cell compartment. Cytokine Growth Factor. Rev. 2007, 18, 73–83. [Google Scholar] [CrossRef]
- Cyster, J.G.; Allen, C.D.C. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 2019, 177, 524–540. [Google Scholar] [CrossRef]
- Dong, C. Cytokine Regulation and Function in T Cells. Annu. Rev. Immunol. 2021, 39, 51–76. [Google Scholar] [CrossRef]
- Zhu, J.; Paul, W.E. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol. Rev. 2010, 238, 247–262. [Google Scholar] [CrossRef]
- Elgueta, R.; Benson, M.J.; de Vries, V.C.; Wasiuk, A.; Guo, Y.; Noelle, R.J. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol. Rev. 2009, 229, 152–172. [Google Scholar] [CrossRef]
- Khanolkar, A.; Muller, W.J.; Simpson, B.M.; Cerullo, J.; Williams, R.; Sowers, S.B.; Matthews, K.; Mercader, S.; Hickman, C.J.; D’Aquila, R.T.; et al. Preservation of lymphocyte functional fitness in perinatally-infected and treated HIV+ pediatric patients displaying sub-optimal viral control. Commun. Med. 2022, 2, 1–15. [Google Scholar] [CrossRef]
- Roifman, C.M. 35—Primary T-Cell Immunodeficiencies. In Clinical Immunology, 5th ed.; Rich, R.R., Fleisher, T.A., Shearer, W.T., Schroeder, H.W., Frew, A.J., Weyand, C.M., Eds.; Elsevier: London, UK, 2019; pp. 489–508.e1. [Google Scholar]
- Khanolkar, A.; Wilks, J.D.; Jennings, L.J.; Davies, J.L.; Zollett, J.A.; Lott, L.L.; Fullmer, E.R.; Bensen, N.E.; Carlson-Leuer, K.M.; Tse, W.T.; et al. Signaling impairments in maternal T cells engrafted in an infant with a novel IL-2 receptor gamma mutation. J. Allergy Clin. Immunol. 2015, 135, 1093–1096.e8. [Google Scholar] [CrossRef]
- Khanolkar, A.; Kirschmann, D.A.; Caparelli, E.A.; Wilks, J.D.; Cerullo, J.M.; Bergerson, J.R.E.; Jennings, L.J.; Fuleihan, R.L. CD4 T cell-restricted IL-2 signaling defect in a patient with a novel IFNGR1 deficiency. J. Allergy Clin. Immunol. 2018, 141, 435–439.e7. [Google Scholar] [CrossRef]
- Spolski, R.; Leonard, W.J. Interleukin-21: A double-edged sword with therapeutic potential. Nat. Rev. Drug Discov. 2014, 13, 379–395. [Google Scholar] [CrossRef]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Saraiva, M.; Vieira, P.; O’Garra, A. Biology and therapeutic potential of interleukin-10. J. Exp. Med. 2020, 217, e20190418. [Google Scholar] [CrossRef]
- Chen, D.; Ireland, S.J.; Remington, G.; Alvarez, E.; Racke, M.K.; Greenberg, B.; Frohman, E.M.; Monson, N.L. CD40-Mediated NF-kappaB Activation in B Cells Is Increased in Multiple Sclerosis and Modulated by Therapeutics. J. Immunol. 2016, 197, 4257–4265. [Google Scholar] [CrossRef]
- Shochat, C.; Tal, N.; Bandapalli, O.R.; Palmi, C.; Ganmore, I.; te Kronnie, G.; Cario, G.; Cazzaniga, G.; Kulozik, A.E.; Stanulla, M.; et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J. Exp. Med. 2011, 208, 901–908. [Google Scholar] [CrossRef]
- Hertzberg, L.; Vendramini, E.; Ganmore, I.; Cazzaniga, G.; Schmitz, M.; Chalker, J.; Shiloh, R.; Iacobucci, I.; Shochat, C.; Zeligson, S.; et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: A report from the International BFM Study Group. Blood 2010, 115, 1006–1017. [Google Scholar] [CrossRef]
- Barata, J.T.; Durum, S.K.; Seddon, B. Flip the coin: IL-7 and IL-7R in health and disease. Nat. Immunol. 2019, 20, 1584–1593. [Google Scholar] [CrossRef]
- Dias, S.; Silva, H., Jr.; Cumano, A.; Vieira, P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 2005, 201, 971–979. [Google Scholar] [CrossRef]
- Clark, M.R.; Mandal, M.; Ochiai, K.; Singh, H. Orchestrating B cell lymphopoiesis through interplay of IL-7 receptor and pre-B cell receptor signalling. Nat. Rev. Immunol. 2014, 14, 69–80. [Google Scholar] [CrossRef]
- Spolski, R.; Leonard, W.J. The Yin and Yang of interleukin-21 in allergy, autoimmunity and cancer. Curr. Opin. Immunol. 2008, 20, 295–301. [Google Scholar] [CrossRef]
- Spolski, R.; Leonard, W.J. IL-21 is an immune activator that also mediates suppression via IL-10. Crit. Rev. Immunol. 2010, 30, 559–570. [Google Scholar] [CrossRef]
- Deenick, E.K.; Avery, D.T.; Chan, A.; Berglund, L.J.; Ives, M.L.; Moens, L.; Stoddard, J.L.; Bustamante, J.; Boisson-Dupuis, S.; Tsumura, M.; et al. Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J. Exp. Med. 2013, 210, 2739–2753. [Google Scholar] [CrossRef]
- Lindner, S.; Dahlke, K.; Sontheimer, K.; Hagn, M.; Kaltenmeier, C.; Barth, T.F.; Beyer, T.; Reister, F.; Fabricius, D.; Lotfi, R.; et al. Interleukin 21-induced granzyme B-expressing B cells infiltrate tumors and regulate T cells. Cancer Res. 2013, 73, 2468–2479. [Google Scholar] [CrossRef]
- Hagn, M.; Schwesinger, E.; Ebel, V.; Sontheimer, K.; Maier, J.; Beyer, T.; Syrovets, T.; Laumonnier, Y.; Fabricius, D.; Simmet, T.; et al. Human B cells secrete granzyme B when recognizing viral antigens in the context of the acute phase cytokine IL-21. J. Immunol. 2009, 183, 1838–1845. [Google Scholar] [CrossRef]
- Bhatt, S.; Sarosiek, K.A.; Lossos, I.S. Interleukin 21—Its potential role in the therapy of B-cell lymphomas. Leuk. Lymphoma 2017, 58, 17–29. [Google Scholar] [CrossRef]
- Zaidi, M.R. The Interferon-Gamma Paradox in Cancer. J. Interferon Cytokine Res. 2019, 39, 30–38. [Google Scholar] [CrossRef]
- Haan, C.; Kreis, S.; Margue, C.; Behrmann, I. Jaks and cytokine receptors—An intimate relationship. Biochem. Pharmacol. 2006, 72, 1538–1546. [Google Scholar] [CrossRef]
- Vonderheide, R.H. CD40 Agonist Antibodies in Cancer Immunotherapy. Annu. Rev. Med. 2020, 71, 47–58. [Google Scholar] [CrossRef]
- Troeger, A.; Glouchkova, L.; Ackermann, B.; Escherich, G.; Meisel, R.; Hanenberg, H.; den Boer, M.L.; Pieters, R.; Janka-Schaub, G.E.; Goebel, U.; et al. High expression of CD40 on B-cell precursor acute lymphoblastic leukemia blasts is an independent risk factor associated with improved survival and enhanced capacity to up-regulate the death receptor CD95. Blood 2008, 112, 1028–1034. [Google Scholar] [CrossRef]
- Law, C.L.; Wormann, B.; LeBien, T.W. Analysis of expression and function of CD40 on normal and leukemic human B cell precursors. Leukemia 1990, 4, 732–738. [Google Scholar]
- Uckun, F.M.; Gajl-Peczalska, K.; Myers, D.E.; Jaszcz, W.; Haissig, S.; Ledbetter, J.A. Temporal association of CD40 antigen expression with discrete stages of human B-cell ontogeny and the efficacy of anti-CD40 immunotoxins against clonogenic B-lineage acute lymphoblastic leukemia as well as B-lineage non-Hodgkin’s lymphoma cells. Blood 1990, 76, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- Saeland, S.; Duvert, V.; Caux, C.; Pandrau, D.; Favre, C.; Valle, A.; Durand, I.; Charbord, P.; de Vries, J.; Banchereau, J. Distribution of surface-membrane molecules on bone marrow and cord blood CD34+ hematopoietic cells. Exp. Hematol. 1992, 20, 24–33. [Google Scholar] [PubMed]
- Ranheim, E.A.; Kipps, T.J. Activated T cells induce expression of B7/BB1 on normal or leukemic B cells through a CD40-dependent signal. J. Exp. Med. 1993, 177, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Yellin, M.J.; Sinning, J.; Covey, L.R.; Sherman, W.; Lee, J.J.; Glickman-Nir, E.; Sippel, K.C.; Rogers, J.; Cleary, A.M.; Parker, M.; et al. T lymphocyte T cell-B cell-activating molecule/CD40-L molecules induce normal B cells or chronic lymphocytic leukemia B cells to express CD80 (B7/BB-1) and enhance their costimulatory activity. J. Immunol. 1994, 153, 666–674. [Google Scholar] [CrossRef]
- Van den Hove, L.E.; Van Gool, S.W.; Vandenberghe, P.; Bakkus, M.; Thielemans, K.; Boogaerts, M.A.; Ceuppens, J.L. CD40 triggering of chronic lymphocytic leukemia B cells results in efficient alloantigen presentation and cytotoxic T lymphocyte induction by up-regulation of CD80 and CD86 costimulatory molecules. Leukemia 1997, 11, 572–580. [Google Scholar] [CrossRef][Green Version]
- Buhmann, R.; Nolte, A.; Westhaus, D.; Emmerich, B.; Hallek, M. CD40-activated B-cell chronic lymphocytic leukemia cells for tumor immunotherapy: Stimulation of allogeneic versus autologous T cells generates different types of effector cells. Blood 1999, 93, 1992–2002. [Google Scholar] [CrossRef]
- Trentin, L.; Perin, A.; Siviero, M.; Piazza, F.; Facco, M.; Gurrieri, C.; Galvan, S.; Adami, F.; Agostini, C.; Pizzolo, G.; et al. B7 costimulatory molecules from malignant cells in patients with b-cell chronic lymphoproliferative disorders trigger t-cell proliferation. Cancer 2000, 89, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; LaPlant, B.; Uy, G.L.; Marcucci, G.; Blum, W.; Larson, R.A.; Stone, R.M.; Mandrekar, S.J. Evaluation of event-free survival as a robust end point in untreated acute myeloid leukemia (Alliance A151614). Blood Adv. 2019, 3, 1714–1721. [Google Scholar] [CrossRef]
- Schultz, K.R.; Pullen, D.J.; Sather, H.N.; Shuster, J.J.; Devidas, M.; Borowitz, M.J.; Carroll, A.J.; Heerema, N.A.; Rubnitz, J.E.; Loh, M.L.; et al. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: A combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood 2007, 109, 926–935. [Google Scholar] [CrossRef]
- Burgler, S.; Nadal, D. Pediatric precursor B acute lymphoblastic leukemia: Are T helper cells the missing link in the infectious etiology theory? Mol. Cell Pediatr. 2017, 4, 6. [Google Scholar] [CrossRef]
- Zaidi, M.R.; Merlino, G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011, 17, 6118–6124. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Algarra, I.; Garcia-Lora, A.; Cabrera, T.; Ruiz-Cabello, F.; Garrido, F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: Implications for tumor immune escape. Cancer Immunol. Immunother. 2004, 53, 904–910. [Google Scholar] [CrossRef]
- Gobin, S.J.; van den Elsen, P.J. Transcriptional regulation of the MHC class Ib genes HLA-E, HLA-F, and HLA-G. Hum. Immunol. 2000, 61, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Wischhusen, J.; Waschbisch, A.; Wiendl, H. Immune-refractory cancers and their little helpers—An extended role for immunetolerogenic MHC molecules HLA-G and HLA-E? Semin. Cancer Biol. 2007, 17, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Schurch, C.; Riether, C.; Amrein, M.A.; Ochsenbein, A.F. Cytotoxic T cells induce proliferation of chronic myeloid leukemia stem cells by secreting interferon-gamma. J. Exp. Med. 2013, 210, 605–621. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S.; Sinha, P. Myeloid-derived suppressor cells: Linking inflammation and cancer. J. Immunol. 2009, 182, 4499–4506. [Google Scholar] [CrossRef]
- Mo, X.; Zhang, H.; Preston, S.; Martin, K.; Zhou, B.; Vadalia, N.; Gamero, A.M.; Soboloff, J.; Tempera, I.; Zaidi, M.R. Interferon-gamma Signaling in Melanocytes and Melanoma Cells Regulates Expression of CTLA-4. Cancer Res. 2018, 78, 436–450. [Google Scholar] [CrossRef]
- Abiko, K.; Matsumura, N.; Hamanishi, J.; Horikawa, N.; Murakami, R.; Yamaguchi, K.; Yoshioka, Y.; Baba, T.; Konishi, I.; Mandai, M. IFN-gamma from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br. J. Cancer 2015, 112, 1501–1509. [Google Scholar] [CrossRef]
- Bellucci, R.; Martin, A.; Bommarito, D.; Wang, K.; Hansen, S.H.; Freeman, G.J.; Ritz, J. Interferon-gamma-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015, 4, e1008824. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Benci, J.L.; Xu, B.; Qiu, Y.; Wu, T.J.; Dada, H.; Twyman-Saint Victor, C.; Cucolo, L.; Lee, D.S.M.; Pauken, K.E.; Huang, A.C.; et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016, 167, 1540–1554.e12. [Google Scholar] [CrossRef] [PubMed]
- Blank, C.U.; Haining, W.N.; Held, W.; Hogan, P.G.; Kallies, A.; Lugli, E.; Lynn, R.C.; Philip, M.; Rao, A.; Restifo, N.P.; et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019, 19, 665–674. [Google Scholar] [CrossRef] [PubMed]
- McLane, L.M.; Abdel-Hakeem, M.S.; Wherry, E.J. CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu. Rev. Immunol. 2019, 37, 457–495. [Google Scholar] [CrossRef]
- Chihara, N.; Madi, A.; Kondo, T.; Zhang, H.; Acharya, N.; Singer, M.; Nyman, J.; Marjanovic, N.D.; Kowalczyk, M.S.; Wang, C.; et al. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 2018, 558, 454–459. [Google Scholar] [CrossRef]
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wolfel, T.; Holzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Young, H.A.; Klinman, D.M.; Reynolds, D.A.; Grzegorzewski, K.J.; Nii, A.; Ward, J.M.; Winkler-Pickett, R.T.; Ortaldo, J.R.; Kenny, J.J.; Komschlies, K.L. Bone marrow and thymus expression of interferon-gamma results in severe B-cell lineage reduction, T-cell lineage alterations, and hematopoietic progenitor deficiencies. Blood 1997, 89, 583–595. [Google Scholar] [CrossRef]
- Wang, Z.; Bunting, K.D. STAT5 activation in B-cell acute lymphoblastic leukemia: Damned if you do, damned if you don’t. Cancer Cell Microenviron. 2016, 3, e1186. [Google Scholar] [CrossRef][Green Version]
- Dai, X.; Chen, Y.; Di, L.; Podd, A.; Li, G.; Bunting, K.D.; Hennighausen, L.; Wen, R.; Wang, D. Stat5 is essential for early B cell development but not for B cell maturation and function. J. Immunol. 2007, 179, 1068–1079. [Google Scholar] [CrossRef]
- Benekli, M.; Baer, M.R.; Baumann, H.; Wetzler, M. Signal transducer and activator of transcription proteins in leukemias. Blood 2003, 101, 2940–2954. [Google Scholar] [CrossRef]
- Katerndahl, C.D.S.; Heltemes-Harris, L.M.; Willette, M.J.L.; Henzler, C.M.; Frietze, S.; Yang, R.; Schjerven, H.; Silverstein, K.A.T.; Ramsey, L.B.; Hubbard, G.; et al. Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival. Nat. Immunol. 2017, 18, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Heltemes-Harris, L.M.; Willette, M.J.; Ramsey, L.B.; Qiu, Y.H.; Neeley, E.S.; Zhang, N.; Thomas, D.A.; Koeuth, T.; Baechler, E.C.; Kornblau, S.M.; et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J. Exp. Med. 2011, 208, 1135–1149. [Google Scholar] [CrossRef]
- Almeida, A.R.M.; Neto, J.L.; Cachucho, A.; Euzebio, M.; Meng, X.; Kim, R.; Fernandes, M.B.; Raposo, B.; Oliveira, M.L.; Ribeiro, D.; et al. Interleukin-7 receptor alpha mutational activation can initiate precursor B-cell acute lymphoblastic leukemia. Nat. Commun. 2021, 12, 7268. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.; Almeida, A.R.M.; Cachucho, A.; Neto, J.L.; Demeyer, S.; de Matos, M.; Hogan, T.; Li, Y.; Meijerink, J.; Cools, J.; et al. Overexpression of wild-type IL-7Ralpha promotes T-cell acute lymphoblastic leukemia/lymphoma. Blood 2021, 138, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Fitch, B.A.; Zhou, M.; Situ, J.; Surianarayanan, S.; Reeves, M.Q.; Hermiston, M.L.; Wiemels, J.L.; Kogan, S.C. Decreased IL-10 accelerates B-cell leukemia/lymphoma in a mouse model of pediatric lymphoid leukemia. Blood Adv. 2022, 6, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Tsuruoka, N.; Arima, M.; Arguni, E.; Saito, T.; Kitayama, D.; Sakamoto, A.; Hatano, M.; Tokuhisa, T. Bcl6 is required for the IL-4-mediated rescue of the B cells from apoptosis induced by IL-21. Immunol. Lett. 2007, 110, 145–151. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Samavati, L.; Rastogi, R.; Du, W.; Huttemann, M.; Fite, A.; Franchi, L. STAT3 tyrosine phosphorylation is critical for interleukin 1 beta and interleukin-6 production in response to lipopolysaccharide and live bacteria. Mol. Immunol. 2009, 46, 1867–1877. [Google Scholar] [CrossRef]
- Bronte-Tinkew, D.M.; Terebiznik, M.; Franco, A.; Ang, M.; Ahn, D.; Mimuro, H.; Sasakawa, C.; Ropeleski, M.J.; Peek, R.M., Jr.; Jones, N.L. Helicobacter pylori cytotoxin-associated gene A activates the signal transducer and activator of transcription 3 pathway in vitro and in vivo. Cancer Res. 2009, 69, 632–639. [Google Scholar] [CrossRef]
- Choudhari, S.R.; Khan, M.A.; Harris, G.; Picker, D.; Jacob, G.S.; Block, T.; Shailubhai, K. Deactivation of Akt and STAT3 signaling promotes apoptosis, inhibits proliferation, and enhances the sensitivity of hepatocellular carcinoma cells to an anticancer agent, Atiprimod. Mol. Cancer Ther. 2007, 6, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Steinberg, B.M. PTEN is a negative regulator of STAT3 activation in human papillomavirus-infected cells. J. Gen. Virol. 2002, 83 Pt 7, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Muromoto, R.; Ikeda, O.; Okabe, K.; Togi, S.; Kamitani, S.; Fujimuro, M.; Harada, S.; Oritani, K.; Matsuda, T. Epstein-Barr virus-derived EBNA2 regulates STAT3 activation. Biochem. Biophys. Res. Commun. 2009, 378, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Kortylewski, M.; Kujawski, M.; Herrmann, A.; Yang, C.; Wang, L.; Liu, Y.; Salcedo, R.; Yu, H. Toll-like receptor 9 activation of signal transducer and activator of transcription 3 constrains its agonist-based immunotherapy. Cancer Res. 2009, 69, 2497–2505. [Google Scholar] [CrossRef]
- Chiron, D.; Bekeredjian-Ding, I.; Pellat-Deceunynck, C.; Bataille, R.; Jego, G. Toll-like receptors: Lessons to learn from normal and malignant human B cells. Blood 2008, 112, 2205–2213. [Google Scholar] [CrossRef]
- Migone, T.S.; Lin, J.X.; Cereseto, A.; Mulloy, J.C.; O’Shea, J.J.; Franchini, G.; Leonard, W.J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science 1995, 269, 79–81. [Google Scholar] [CrossRef]
- Kortylewski, M.; Kujawski, M.; Wang, T.; Wei, S.; Zhang, S.; Pilon-Thomas, S.; Niu, G.; Kay, H.; Mule, J.; Kerr, W.G.; et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005, 11, 1314–1321. [Google Scholar] [CrossRef]
- Lee, H.; Herrmann, A.; Deng, J.H.; Kujawski, M.; Niu, G.; Li, Z.; Forman, S.; Jove, R.; Pardoll, D.M.; Yu, H. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell 2009, 15, 283–293. [Google Scholar] [CrossRef]
- Kujawski, M.; Kortylewski, M.; Lee, H.; Herrmann, A.; Kay, H.; Yu, H. Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J. Clin. Investig. 2008, 118, 3367–3377. [Google Scholar] [CrossRef]
- Kortylewski, M.; Xin, H.; Kujawski, M.; Lee, H.; Liu, Y.; Harris, T.; Drake, C.; Pardoll, D.; Yu, H. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 2009, 15, 114–123. [Google Scholar] [CrossRef]
- Wang, L.; Yi, T.; Kortylewski, M.; Pardoll, D.M.; Zeng, D.; Yu, H. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J. Exp. Med. 2009, 206, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jove, R. The STATs of cancer--new molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Kortylewski, M.; Pardoll, D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007, 7, 41–51. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, H.W.; Cuenca, A.; Huang, M.; Ghansah, T.; Brayer, J.; Kerr, W.G.; Takeda, K.; Akira, S.; Schoenberger, S.P.; et al. A critical role for Stat3 signaling in immune tolerance. Immunity 2003, 19, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Corzo, C.A.; Luetteke, N.; Yu, B.; Nagaraj, S.; Bui, M.M.; Ortiz, M.; Nacken, W.; Sorg, C.; Vogl, T.; et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J. Exp. Med. 2008, 205, 2235–2249. [Google Scholar] [CrossRef]
- Ho, H.H.; Ivashkiv, L.B. Role of STAT3 in type I interferon responses. Negative regulation of STAT1-dependent inflammatory gene activation. J. Biol. Chem. 2006, 281, 14111–14118. [Google Scholar] [CrossRef]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D.; et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef]
- Catlett-Falcone, R.; Landowski, T.H.; Oshiro, M.M.; Turkson, J.; Levitzki, A.; Savino, R.; Ciliberto, G.; Moscinski, L.; Fernandez-Luna, J.L.; Nunez, G.; et al. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity 1999, 10, 105–115. [Google Scholar] [CrossRef]
- Grivennikov, S.; Karin, E.; Terzic, J.; Mucida, D.; Yu, G.Y.; Vallabhapurapu, S.; Scheller, J.; Rose-John, S.; Cheroutre, H.; Eckmann, L.; et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Shain, K.H.; Yarde, D.N.; Meads, M.B.; Huang, M.; Jove, R.; Hazlehurst, L.A.; Dalton, W.S. Beta1 integrin adhesion enhances IL-6-mediated STAT3 signaling in myeloma cells: Implications for microenvironment influence on tumor survival and proliferation. Cancer Res. 2009, 69, 1009–1015. [Google Scholar] [CrossRef]
- Karin, M.; Greten, F.R. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005, 5, 749–759. [Google Scholar] [CrossRef]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef]
- Baud, V.; Karin, M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 2009, 8, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. Stat3 as an oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Cao, Y.; Greten, F.R.; Li, Z.W. NF-kappaB in cancer: From innocent bystander to major culprit. Nat. Rev. Cancer 2002, 2, 301–310. [Google Scholar] [CrossRef]
- Ghosh, S.; May, M.J.; Kopp, E.B. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998, 16, 225–260. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.F.; Greene, W.C. Shaping the nuclear action of NF-kappaB. Nat. Rev. Mol. Cell Biol. 2004, 5, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.M.; Myklebust, J.H.; Alizadeh, A.A.; Houot, R.; Sharman, J.P.; Czerwinski, D.K.; Nolan, G.P.; Levy, R. B-cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 12747–12754. [Google Scholar] [CrossRef]
- Wu, S.; Jin, L.; Vence, L.; Radvanyi, L.G. Development and application of ‘phosphoflow’ as a tool for immunomonitoring. Expert. Rev. Vaccines 2010, 9, 631–643. [Google Scholar] [CrossRef]
- McKenna, R.W.; Asplund, S.L.; Kroft, S.H. Immunophenotypic analysis of hematogones (B-lymphocyte precursors) and neoplastic lymphoblasts by 4-color flow cytometry. Leuk. Lymphoma 2004, 45, 277–285. [Google Scholar] [CrossRef]
- McKenna, R.W.; Washington, L.T.; Aquino, D.B.; Picker, L.J.; Kroft, S.H. Immunophenotypic analysis of hematogones (B-lymphocyte precursors) in 662 consecutive bone marrow specimens by 4-color flow cytometry. Blood 2001, 98, 2498–2507. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, D.W.; Colovai, A.I.; Emmons, F.N.; Bhagat, G.; Alobeid, B. Hematogones: A review and update. Leuk. Lymphoma 2010, 51, 10–19. [Google Scholar] [CrossRef]
- Lucio, P.; Parreira, A.; van den Beemd, M.W.; van Lochem, E.G.; van Wering, E.R.; Baars, E.; Porwit-MacDonald, A.; Bjorklund, E.; Gaipa, G.; Biondi, A.; et al. Flow cytometric analysis of normal B cell differentiation: A frame of reference for the detection of minimal residual disease in precursor-B-ALL. Leukemia 1999, 13, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Sedek, L.; Bulsa, J.; Sonsala, A.; Twardoch, M.; Wieczorek, M.; Malinowska, I.; Derwich, K.; Niedzwiecki, M.; Sobol-Milejska, G.; Kowalczyk, J.R.; et al. The immunophenotypes of blast cells in B-cell precursor acute lymphoblastic leukemia: How different are they from their normal counterparts? Cytometry B Clin. Cytom. 2014, 86, 329–339. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khanolkar, A.; Liu, G.; Simpson Schneider, B.M. Defining the Basal and Immunomodulatory Mediator-Induced Phosphoprotein Signature in Pediatric B Cell Acute Lymphoblastic Leukemia (B-ALL) Diagnostic Samples. Int. J. Mol. Sci. 2023, 24, 13937. https://doi.org/10.3390/ijms241813937
Khanolkar A, Liu G, Simpson Schneider BM. Defining the Basal and Immunomodulatory Mediator-Induced Phosphoprotein Signature in Pediatric B Cell Acute Lymphoblastic Leukemia (B-ALL) Diagnostic Samples. International Journal of Molecular Sciences. 2023; 24(18):13937. https://doi.org/10.3390/ijms241813937
Chicago/Turabian StyleKhanolkar, Aaruni, Guorong Liu, and Bridget M. Simpson Schneider. 2023. "Defining the Basal and Immunomodulatory Mediator-Induced Phosphoprotein Signature in Pediatric B Cell Acute Lymphoblastic Leukemia (B-ALL) Diagnostic Samples" International Journal of Molecular Sciences 24, no. 18: 13937. https://doi.org/10.3390/ijms241813937
APA StyleKhanolkar, A., Liu, G., & Simpson Schneider, B. M. (2023). Defining the Basal and Immunomodulatory Mediator-Induced Phosphoprotein Signature in Pediatric B Cell Acute Lymphoblastic Leukemia (B-ALL) Diagnostic Samples. International Journal of Molecular Sciences, 24(18), 13937. https://doi.org/10.3390/ijms241813937




