Photonics of Some Monomethine Cyanine Dyes in Solutions and in Complexes with Biomolecules
Abstract
1. Introduction
2. Results and Discussion
2.1. Solvatofluorochromism of MCD 1–4 in Organic Solvents and Aqueous Medium
2.2. Influence of Biopolymers on Spectral Fluorescent Properties of Cyanine Dyes
2.3. Molecular Docking of MCD 1–4 with dsDNA and HSA
2.4. Thermal Dissociation of dsDNA in the Presence of MCD 1–4
2.5. Study of the Interaction of MCD 1–4 with HSA by the Method of Synchronous Fluorescence Scanning
2.6. Binding Constants, Limits of Detection/Quantification
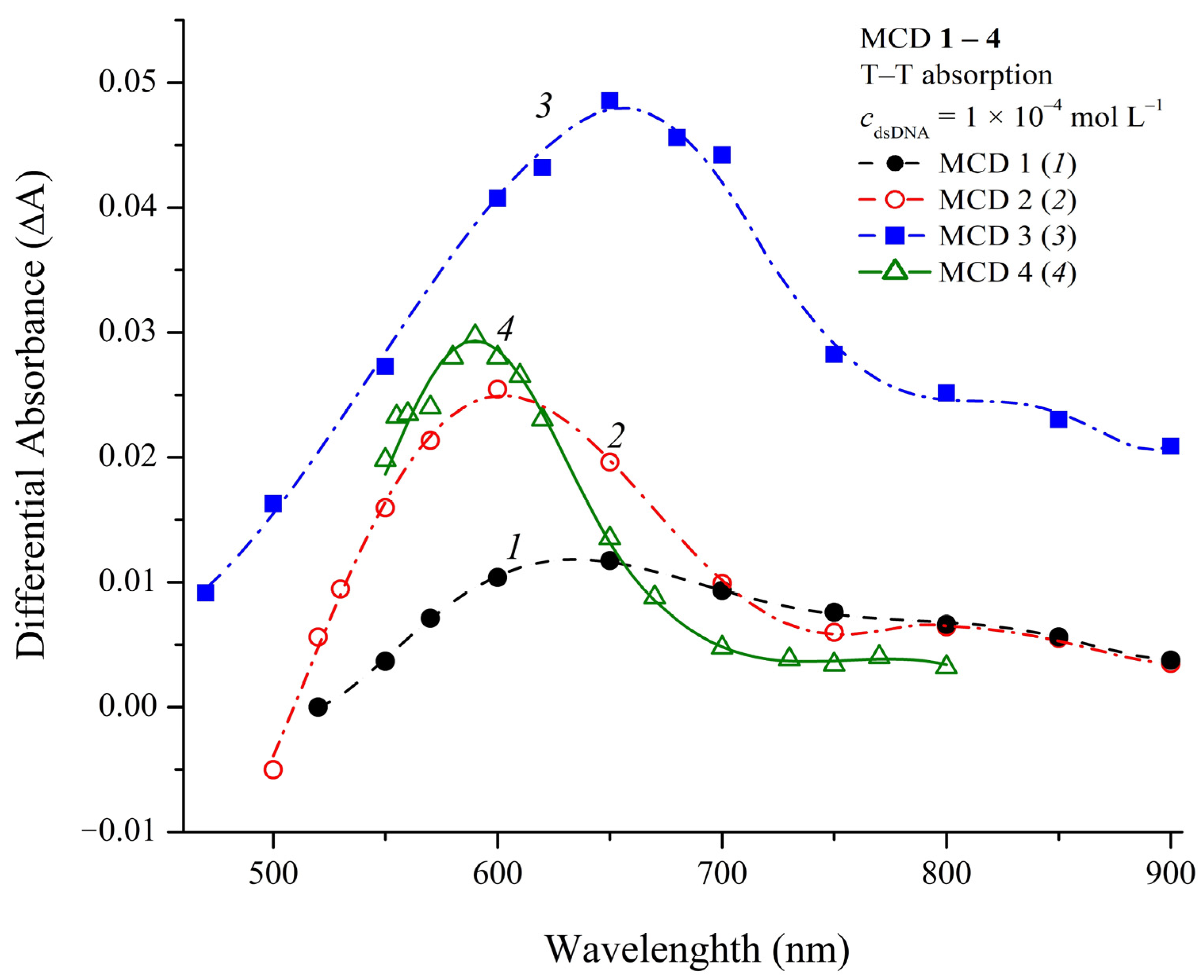
2.7. Photochemical Properties of MCD 1–4 in the Presence of DNA
3. Materials and Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peng, C.-L.; Shih, Y.-H.; Chiang, P.-F.; Chen, C.-T.; Chang, M.-C. Multifunctional cyanine-based theranostic probe for cancer imaging and therapy. Int. J. Mol. Sci. 2021, 22, 12214. [Google Scholar] [CrossRef]
- Schirripa Spagnolo, C.; Luin, S. Choosing the probe for single-molecule fluorescence microscopy. Int. J. Mol. Sci. 2022, 23, 14949. [Google Scholar] [CrossRef] [PubMed]
- Le Guern, F.; Mussard, V.; Gaucher, A.; Rottman, M.; Prim, D. Fluorescein derivatives as fluorescent probes for pH monitoring along recent biological applications. Int. J. Mol. Sci. 2020, 21, 9217. [Google Scholar] [CrossRef] [PubMed]
- Fluorescent Probes. ThermoFisher Scientific. Available online: https://www.thermofisher.com/ru/ru/home/life-science/protein-biology/protein-biology-learning-center/protein-biology-resource-library/pierce-protein-methods/fluorescent-probes.html#3 (accessed on 2 August 2023).
- Tatikolov, A.S. Polymethine dyes as spectral-fluorescent probes for biomacromolecules. J. Photochem. Photobiol. C 2012, 13, 55–90. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Tatikolov, A.S. Photonics of trimethine cyanine dyes as probes for biomolecules. Molecules 2022, 27, 6367. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.G.; Chen, C.H.; Chiu, L.A. Thiazole orange: A new dye for reticulocyte analysis. Cytometry 1986, 7, 508–517. [Google Scholar] [CrossRef]
- Larsson, A.; Carlsson, C.; Jonsson, M. Characterization of the binding of YO to [poly(dA-dT)]2 and [poly(dG-dC)]2, and of the fluorescent properties of YO and YOYO complexed with the polynucleotides and double-stranded DNA. Biopolymers 1995, 36, 153–167. [Google Scholar] [CrossRef]
- SYBR Products for Real Time PCR & Nucleic Acid Staining. ThermoFisher Scientific. Available online: https://www.thermofisher.com/ro/en/home/brands/product-brand/sybr.html (accessed on 2 August 2023).
- Dragan, A.I.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. Characterization of PicoGreen interaction with dsDNA and the origin of its fluorescence enhancement upon binding. Biophys. J. 2010, 99, 3010–3019. [Google Scholar] [CrossRef]
- Wang, Y.; Schellenberg, H.; Walhorn, V.; Toensing, K.; Anselmetti, D. Binding mechanism of PicoGreen to DNA characterized by magnetic tweezers and fluorescence spectroscopy. Eur. Biophys. J. 2017, 46, 561–566. [Google Scholar] [CrossRef]
- Tse, C.; Capeau, J. Quantification des acides nucléiques par PCR quantitative en temps réel [Real time PCR methodology for quantification of nucleic acids]. Ann. Biol. Clin. 2003, 61, 279–293. [Google Scholar]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, J.; Pingoud, A.; Hahn, M. Real-time PCR-based method for the estimation of genome sizes. Nucl. Acids Res. 2003, 31, e56. [Google Scholar] [CrossRef] [PubMed]
- ThermoFisher Scientific. Real-Time PCR Reagents and Kits. Available online: https://www.thermofisher.com/ru/ru/home/life-science/pcr/real-time-pcr.html (accessed on 28 August 2023).
- Vus, K.; Tarabara, U.; Balklava, Z.; Nerukh, D.; Stich, M.; Laguta, A.; Vodolazkaya, N.; Mchedlov-Petrossyan, N.O.; Farafonov, V.; Kriklya, N.; et al. Association of novel monomethine cyanine dyes with bacteriophage MS2: A fluorescence study. J. Mol. Liq. 2020, 302, 112569. [Google Scholar] [CrossRef]
- Ishkitiev, N.; Miteva, M.; Micheva, M.; Stoyanova, T.; Lozanova, V.V.; Lozanov, V.S.; Mihaylova, Z.; Cheshmedzhieva, D.V.; Kandinska, M.; Rangelov, M.; et al. Aggregation induced nucleic acids recognition by homodimeric asymmetric monomethyne cyanine fluorochromes in mesenchymal stem cells. Int. J. Biol. Macromol. 2023, 250, 126094. [Google Scholar] [CrossRef]
- Kurutos, A.; Balabanov, I.; Kamounah, F.S.; Nikolova-Ganeva, K.; Borisova, D.; Gadjev, N.; Deligeorgiev, T.; Tchorbanov, A. Bright green-emitting ds-DNA labeling employed by dicationic monomethine cyanine dyes: Apoptosis assay and fluorescent bio-imaging. Dye. Pigment. 2018, 157, 267–277. [Google Scholar] [CrossRef]
- Yarmoluk, S.M.; Lukashov, S.S.; Ogul’chansky, T.Y.; Losytskyy, M.Y.; Kornyushyna, O.S. Interaction of cyanine dyes with nucleic acids. XXI. Arguments for half-intercalation model of interaction. Biopolym. Orig. Res. Biomol. 2001, 62, 219–227. [Google Scholar] [CrossRef]
- Ogul’chansky, T.Y.; Losytskyy, M.Y.; Kovalska, V.B.; Yashchuk, V.M.; Yarmoluk, S.M. Interactions of cyanine dyes with nucleic acids. XXIV. Aggregation of monomethine cyanine dyes in presence of DNA and its manifestation in absorption and fluorescence spectra. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2001, 57, 1525–1532. [Google Scholar] [CrossRef]
- Deligeorgiev, T.; Vasilev, A.; Drexhage, K.-H. Synthesis of novel monomeric cyanine dyes containing 2-hydroxypropyl and 3-chloro-2-hydroxypropyl substituents–noncovalent labels for nucleic acids. Dye. Pigment. 2007, 73, 69–75. [Google Scholar] [CrossRef]
- Deligeorgiev, T.; Vasilev, A.; Tsvetkova, T.; Drexhage, K.-H. Synthesis of novel monomeric asymmetric tri- and tetracationic monomethine cyanine dyes as fluorescent non-covalent nucleic acid labels. Dye. Pigment. 2007, 75, 658–663. [Google Scholar] [CrossRef]
- Deligeorgiev, T.; Vasilev, A.; Drexhage, K.-H. Synthesis of novel cyanine dyes containing carbamoylethyl component—Noncovalent labels for nucleic acids detection. Dye. Pigment. 2007, 74, 320–328. [Google Scholar] [CrossRef]
- Saarnio, V.K.; Salorinne, K.; Ruokolainen, V.P.; Nilsson, J.R.; Tero, T.-R.; Oikarinen, S.; Wilhelmsson, L.M.; Lahtinen, T.M.; Marjomäki, V.S. Development of functionalized SYBR green II related cyanine dyes for viral RNA detection. Dye. Pigment. 2020, 177, 108282. [Google Scholar] [CrossRef]
- Kurutos, A.; Nikodinovic-Runic, J.; Veselinovic, A.; Veselinović, J.B.; Kamounahd, F.S.; Ilic-Tomic, T. RNA-targeting low-molecular-weight fluorophores for nucleoli staining: Synthesis, in silico modelling and cellular imaging. New J. Chem. 2021, 45, 12818–12829. [Google Scholar] [CrossRef]
- Aristova, D.; Kosach, V.; Chernii, S.; Slominsky, Y.; Balanda, A.; Filonenko, V.; Yarmoluk, S.; Rotaru, A.; Özkan, H.G.; Mokhir, A.; et al. Monomethine cyanine probes for visualization of cellular RNA by fluorescence microscopy. Methods Appl. Fluor. 2021, 9, 045002. [Google Scholar] [CrossRef]
- Soriano, E.; Holder, C.; Levitz, A.; Henary, M. Benz[c,d]indolium-containing monomethine cyanine dyes: Synthesis and photophysical properties. Molecules 2016, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Kurutos, A.; Ilic-Tomic, T.; Kamounah, F.S.; Vasilev, A.A.; Nikodinovic-Runic, J. Non-cytotoxic photostable monomethine cyanine platforms: Combined paradigm of nucleic acid staining and in vivo imaging. J. Photochem. Photobiol. A Chem. 2020, 397, 112598. [Google Scholar] [CrossRef]
- Kurutos, A.; Orehovec, I.; Tomašić Paić, A.; Crnolatac, I.; Horvat, L.; Gadjev, N.; Piantanida, I.; Deligeorgiev, T. New series of non-toxic DNA intercalators, mitochondria targeting fluorescent dyes. Dye. Pigment. 2018, 148, 452–459. [Google Scholar] [CrossRef]
- Fei, X.; Gu, Y.; Ban, Y.; Liu, Z.; Zhang, B. Thiazole Orange derivatives: Synthesis, fluorescence properties, and labeling cancer cells. Bioorg. Med. Chem. 2009, 17, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Fabijanić, I.; Kurutos, A.; Tomašić Paić, A.; Tadić, V.; Kamounah, F.S.; Horvat, L.; Brozovic, A.; Crnolatac, I.; Radić Stojković, M. Selenium-substituted monomethine cyanine dyes as selective G-quadruplex spectroscopic probes with theranostic potential. Biomolecules 2023, 13, 128. [Google Scholar] [CrossRef] [PubMed]
- Deligeorgiev, T.G.; Gadjev, N.I.; Timtcheva, I.I.; Maximova, V.A.; Katerinopoulos, H.E.; Foukaraki, E. Synthesis of homodimeric monomethine cyanine dyes as noncovalent nucleic acid labels and their absorption and fluorescence spectral characteristics. Dye. Pigment. 2000, 44, 131–136. [Google Scholar] [CrossRef]
- Deligeorgiev, T.; Timtcheva, I.; Maximova, V.; Gadjev, N.; Drexhage, K.-H. Fluorescence characteristics of variously charged asymmetric monomethine cyanine dyes in the presence of nucleic acids. J. Fluoresc. 2002, 12, 225–229. [Google Scholar] [CrossRef]
- Karlson, H.J.; Bergqvist, M.H.; Lincoln, P.; Westman, G. Syntheses and DNA-binding studies of a series of unsymmetrical cyanine dyes: Structural influence on the degree of minor groove binding to natural DNA. Bioorg. Med. Chem. 2004, 12, 2369–2384. [Google Scholar] [CrossRef] [PubMed]
- Åberg, U.; Åkesson, E.; Sundström, V. Excited state dynamics of barrierless isomerization in solution. Chem. Phys. Lett. 1993, 215, 388–394. [Google Scholar] [CrossRef]
- Åberg, U.; Åkesson, E.; Alvarez, J.-L.; Fedchenia, I.; Sundström, V. Femtosecond spectral evolution monitoring the bond-twisting event in barrierless isomerization in solution. Chem. Phys. 1994, 183, 269–288. [Google Scholar] [CrossRef]
- Sahyun, M.R.V.; Blair, J.T. Photophysics of a “simple” cyanide dye. J. Photochem. Photobiol. A Chem. 1997, 104, 179–187. [Google Scholar] [CrossRef]
- Fürstenberg, A.; Julliard, M.D.; Deligeorgiev, T.G.; Gadjev, N.I.; Vasilev, A.A.; Vauthey, E. Ultrafast excited-state dynamics of DNA fluorescent intercalators: New insight into the fluorescence enhancement mechanism. J. Am. Chem. Soc. 2006, 128, 7661–7669. [Google Scholar] [CrossRef] [PubMed]
- Dietzek, B.; Yartsev, A.; Tarnovsky, A.N. Watching ultrafast barrierless excited-state isomerization of pseudocyanine in real time. J. Phys. Chem. B 2007, 111, 4520–4526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Piontkowski, Z.; Mark, D.J.; Bedics, M.A.; Sabatini, R.P.; Mark, M.F.; Detty, M.R.; McCamant, D.W. Excited state torsional processes in chalcogenopyrylium monomethine dyes. J. Phys. Chem. A 2019, 123, 8807–8822. [Google Scholar] [CrossRef] [PubMed]
- Pronkin, P.G.; Tatikolov, A.S. Fluorescent probes for biomacromolecules based on monomethine cyanine dyes. Chemosensors 2023, 11, 280. [Google Scholar] [CrossRef]
- Serrano, J.L.; Maia, A.; Santos, A.O.; Lima, E.; Reis, L.V.; Nunes, M.J.; Boto, R.E.F.; Silvestre, S.; Almeida, P. An insight into symmetrical cyanine dyes as promising selective antiproliferative agents in Caco-2 colorectal cancer cells. Molecules 2022, 27, 5779. [Google Scholar] [CrossRef]
- Tatikolov, A.S.; Dzhulibekov, K.S.; Shvedova, L.A.; Kuzmin, V.A.; Ishchenko, A.A. Influence of “inert” counterions on the photochemistry of some cationic polymethine dyes. J. Phys. Chem. 1995, 99, 6525–6529. [Google Scholar] [CrossRef]
- Marcus, Y. The properties of organic liquids that are relevant to their use as solvating solvents. Chem. Soc. Rev. 1993, 22, 409–416. [Google Scholar] [CrossRef]
- Derevyanko, N.A.; Dyadyusha, G.G.; Ishchenko, A.A.; Tolmachev, A.I. Influence of nature of solvent on position, intensity, and shape of absorption bands of polymethine dyes. Theor. Exp. Chem. 1983, 19, 150–157. [Google Scholar] [CrossRef]
- Dyadyusha, G.G.; Ishchenko, A.A. Application of the method of moments to the study of the electronic spectra of organic dyes. J. Appl. Spectrosc. 1979, 30, 746–750. [Google Scholar] [CrossRef]
- Mackay, D. Multimedia Environmental Models: The Fugacity Approach, 3rd ed.; Mark Parnis, J., Ed.; CRC press: Boca Raton, FL, USA, 2021; ISBN 978-1-000-09499-2. [Google Scholar]
- Molinspiration. Calculation of Molecular Properties and Bioactivity Score. 2015. Available online: http://www.molinspiration.com (accessed on 2 August 2023).
- Valdes-Tresanco, M.S.; Valdes-Tresanco, M.E.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct 2020, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin., T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Rehman, S.U.; Sarwar, T.; Ishqi, H.M.; Husain, M.A.; Hasan, Z.; Tabish, M. Deciphering the interactions between chlorambucil and calf thymus DNA: A multi-spectroscopic and molecular docking study. Arch. Biochem. Biophys. 2015, 566, 7–14. [Google Scholar] [CrossRef]
- Jana, B.; Senapati, S.; Ghosh, D.; Bose, D.; Chattopadhyay, N. Spectroscopic exploration of mode of binding of ctDNA with 3-hydroxyflavone: A contrast to the mode of binding with flavonoids having additional hydroxyl groups. J. Chem. Phys. B 2012, 116, 639–645. [Google Scholar] [CrossRef]
- Miller, J.N. Recent developments in fluorescence and chemiluminescence analysis. Plenary lecture. Analyst 1984, 109, 191–198. [Google Scholar] [CrossRef]
- Abert, W.C.; Gregory, W.M.; Allan, G.S. The binding interaction of Coomassie Blue with proteins. Anal. Biochem. 1993, 213, 407–413. [Google Scholar] [CrossRef]
- Samanta, A.K.; Paul, B.K.; Guchhait, N. Spectroscopic probe analysis for exploring probe-protein interaction: A mapping of native, unfolding and refolding of protein bovine serum albumin by extrinsic fluorescence probe. Biophys. Chem. 2011, 156, 128–139. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Hardcover; Springer: Boston, MA, USA, 2006; 954p, ISBN 978-0-387-31278-1. [Google Scholar]
- Lehrer, S.S.; Leavis, P.C. Solute quenching of protein fluorescence. Methods Enzymol. 1978, 49, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D. Measurement of ligand binding to proteins by fluorescence spectroscopy. Methods Enzymol. 1985, 117, 400–414. [Google Scholar] [CrossRef]
- Benesi, H.; Hildebrand, J. A spectrophotometric investigation of the interaction of iodine with aromatic hydrocarbons. J. Am. Chem. Soc. 1949, 71, 2703–2707. [Google Scholar] [CrossRef]
- Gesztelyi, R.; Zsuga, J.; Kemeny-Beke, A.; Varga, B.; Juhasz, B.; Tosaki, A. The Hill equation and the origin of quantitative pharmacology. Arch. Hist. Exact Sci. 2012, 66, 427–438. [Google Scholar] [CrossRef]
- Scatchard, G. The attractions of proteins for small molecules and ions. Annu. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Zipper, H.; Brunner, H.; Bernhagen, J.; Vitzthum, F. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucl. Acids Res. 2004, 32, e103. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, C.; Scaiano, J.C. Selective binding and local photophysics of the fluorescent cyanine dye PicoGreen in double-stranded and single-stranded DNA. Phys. Chem. Chem. Phys. 2003, 5, 4911–4917. [Google Scholar] [CrossRef]
- Tatikolov, A.S.; Costa, S.M.B. Complexation of polymethine dyes with human serum albumin: A spectroscopic study. Biophys. Chem. 2004, 107, 33–49. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Tatikolov, A.S. Spectral fluorescence properties of an anionic oxacarbocyanine dye in complexes with human serum albumin. J. Appl. Spectrosc. 2015, 82, 438–444. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Shvedova, L.A.; Tatikolov, A.S. Comparative study of the interaction of some meso-substituted anionic cyanine dyes with human serum albumin. Biophys. Chem. 2020, 261, 106378. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Shvedova, L.A.; Tatikolov, A.S. Hydrophilic meso-substituted cyanine dyes in solution and in complexes with serum albumins: Spectral properties and molecular docking study. J. Chem. Sci. 2020, 132, 152. [Google Scholar] [CrossRef]
- Yarmoluk, S.M.; Kryvorotenko, D.V.; Balanda, A.O.; Losytskyy, M.Y.; Kovalska, V.B. Proteins and cyanine dyes. Part III. Synthesis and spectroscopic studies of benzothiazolo-4-[1,2,6-trimethylpyridinium] monomethine cyanine dyes for fluorescent detection of bovine serum albumin in solutions. Dye. Pigment. 2001, 51, 41–49. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Tatikolov, A.S. Meso-aryl-substituted thiacarbocyanine dyes as spectral-fluorescent probes for DNA. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2022, 269, 120744. [Google Scholar] [CrossRef] [PubMed]
- Tatikolov, A.S.; Pronkin, P.G.; Shvedova, L.A.; Panova, I.G. Meso-substituted carbocyanines as effective spectral-fluorescent and photochemical probes for structurally organized systems based on macromolecules. Russ. J. Phys. Chem. B 2019, 13, 900–906. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Tatikolov, A.S. Photonics of meso-substituted carbocyanine dyes in solutions and complexes with DNA. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 263, 120171. [Google Scholar] [CrossRef]
- Pronkin, P.G.; Tatikolov, A.S. Influence of the interaction with DNA on the spectral-fluorescent and photochemical properties of some meso-substituted polymethine dyes. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 202, 269–275. [Google Scholar] [CrossRef]
- Alganzory, H.H.; El-Sayed, W.A.; Arief, M.H.; Amine, M.S.; Ebeid, E.-Z.M. Microwave synthesis and fluorescence properties of homo- and heterodimeric monomethine cyanine dyes TOTO and their precursors. Green Chem. Lett. Rev. 2017, 10, 10–22. [Google Scholar] [CrossRef]
- Song, G.W.; He, Y.; Cai, Z.X. The interaction between levofloxacine hydrochloride and DNA mediated by Cu2+. J. Fluoresc. 2004, 14, 705–710. [Google Scholar] [CrossRef]
- Baguley, B.C.; Falkenhang, E.-M. The interaction of ethidium with synthetic double-stranded polynucleotides at low ionic strength. Nucl. Acids Res. 1978, 5, 161–171. [Google Scholar] [CrossRef]
- Tataurov, A.V.; You, Y.; Owczarzy, R. Predicting ultraviolet spectrum of single stranded and double stranded deoxyribonucleic acids. Biophys. Chem. 2008, 133, 66–70. [Google Scholar] [CrossRef]
- Mergny, J.-L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Huang, X.Z.; Xu, J.G. Synchronous fluorescence spectrometric methodology in the wavelength domain. J. Fluoresc. 1999, 9, 173–179. [Google Scholar] [CrossRef]
- Saeidifar, M.; Mansouri-Torshizi, H.; Saboury, A.A. Biophysical study on the interaction between two palladium(II) complexes and human serum albumin by multispectroscopic methods. J. Luminescence 2015, 167, 391–398. [Google Scholar] [CrossRef]
- Magde, D.; Wong, R.; Seybold, P.G. Fluorescence quantum yields and their relation to lifetimes of rhodamine 6G and fluorescein in nine solvents: Improved absolute standards for quantum yields. Photochem. Photobiol. 2002, 75, 327–334. [Google Scholar] [CrossRef]
- Olsen, A.L.; Washburn, E.R. An interpolation table for refractive index-normality relationship for solutions of hydrochloric acid and sodium hydroxide. Trans. Kansas Acad. Sci. 1937, 40, 117–126. [Google Scholar] [CrossRef]
- Parker, C.A. Photoluminescence of Solutions with Applications to Photochemistry and Analytical Chemistry; Hardcover; Elsevier: Amsterdam, The Netherland, 1968; 544p, ISBN 13 978-0444407634. [Google Scholar]
- Treinin, A.; Hayon, E. Quenching of triplet states by inorganic ions. Energy transfer and charge transfer mechanisms. J. Am. Chem. Soc. 1976, 98, 3884–3891. [Google Scholar] [CrossRef]
- Penzkofer, A.; Beidoun, A.; Daiber, M. Intersystem-crossing and excited-state absorption in eosin Y solutions determined by picosecond double pulse transient absorption measurements. J. Luminescence 1992, 51, 297–314. [Google Scholar] [CrossRef]
- Chibisov, A.K. Triplet states of cyanine dyes and reactions of electron transfer with their participation. J. Photochem. 1976–1977, 6, 199–214. [Google Scholar] [CrossRef]
- MacDougall, D.; Crummett, W.B. Guidelines for data acquisition and data quality evaluation in environmental chemistry. Anal. Chem. 1980, 52, 2242–2249. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA dodecamer: Conformation and dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Dautant, A.; Langlois d’Estaintot, B.; Gallois, B.; Brown, T.; Hunter, W.N. A trigonal form of the idarubicin: D(CGATCG) complex; crystal and molecular structure at 2.0 A resolution. Nucl. Acids Res. 1995, 23, 1710–1716. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. D Biol. Crystallogr. 2012, 68 Pt 10, 1278–1289. [Google Scholar] [CrossRef]
- Demas, J.N.; Crosby, G.A. Measurement of photoluminescence quantum yields. Review. J. Phys. Chem. 1971, 75, 991–1024. [Google Scholar] [CrossRef]
- Hill, A.V. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J. Physiol. 1910, 40, iv–vii. [Google Scholar]
- Nørby, J.G.; Ottolenghi, P.; Jensen, J. Scatchard plot: Common misinterpretation of binding experiments. Anal. Biochem. 1980, 102, 318–320. [Google Scholar] [CrossRef]


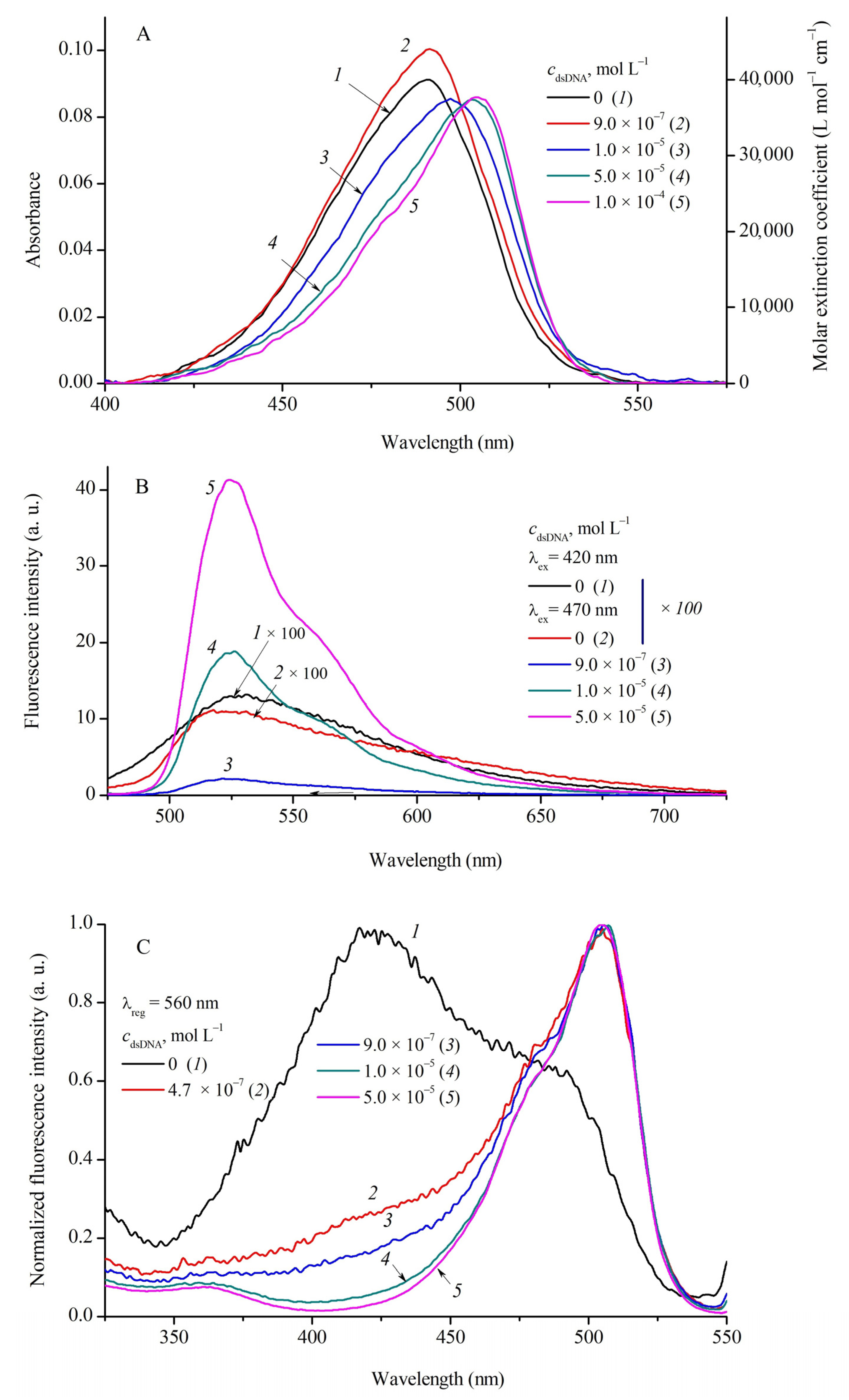

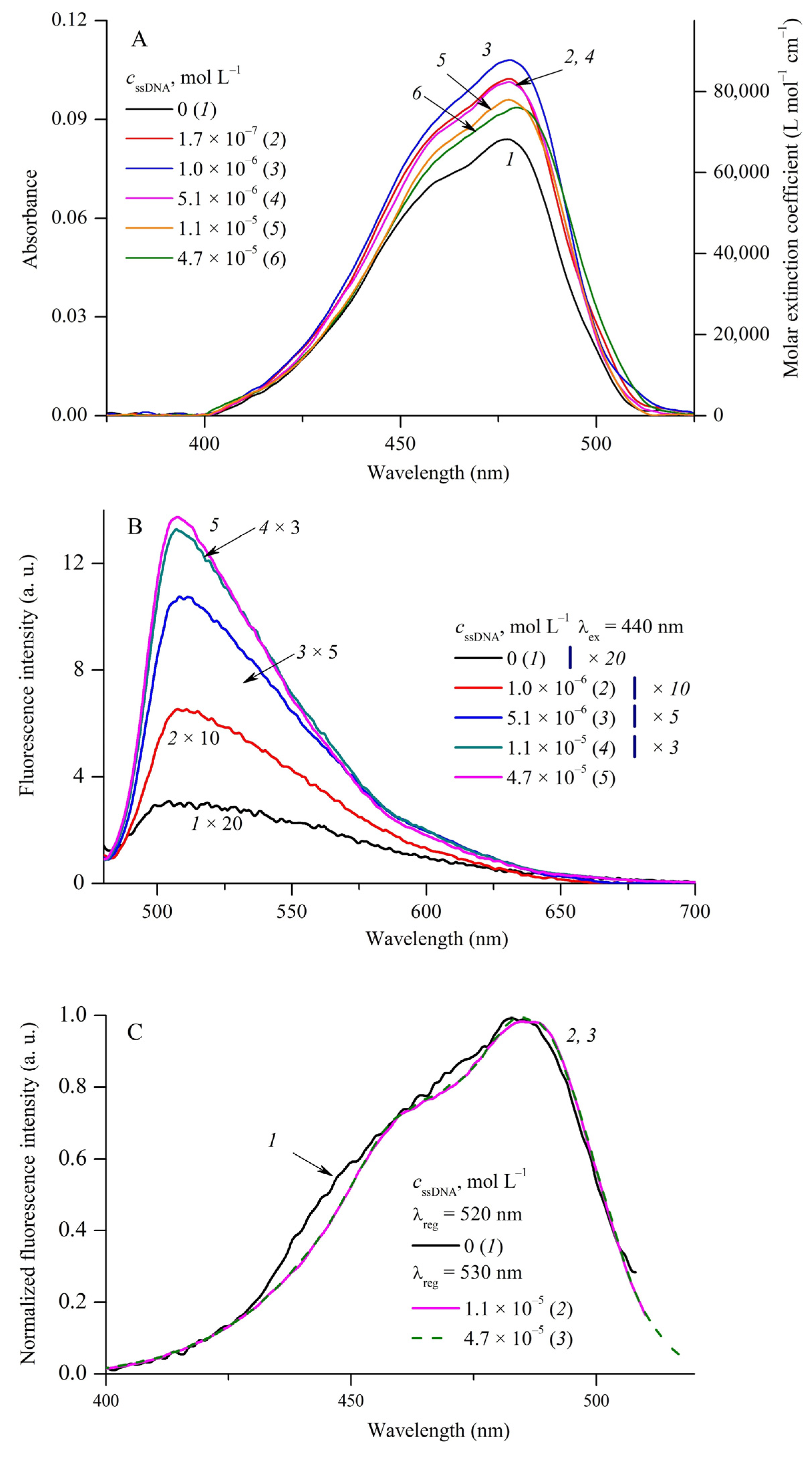

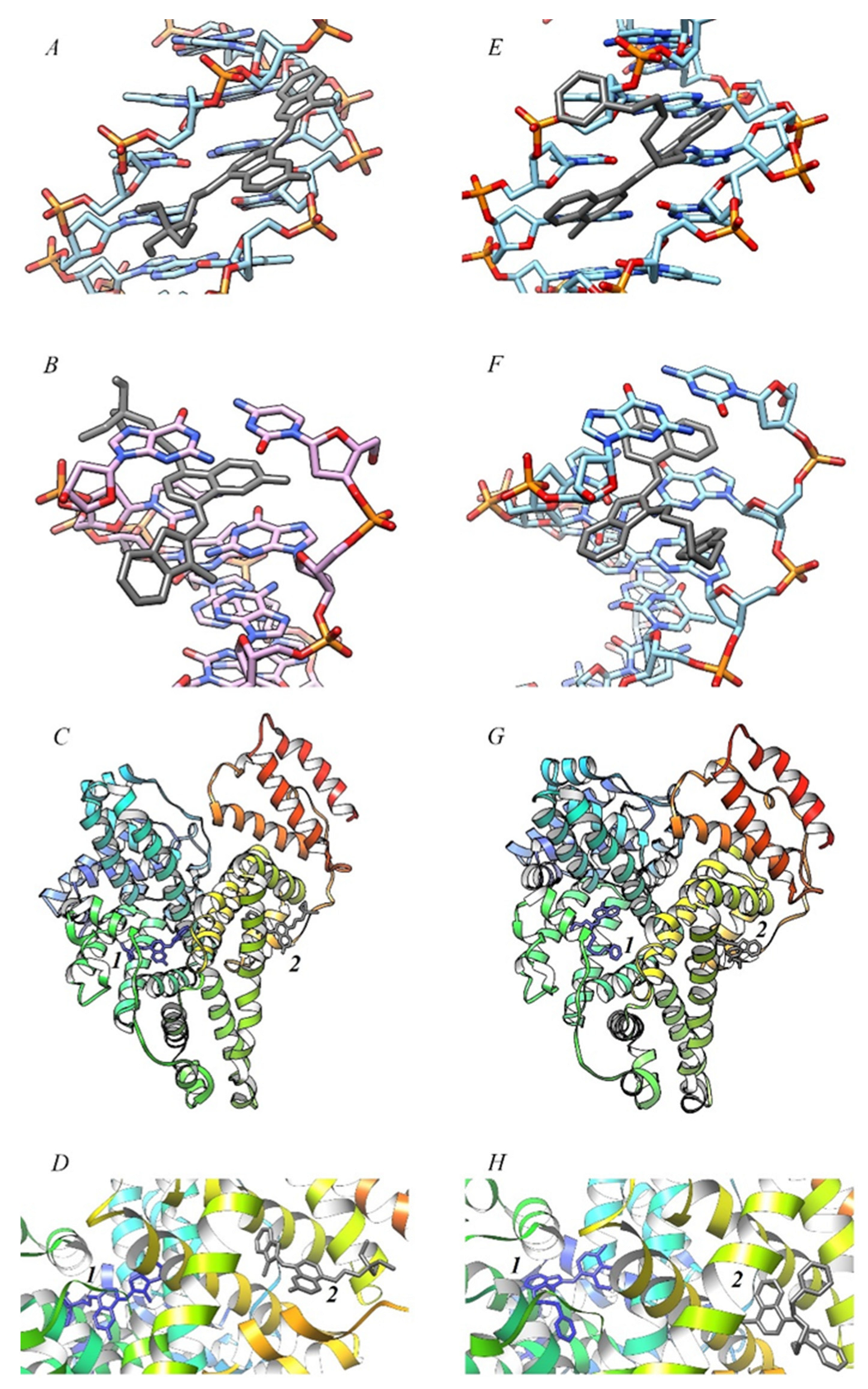

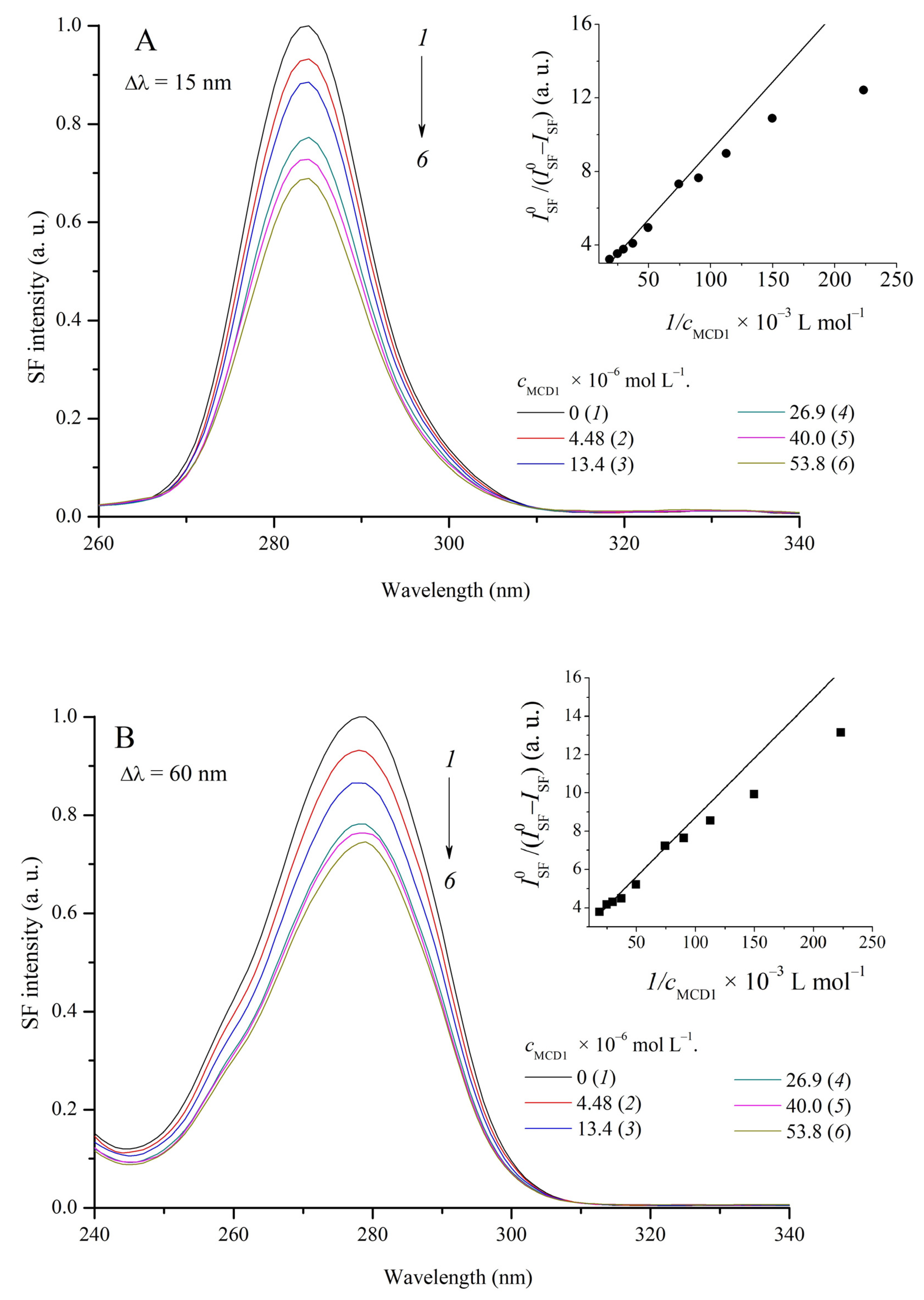
| MCD | 1 | 2 | 3 | 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Solvent | λabs | λfl | λex | λabs | λfl | λex | λabs | λfl | λex | λabs | λfl | λex |
| nm | ||||||||||||
| Ethanol | 492 | 526 | 487 | 477 | 496 | 475 | 424 | 455 | 359, 422 | 492, 523 | 568 | 525 |
| Acetone | 492 | 559 | 498 | 478 | 501 | 475 | 424 | 481 | 421 | 493, 523 | 559 | 524 |
| DMSO | 501 | 535 | 496 | 481 | 504 | 479 | 427 | 476 | 424 | 493, 530 | 553 | 536 |
| Acetonitrile | 497 | 555 | 495 | 477 | 501 | 471 | 422 | 457 | 422 | 500, 523 | 548 | 522 |
| 1,4-Dioxane | 481 | 582 | 478 | 484 | 527 | 472 | 428 | 475 | 425 | 502, 530 | 524, 570 | 484, 517 |
| Chloroform | 492 | 543 | 491 | 486 | 504 | 481 | 428 | 483 | 428 | 497, 529 | 581 | 526 |
| Ethyl acetate | 476 | 619 | 459 | 480 | 513 | 471 | 426 | 478 | 426 | 496, 524 | 559, 670 | 484, 516 |
| PBS buffer | 491 | 520 | 422, 492 | 478 | 509 | 485 | 422 | 463–489 1 | 400, 423 | 494, 523 | 512, 554 | 490, 525 |
| Molar extinction coefficients (ε, L mol−1 cm−1) 2 | ||||||||||||
| Ethanol | 58,400 | 78,800 | 69,200 | 49,000 | ||||||||
| Acetone | 44,300 | 94,000 | 50,900 | 34,000 | ||||||||
| DMSO | 58,500 | 60,200 | 55,100 | 28,000 | ||||||||
| Acetonitrile | 48,700 | 91,200 | 61,900 | 44,700 | ||||||||
| 1,4-Dioxane | 19,500 | 55,400 | 50,300 | 28,000 | ||||||||
| Chloroform | 31,100 | 85,000 | 59,200 | 47,000 | ||||||||
| Ethyl acetate | 16,000 | 45,700 | 44,000 | 32,600 | ||||||||
| PBS buffer | 40,000 | 68,000 | 61,800 | 44,500 | ||||||||
| Solvent | Ethanol | Acetonitrile | 1.4-Dioxane | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dye | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 |
| φfl, % | 0.11 | 0.06 | 0.7 | ~0.06 | 0.14 | 0.02 | 0.6 | ~0.05 | 0.4 | 0.16 | 1.3 | ~0.06 |
| kr × 10−8, s−1 | 1.9 | 3.4 | 4.7 | 2.3 | 1.8 | 3.9 | 4.1 | 2.9 | 0.8 | 2.4 | 2.3 | 1.6 |
| τr, ns | 5.2 | 2.9 | 2.1 | 4.4 | 5.7 | 2.5 | 2.4 | 3.4 | 12 | 4.2 | 4.4 | 6.2 |
| knr × 10−11, s−1 | 1.8 | 5.7 | 0.67 | 3.8 | 1.2 | 1.6 | 0.68 | 5.0 | 0.2 | 1.5 | 0.17 | 2.7 |
| Solvent | εr | n | P* | ET(30) | MCD 1 | MCD 2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ν1, µm−1 | σ, 103 cm−1 | Ifl, a.u | ν1, µm−1 | σ,103 cm−1 | Ifl, a.u | |||||
| Acetonitrile | 36.2 | 1.3441 | 0.2119 | 45.6 | 2.095 | 1.384 | 0.63 | 2.158 | 1.092 | 0.24 |
| Acetone | 20.7 | 1.3586 | 0.2199 | 42.2 | 2.076 | 1.194 | 1.12 | 2.146 | 1.015 | 0.23 |
| Ethanol | 24.3 | 1.3614 | 0.2215 | 51.9 | 2.086 | 1.188 | 0.49 | 2.159 | 1.102 | 0.56 |
| Ethyl acetate | 6.02 | 1.3724 | 0.2275 | 38.1 | 2.104 | 1.526 | 0.46 | 2.144 | 1.043 | 0.42 |
| Isopentyl alcohol | 15.2 | 1.4075 | 0.2275 | 49 | 2.126 | 1.472 | 2.17 | 2.138 | 0.996 | 1.32 |
| 1.4-Dioxane | 2.22 | 1.4224 | 0.2464 | 36 | 2.114 | 1.322 | 1.09 | 2.127 | 1.064 | 1.39 |
| Chloroform | 4.81 | 1.4459 | 0.2586 | 39.1 | 2.069 | 0.977 | 0.58 | 2.122 | 1.006 | 0.69 |
| DMSO | 49 | 1.4793 | 0.2666 | 45.1 | 2.039 | 1.073 | 0.36 | 2.136 | 1.095 | 1.06 |
| Benzyl alcohol | 13.5 | 1.5396 | 0.2837 | 50.4 | 2.026 | 1.066 | 0.98 | 2.117 | 1.046 | 2.39 |
| MCD | λabsb | Δλabs | ν1 | M−1 | ΔM−1 | σ | Δσ | λflb | Δλfl | λexb | φflb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| nm | cm−1 | nm | cm−1 | % | nm | % | |||||
| cdsDNA~5 × 10−5 mol L−1 | |||||||||||
| 1 | 503 | 12 | 20,450 | 489 | 8 | 1100 | −4 | 525 | 5 | 505 | 13 ± 1.5 |
| 2 | 485 | 7 | 21,146 | 473 | 10 | 1130 | −3 | 505 | −4 | 485 | 14 ± 1.7 |
| 3 | 423 | 1 | 24,030 | 416 | 6 | 1170 | −2 | 460 | −16 | 430 | 4.8 ± 0.5 |
| 4 | 522 | −1 | 19,880 | 503 | 0 | 1242 | 1 | 522 | 10 | 506 | ~1.5 ± 0.2 |
| cssDNA~5 × 10−5 mol L−1 | |||||||||||
| 1 | 499 | 8 | 20,550 | 486 | 5 | 1074 | −7 | 525 | 5 | 507 | 12 ± 1.5 |
| 2 | 480 | 2 | 21,490 | 465 | 2 | 1088 | −6 | 508 | 2 | 485 | 14 ± 1.7 |
| 3 | 424 | 2 | 23,940 | 418 | 8 | 1060 | −11 | 460 | −16 | 430 | 4.7 ± 0.5 |
| 4 | 526 | 3 | 19,644 | 509 | 6 | 1038 | −16 | 508 | −4 | 488 | ~1.4 ± 0.2 |
| cHSA~5 × 10−5 mol L−1 | |||||||||||
| 1 | 493 | 2 | 20,756 | 482 | 1 | 1101 | −4 | 523 | 3 | 498 | ~1 ± 0.1 |
| 2 | 478 | 0 | 21,600 | 463 | 0 | 1278 | 10 | 504 | −5 | 486 | 4 ± 0.8 |
| 3 | 379, 423 | 2 | 24,760 | 404 | −6 | 1450 | 22 | 472 | −4 | 360, 427 | ~50 ± 6 |
| 4 | 498, 521 | 2 | 20,050 | 499 | −4 | 1988 | 61 | 515 | 3 | 362 | ~0.5 ± 0.1 |
| MCD | Run | ΔGest | Eintmol | EVdWHD | Eel | Etint | Etor |
|---|---|---|---|---|---|---|---|
| kcal mol−1 | |||||||
| DNA (minor groove) | |||||||
| 1 | 1 | −10.18 | −12.57 | −11.28 | −1.29 | −0.65 | +2.39 |
| 2 | 3 | −9.01 | −11.40 | −11.39 | −0.01 | −1.28 | +2.39 |
| 3 | 10 | −9.70 | −10.59 | −10.56 | −0.04 | −0.04 | +0.89 |
| 4 | 4 | −10.40 | −11.30 | −11.22 | −0.08 | −0.62 | +0.89 |
| DNA (intercalation) | |||||||
| 1 | 4 | −8.07 | −10.46 | −10.16 | −0.3 | −1.14 | +2.39 |
| 2 | 6 | −8.58 | −10.96 | −10.86 | −0.11 | −1.88 | +2.39 |
| 3 | 6 | −7.83 | −8.73 | −8.63 | −0.09 | −0.75 | +0.89 |
| 4 | 3 | −8.40 | −8.70 | −8.67 | −0.03 | −0.13 | +0.30 |
| HSA (Sudlow I) | |||||||
| 1 | 2 | −7.85 | −10.24 | −10.15 | −0.09 | −0.88 | +2.39 |
| 2 | 2 | −8.79 | −11.18 | −11.08 | −0.1 | −1.19 | +2.39 |
| 3 | 5 | −7.47 | −8.36 | −8.33 | -0.04 | −0.73 | +0.89 |
| 4 | 1 | −8.20 | −9.09 | −9.09 | −0.01 | −0.90 | +0.89 |
| HSA (Sudlow II) | |||||||
| 1 | 5 | −7.07 | −9.45 | −9.17 | −0.28 | −0.90 | +2.39 |
| 2 | 2 | −7.44 | −9.82 | −9.65 | −0.18 | −1.45 | +2.39 |
| 3 | 8 | −5.75 | −6.65 | −6.57 | −0.08 | −0.75 | +0.89 |
| 4 | 8 | −7.61 | −8.51 | −8.52 | +0.01 | −0.91 | +0.89 |
| Fluorophore/ SFS Δλ, nm | MCD | |||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| KSV, L mol−1; α | ||||
| Tyr/15 nm | 2.18 × 104; 0.61 | 2.16 × 104; 0.45 | 1.12 × 105; 0.15 | 3.82 × 104; 0.33 |
| Trp/60 nm | 4.04 × 104; 0.40 | 2.53 × 104; 0.55 | 6.12 × 107; 0.82 | 2.10 × 104; 0.60 |
| MCD | εf/εb | KeffBH ×10−4, L mol−1 | αb, 1 % | KeffHill ×10−4, L mol−1 | nHill | K1Sc | K2Sc | nSc | LR, µmol L−1 | LOD | LOQ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ×10−5, L mol−1 | × 108, mol L−1 | |||||||||||
| dsDNA | ||||||||||||
| 1 | 1.05 | 5.4 | 460 | 80% | 5.34 | 1.04 | 7.93 | 1.69 | 2 | 0–3 | 0.72 | 2.1 |
| 2 | 1.36 | 3.94 | 198 | 72% | 3.8 | 0.99 | 4.35 | 24.5 | 56 | 0–~2 | 1.2 | 3.5 |
| 3 | 3.90 | 1.35 | 136 | 40% | 0.69 | 0.88 | 1.35 | 4.8 | 35 | 0–25 | 4.1 | 12.6 |
| 4 | 1.83 | 1.95 | 169 | 46% | 0.185 | 0.62 | 1.86 | 0.37 | 2 | 1.5–13 2 | 9.4 | 21 |
| ssDNA | ||||||||||||
| 1 | 1.73 | 2.42 | 60.9 | 60% | 0.283 | 0.7 | 2.28 | 1.59 | 2 | 0–3.5 | 4.0 | 12 |
| 2 | 0.72 | 1.64 | 178 | 54% | 0.31 | 0.95 | 1.86 | 3.9 | 21 | 0–~12 | 10.7 | 32 |
| 3 | 2.88 | 1.16 | 71 | 37% | 0.294 | 0.76 | 1.12 | 4.3 | 37 | 0–10 | 8.7 | 26 |
| 4 | – | 0.68 | 61 | 25% | 0.035 | 0.57 | 0.65 | 0.195 | 3 | 1–140 2 | 76 | 230 |
| HSA | ||||||||||||
| 1 | 1.59 | 0.80 | 65 | 27% | 0.335 | 0.89 | – | 0.073 | 1 | 0–0.6 3 | 27 | 89 |
| 2 | 2.19 | 6.34 | 47 | 74% | 6.33 | 0.95 | – | 0.60 | 0–10 | 3.4 | 11.3 | |
| 3 | – | 1.76 | 26 | 50% | 2.78 | 0.59 | – | 0.34 | 0–1 | 7.7 | 23 | |
| 4 | – | 0.482 | 109 | 20% | 0.183 | 0.61 | – | 0.17 | 0.7–7.5 2 | 73 | 220 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pronkin, P.G.; Tatikolov, A.S. Photonics of Some Monomethine Cyanine Dyes in Solutions and in Complexes with Biomolecules. Int. J. Mol. Sci. 2023, 24, 13954. https://doi.org/10.3390/ijms241813954
Pronkin PG, Tatikolov AS. Photonics of Some Monomethine Cyanine Dyes in Solutions and in Complexes with Biomolecules. International Journal of Molecular Sciences. 2023; 24(18):13954. https://doi.org/10.3390/ijms241813954
Chicago/Turabian StylePronkin, Pavel G., and Alexander S. Tatikolov. 2023. "Photonics of Some Monomethine Cyanine Dyes in Solutions and in Complexes with Biomolecules" International Journal of Molecular Sciences 24, no. 18: 13954. https://doi.org/10.3390/ijms241813954
APA StylePronkin, P. G., & Tatikolov, A. S. (2023). Photonics of Some Monomethine Cyanine Dyes in Solutions and in Complexes with Biomolecules. International Journal of Molecular Sciences, 24(18), 13954. https://doi.org/10.3390/ijms241813954








