Lactiplantibacillus plantarum LPJZ-658 Improves Non-Alcoholic Steatohepatitis by Modulating Bile Acid Metabolism and Gut Microbiota in Mice
Abstract
:1. Introduction
2. Results
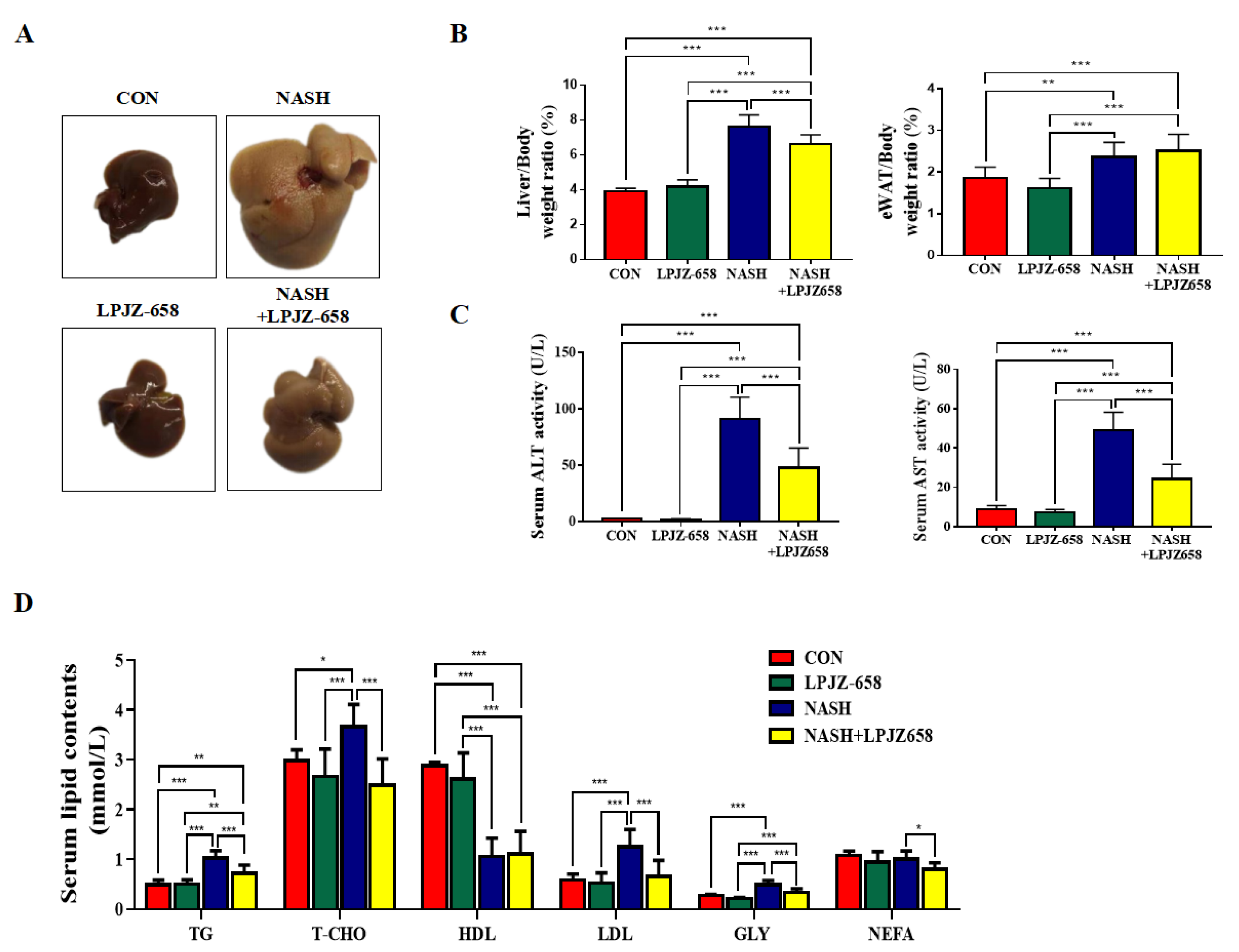
2.1. LPJZ-658 Attenuates Hepatic Injury and Serum Lipid Metabolic Disorders in NASH Mice
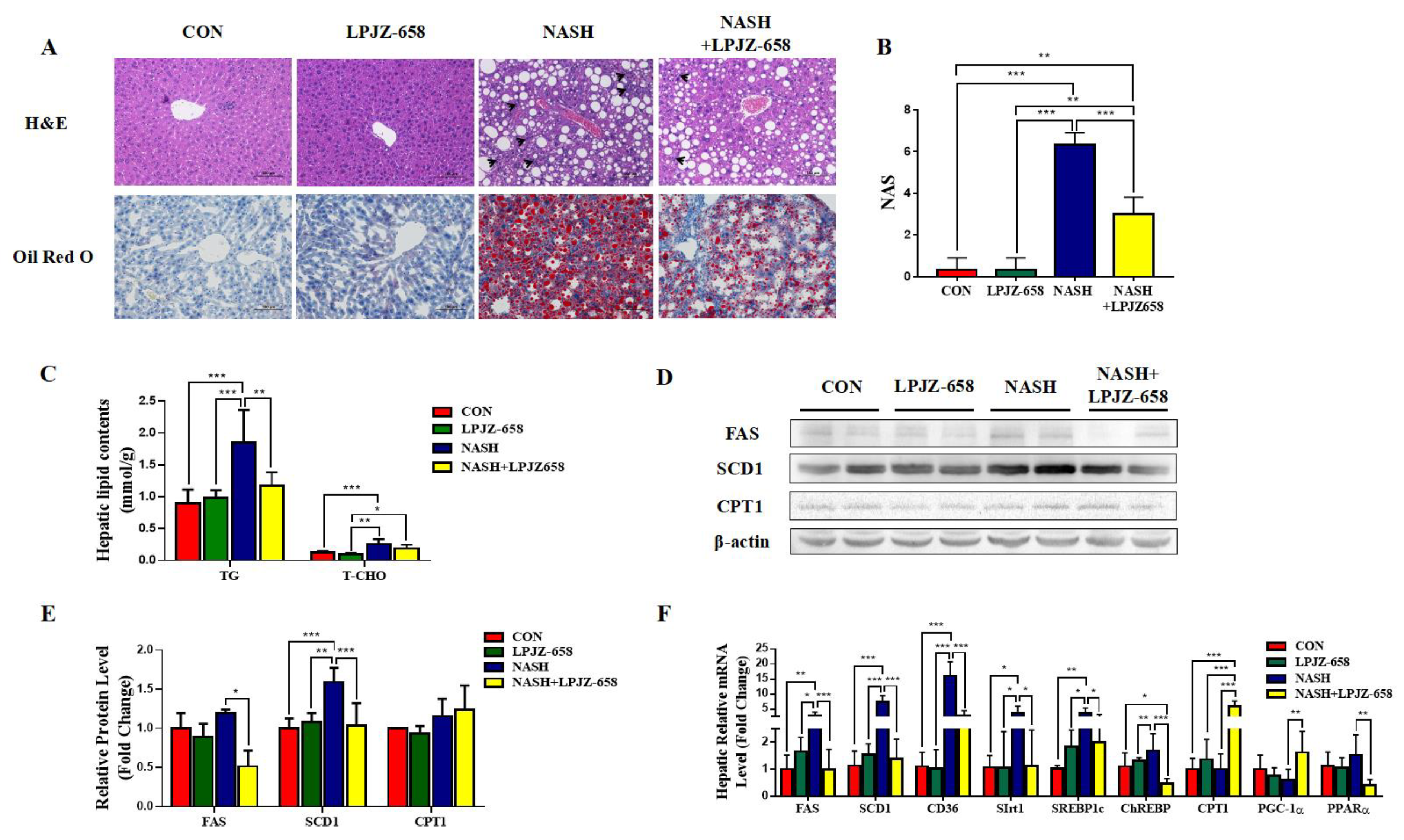
2.2. LPJZ-658 Prevents the Development of Hepatic Steatosis in NASH Mice
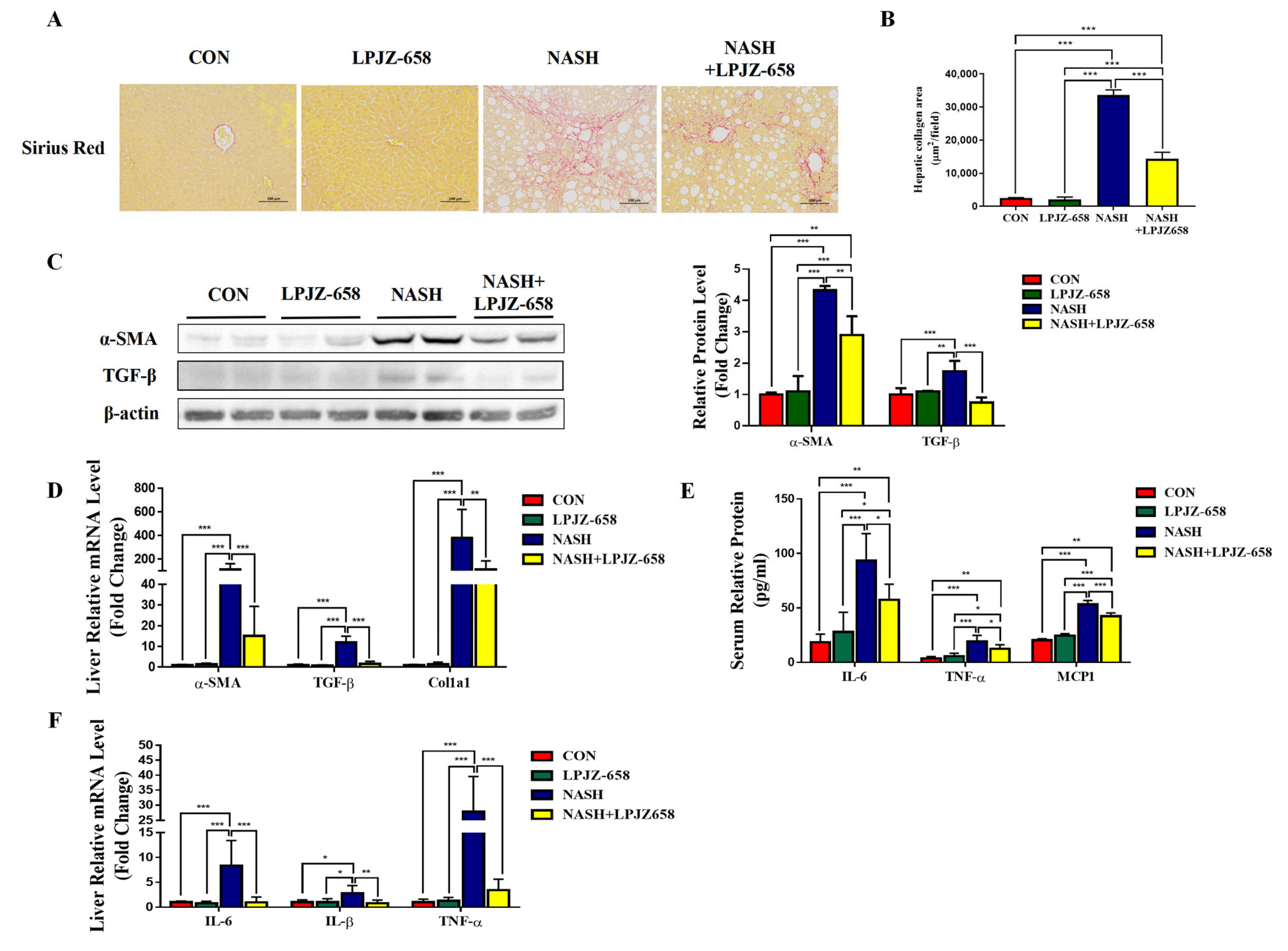
2.3. LPJZ-658 Attenuates Hepatic Fibrosis and Inflammation in NASH Mice
2.4. Serum Metabolome Profile and Biomarker Annotation
2.5. Metabolic Pathway Analysis
2.6. LPJZ-658 Altered the BAs Metabolism Profiles in NASH Mice Serum
2.7. Effect of LPJZ-658 Gut Microbiota in NASH Mice
2.8. Correlations between BAs Metabolism Profiles and Gut Bacteria
3. Discussion
4. Materials and Methods
4.1. Probiotic Strain
4.2. Animals and Treatments
4.3. Statistical Analysis and Biomarker Screening
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Paik, J.M.; Golabi, P.; Younossi, Y.; Mishra, A.; Younossi, Z.M. Changes in the Global Burden of Chronic Liver Diseases From 2012 to 2017: The Growing Impact of NAFLD. Hepatology 2020, 72, 1605–1616. [Google Scholar] [CrossRef] [PubMed]
- Powell, E.E.; Wong, V.W.; Rinella, M. Non-alcoholic fatty liver disease. Lancet 2021, 397, 2212–2224. [Google Scholar] [CrossRef] [PubMed]
- Brunt, E.M. Nonalcoholic steatohepatitis: Definition and pathology. Semin. Liver Dis. 2001, 21, 3–16. [Google Scholar] [CrossRef]
- Machado, M.V.; Diehl, A.M. Pathogenesis of Nonalcoholic Steatohepatitis. Gastroenterology 2016, 150, 1769–1777. [Google Scholar] [CrossRef]
- Chiang, J.Y.L. Targeting bile acids and lipotoxicity for NASH treatment. Hepatol. Commun. 2017, 1, 1002–1004. [Google Scholar] [CrossRef]
- Forlano, R.; Sivakumar, M.; Mullish, B.H.; Manousou, P. Gut Microbiota-A Future Therapeutic Target for People with Non-Alcoholic Fatty Liver Disease: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 8307. [Google Scholar] [CrossRef]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef]
- Lin, H.Y.; Yang, Y.L.; Wang, P.W.; Wang, F.S.; Huang, Y.H. The Emerging Role of MicroRNAs in NAFLD: Highlight of MicroRNA-29a in Modulating Oxidative Stress, Inflammation, and Beyond. Cells 2020, 9, 1041. [Google Scholar] [CrossRef]
- Masoodi, M.; Gastaldelli, A.; Hyotylainen, T.; Arretxe, E.; Alonso, C.; Gaggini, M.; Brosnan, J.; Anstee, Q.M.; Millet, O.; Ortiz, P.; et al. Metabolomics and lipidomics in NAFLD: Biomarkers and non-invasive diagnostic tests. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 835–856. [Google Scholar] [CrossRef]
- Schneider, K.M.; Albers, S.; Trautwein, C. Role of bile acids in the gut-liver axis. J. Hepatol. 2018, 68, 1083–1085. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.W.; Ge, T.T.; Chen, S.Z.; Wang, G.; Yang, Q.; Huang, C.H.; Xu, L.C.; Chen, Z. Role of bile acids in liver diseases mediated by the gut microbiome. World J. Gastroenterol. 2021, 27, 3010–3021. [Google Scholar] [CrossRef] [PubMed]
- Dasarathy, S.; Yang, Y.; McCullough, A.J.; Marczewski, S.; Bennett, C.; Kalhan, S.C. Elevated hepatic fatty acid oxidation, high plasma fibroblast growth factor 21, and fasting bile acids in nonalcoholic steatohepatitis. Eur. J. Gastroenterol. Hepatol. 2011, 23, 382–388. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Hu, S.; Xu, Y.; Stroup, D.; Pan, X.; Bawa, F.C.; Chen, S.; Gopoju, R.; Yin, L.; et al. Hepatocyte Nuclear Factor 4alpha Prevents the Steatosis-to-NASH Progression by Regulating p53 and Bile Acid Signaling (in mice). Hepatology 2021, 73, 2251–2265. [Google Scholar] [CrossRef] [PubMed]
- Grzych, G.; Chavez-Talavera, O.; Descat, A.; Thuillier, D.; Verrijken, A.; Kouach, M.; Legry, V.; Verkindt, H.; Raverdy, V.; Legendre, B.; et al. NASH-related increases in plasma bile acid levels depend on insulin resistance. JHEP Rep. 2021, 3, 100222. [Google Scholar] [CrossRef]
- Villanueva, M.T. Liver disease: Conscious uncoupling in NASH. Nat. Rev. Drug Discov. 2017, 16, 238–239. [Google Scholar] [CrossRef]
- Pengyue, Z.; Tao, G.; Hongyun, H.; Liqiang, Y.; Yihao, D. Breviscapine confers a neuroprotective efficacy against transient focal cerebral ischemia by attenuating neuronal and astrocytic autophagy in the penumbra. Biomed. Pharmacother. 2017, 90, 69–76. [Google Scholar] [CrossRef]
- Esposito, E.; Iacono, A.; Bianco, G.; Autore, G.; Cuzzocrea, S.; Vajro, P.; Canani, R.B.; Calignano, A.; Raso, G.M.; Meli, R. Probiotics reduce the inflammatory response induced by a high-fat diet in the liver of young rats. J. Nutr. 2009, 139, 905–911. [Google Scholar] [CrossRef]
- Mohamad Nor, M.H.; Ayob, N.; Mokhtar, N.M.; Raja Ali, R.A.; Tan, G.C.; Wong, Z.; Shafiee, N.H.; Wong, Y.P.; Mustangin, M.; Nawawi, K.N.M. The Effect of Probiotics (MCP((R)) BCMC((R)) Strains) on Hepatic Steatosis, Small Intestinal Mucosal Immune Function, and Intestinal Barrier in Patients with Non-Alcoholic Fatty Liver Disease. Nutrients 2021, 13, 3192. [Google Scholar] [CrossRef]
- Mofidi, F.; Poustchi, H.; Yari, Z.; Nourinayyer, B.; Merat, S.; Sharafkhah, M.; Malekzadeh, R.; Hekmatdoost, A. Synbiotic supplementation in lean patients with non-alcoholic fatty liver disease: A pilot, randomised, double-blind, placebo-controlled, clinical trial. Br. J. Nutr. 2017, 117, 662–668. [Google Scholar] [CrossRef]
- Xin, J.; Zeng, D.; Wang, H.; Ni, X.; Yi, D.; Pan, K.; Jing, B. Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl. Microbiol. Biotechnol. 2014, 98, 6817–6829. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, L.; Liu, Q.; Li, F.; Zhang, L.; Zhu, F.; Shao, T.; Barve, S.; Chen, Y.; Li, X.; et al. Fibroblast growth factor 21 is required for the therapeutic effects of Lactobacillus rhamnosus GG against fructose-induced fatty liver in mice. Mol. Metab. 2019, 29, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, C.; Dong, Y.; Zhang, M.; Wang, Y.; Li, F.; Li, X.; McClain, C.; Yang, S.; Feng, W. Inhibition of miR122a by Lactobacillus rhamnosus GG culture supernatant increases intestinal occludin expression and protects mice from alcoholic liver disease. Toxicol. Lett. 2015, 234, 194–200. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, C.; Wang, C.; Zhao, H.; Zhao, C.; Chen, Y.; Wang, Y.; McClain, C.; Feng, W. Enhanced AMPK phosphorylation contributes to the beneficial effects of Lactobacillus rhamnosus GG supernatant on chronic-alcohol-induced fatty liver disease. J. Nutr. Biochem. 2015, 26, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Wei, X.; Yin, X.; Wang, Y.; Zhang, M.; Zhao, C.; Zhao, H.; McClain, C.J.; Feng, W.; Zhang, X. Hepatic and fecal metabolomic analysis of the effects of Lactobacillus rhamnosus GG on alcoholic fatty liver disease in mice. J. Proteome Res. 2015, 14, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Liu, L.; Fu, T.; Li, C.; Jin, N.; Zhang, H.; Li, C.; Liu, Y.; Zhao, C. Genome Sequence and Evaluation of Safety and Probiotic Potential of Lactiplantibacillus plantarum LPJZ-658. Microorganisms 2023, 11, 1620. [Google Scholar] [CrossRef]
- Liu, L.; Li, L.; Li, C.; Wang, H.; Zhang, X.; Ren, Q.; Zhang, H.; Jin, N.; Li, C.; Zhao, C. Effects of Lactiplantibacillus plantarum LPJZ-658 Supplementation on the Production, Meat Quality, Intestinal Morphology, and Cecal Microbiota of Broilers Chickens. Microorganisms 2023, 11, 1549. [Google Scholar] [CrossRef]
- Weiss, J.; Rau, M.; Geier, A. Non-alcoholic fatty liver disease: Epidemiology, clinical course, investigation, and treatment. Dtsch. Arztebl. Int. 2014, 111, 447–452. [Google Scholar] [CrossRef]
- Liu, W.; Baker, R.D.; Bhatia, T.; Zhu, L.; Baker, S.S. Pathogenesis of nonalcoholic steatohepatitis. Cell. Mol. Life Sci. 2016, 73, 1969–1987. [Google Scholar] [CrossRef]
- Castillo, V.; Figueroa, F.; Gonzalez-Pizarro, K.; Jopia, P.; Ibacache-Quiroga, C. Probiotics and Prebiotics as a Strategy for Non-Alcoholic Fatty Liver Disease, a Narrative Review. Foods 2021, 10, 1719. [Google Scholar] [CrossRef]
- Hany, N.M.; Eissa, S.; Basyouni, M.; Hasanin, A.H.; Aboul-Ela, Y.M.; Elmagd, N.M.A.; Montasser, I.F.; Ali, M.A.; Skipp, P.J.; Matboli, M. Modulation of hepatic stellate cells by Mutaflor((R)) probiotic in non-alcoholic fatty liver disease management. J. Transl. Med. 2022, 20, 342. [Google Scholar] [CrossRef] [PubMed]
- Segers, M.E.; Lebeer, S. Towards a better understanding of Lactobacillus rhamnosus GG-host interactions. Microb. Cell Fact. 2014, 13 (Suppl. S1), S7. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, K.; Li, F.; Gu, Z.; Liu, Q.; He, L.; Shao, T.; Song, Q.; Zhu, F.; Zhang, L.; et al. Probiotic Lactobacillus rhamnosus GG Prevents Liver Fibrosis Through Inhibiting Hepatic Bile Acid Synthesis and Enhancing Bile Acid Excretion in Mice. Hepatology 2020, 71, 2050–2066. [Google Scholar] [CrossRef] [PubMed]
- Dowman, J.K.; Tomlinson, J.W.; Newsome, P.N. Pathogenesis of non-alcoholic fatty liver disease. QJM Int. J. Med. 2010, 103, 71–83. [Google Scholar] [CrossRef]
- Lee, H.S.; Nam, Y.; Chung, Y.H.; Kim, H.R.; Park, E.S.; Chung, S.J.; Kim, J.H.; Sohn, U.D.; Kim, H.C.; Oh, K.W.; et al. Beneficial effects of phosphatidylcholine on high-fat diet-induced obesity, hyperlipidemia and fatty liver in mice. Life Sci. 2014, 118, 7–14. [Google Scholar] [CrossRef]
- Esler, W.P.; Bence, K.K. Metabolic Targets in Nonalcoholic Fatty Liver Disease. Cell Mol. Gastroenterol. Hepatol. 2019, 8, 247–267. [Google Scholar] [CrossRef]
- Hao, L.; Chai, Z. Nonalcoholic fatty liver disease: Pathogenesis and intervention. Precis. Nutr. 2023, 2, e00029. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Zhang, C.; Huang, K.; Zhao, J.; Le, S.; Jiang, L.; Liu, H.; Yang, P.; Xiao, X.; et al. B-cell lymphoma 6 alleviates nonalcoholic fatty liver disease in mice through suppression of fatty acid transporter CD36. Cell Death Dis. 2022, 13, 359. [Google Scholar] [CrossRef]
- Gao, Y.; Hua, R.; Peng, K.; Yin, Y.; Zeng, C.; Guo, Y.; Wang, Y.; Li, L.; Li, X.; Qiu, Y.; et al. High-starchy carbohydrate diet aggravates NAFLD by increasing fatty acids influx mediated by NOX2. Food Sci. Hum. Wellness 2023, 12, 1081–1101. [Google Scholar] [CrossRef]
- Lee, E.; Korf, H.; Vidal-Puig, A. An adipocentric perspective on the development and progression of non-alcoholic fatty liver disease. J. Hepatol. 2023, 78, 1048–1062. [Google Scholar] [CrossRef]
- Weiss, J.M.; Palmieri, E.M.; Gonzalez-Cotto, M.; Bettencourt, I.A.; Megill, E.L.; Snyder, N.W.; McVicar, D.W. Itaconic acid underpins hepatocyte lipid metabolism in non-alcoholic fatty liver disease in male mice. Nat. Metab. 2023, 5, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fang, W.; Sun, L.; Lu, Y.; Dou, L.; Huang, X.; Tang, W.; Yu, L.; Li, J. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat. Commun. 2018, 9, 612. [Google Scholar] [CrossRef]
- Iizuka, K. The transcription factor carbohydrate-response element-binding protein (ChREBP): A possible link between metabolic disease and cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Hua, J.; Li, Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J. Hepatol. 2008, 49, 821–830. [Google Scholar] [CrossRef]
- Li, Z.; Yang, S.; Lin, H.; Huang, J.; Watkins, P.A.; Moser, A.B.; Desimone, C.; Song, X.Y.; Diehl, A.M. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology 2003, 37, 343–350. [Google Scholar] [CrossRef]
- Wieckowska, A.; Papouchado, B.G.; Li, Z.; Lopez, R.; Zein, N.N.; Feldstein, A.E. Increased hepatic and circulating interleukin-6 levels in human nonalcoholic steatohepatitis. Am. J. Gastroenterol. 2008, 103, 1372–1379. [Google Scholar] [CrossRef]
- Kakino, S.; Ohki, T.; Nakayama, H.; Yuan, X.; Otabe, S.; Hashinaga, T.; Wada, N.; Kurita, Y.; Tanaka, K.; Hara, K.; et al. Pivotal Role of TNF-alpha in the Development and Progression of Nonalcoholic Fatty Liver Disease in a Murine Model. Horm. Metab. Res. 2018, 50, 80–87. [Google Scholar] [CrossRef]
- Tosello-Trampont, A.C.; Landes, S.G.; Nguyen, V.; Novobrantseva, T.I.; Hahn, Y.S. Kuppfer cells trigger nonalcoholic steatohepatitis development in diet-induced mouse model through tumor necrosis factor-alpha production. J. Biol. Chem. 2012, 287, 40161–40172. [Google Scholar] [CrossRef]
- Deng, F.; Tang, S.; Zhao, H.; Zhong, R.; Liu, L.; Meng, Q.; Zhang, H.; Chen, L. Combined effects of sodium butyrate and xylo-oligosaccharide on growth performance, anti-inflammatory and antioxidant capacity, intestinal morphology and microbiota of broilers at early stage. Poult. Sci. 2023, 102, 102585. [Google Scholar] [CrossRef]
- Fujii, H.; Kawada, N. Inflammation and fibrogenesis in steatohepatitis. J. Gastroenterol. 2012, 47, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Dooley, S.; ten Dijke, P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012, 347, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Radun, R.; Trauner, M. Role of FXR in Bile Acid and Metabolic Homeostasis in NASH: Pathogenetic Concepts and Therapeutic Opportunities. Semin. Liver Dis. 2021, 41, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Geier, A.; Tiniakos, D.; Denk, H.; Trauner, M. From the origin of NASH to the future of metabolic fatty liver disease. Gut 2021, 70, 1570–1579. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; Moschetta, A. Metabolic Messengers: Fibroblast growth factor 15/19. Nat. Metab. 2019, 1, 588–594. [Google Scholar] [CrossRef]
- Chiang, J.Y. Bile acids: Regulation of synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef]
- Gottlieb, A.; Canbay, A. Why Bile Acids Are So Important in Non-Alcoholic Fatty Liver Disease (NAFLD) Progression. Cells 2019, 8, 1358. [Google Scholar] [CrossRef]
- Puri, P.; Daita, K.; Joyce, A.; Mirshahi, F.; Santhekadur, P.K.; Cazanave, S.; Luketic, V.A.; Siddiqui, M.S.; Boyett, S.; Min, H.K.; et al. The presence and severity of nonalcoholic steatohepatitis is associated with specific changes in circulating bile acids. Hepatology 2018, 67, 534–548. [Google Scholar] [CrossRef]
- Perez, M.J.; Briz, O. Bile-acid-induced cell injury and protection. World J. Gastroenterol. 2009, 15, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Sanchez, N.; Cruz-Ramon, V.C.; Ramirez-Perez, O.L.; Hwang, J.P.; Barranco-Fragoso, B.; Cordova-Gallardo, J. New Aspects of Lipotoxicity in Nonalcoholic Steatohepatitis. Int. J. Mol. Sci. 2018, 19, 2034. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Ratziu, V.; Loomba, R.; Rinella, M.; Anstee, Q.M.; Goodman, Z.; Bedossa, P.; Geier, A.; Beckebaum, S.; Newsome, P.N.; et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: Interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2019, 394, 2184–2196. [Google Scholar] [CrossRef]
- Jena, P.K.; Sheng, L.; Li, Y.; Wan, Y.Y. Probiotics VSL#3 are effective in reversing non-alcoholic steatohepatitis in a mouse model. Hepatobiliary Surg. Nutr. 2020, 9, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Sayin, S.I.; Wahlstrom, A.; Felin, J.; Jantti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyotylainen, T.; Oresic, M.; Backhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Geng, W.; Lin, J. Bacterial bile salt hydrolase: An intestinal microbiome target for enhanced animal health. Anim. Health Res. Rev. 2016, 17, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef]
- Zhao, D.; Cao, J.; Jin, H.; Shan, Y.; Fang, J.; Liu, F. Beneficial impacts of fermented celery (Apium graveolens L.) juice on obesity prevention and gut microbiota modulation in high-fat diet fed mice. Food Funct. 2021, 12, 9151–9164. [Google Scholar] [CrossRef]
- Chen, M.; Hui, S.; Lang, H.; Zhou, M.; Zhang, Y.; Kang, C.; Zeng, X.; Zhang, Q.; Yi, L.; Mi, M. SIRT3 Deficiency Promotes High-Fat Diet-Induced Nonalcoholic Fatty Liver Disease in Correlation with Impaired Intestinal Permeability through Gut Microbial Dysbiosis. Mol. Nutr. Food Res. 2019, 63, e1800612. [Google Scholar] [CrossRef]
- Claesson, M.J.; O’Sullivan, O.; Wang, Q.; Nikkila, J.; Marchesi, J.R.; Smidt, H.; de Vos, W.M.; Ross, R.P.; O’Toole, P.W. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE 2009, 4, e6669. [Google Scholar] [CrossRef]
- Tsuchida, T.; Lee, Y.A.; Fujiwara, N.; Ybanez, M.; Allen, B.; Martins, S.; Fiel, M.I.; Goossens, N.; Chou, H.I.; Hoshida, Y.; et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018, 69, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Deng, L.; Wei, W.; Li, C.; Lu, Y.; Bai, J.; Li, L.; Zhang, H.; Jin, N.; Li, C.; et al. Lactiplantibacillus plantarum LPJZ-658 Improves Non-Alcoholic Steatohepatitis by Modulating Bile Acid Metabolism and Gut Microbiota in Mice. Int. J. Mol. Sci. 2023, 24, 13997. https://doi.org/10.3390/ijms241813997
Liu L, Deng L, Wei W, Li C, Lu Y, Bai J, Li L, Zhang H, Jin N, Li C, et al. Lactiplantibacillus plantarum LPJZ-658 Improves Non-Alcoholic Steatohepatitis by Modulating Bile Acid Metabolism and Gut Microbiota in Mice. International Journal of Molecular Sciences. 2023; 24(18):13997. https://doi.org/10.3390/ijms241813997
Chicago/Turabian StyleLiu, Liming, Liquan Deng, Wei Wei, Chunhua Li, Yuting Lu, Jieying Bai, Letian Li, Heping Zhang, Ningyi Jin, Chang Li, and et al. 2023. "Lactiplantibacillus plantarum LPJZ-658 Improves Non-Alcoholic Steatohepatitis by Modulating Bile Acid Metabolism and Gut Microbiota in Mice" International Journal of Molecular Sciences 24, no. 18: 13997. https://doi.org/10.3390/ijms241813997
APA StyleLiu, L., Deng, L., Wei, W., Li, C., Lu, Y., Bai, J., Li, L., Zhang, H., Jin, N., Li, C., & Zhao, C. (2023). Lactiplantibacillus plantarum LPJZ-658 Improves Non-Alcoholic Steatohepatitis by Modulating Bile Acid Metabolism and Gut Microbiota in Mice. International Journal of Molecular Sciences, 24(18), 13997. https://doi.org/10.3390/ijms241813997






