Mechanism of Escherichia coli Lethality Caused by Overexpression of flhDC, the Flagellar Master Regulator Genes, as Revealed by Transcriptome Analysis
Abstract
:1. Introduction
2. Results
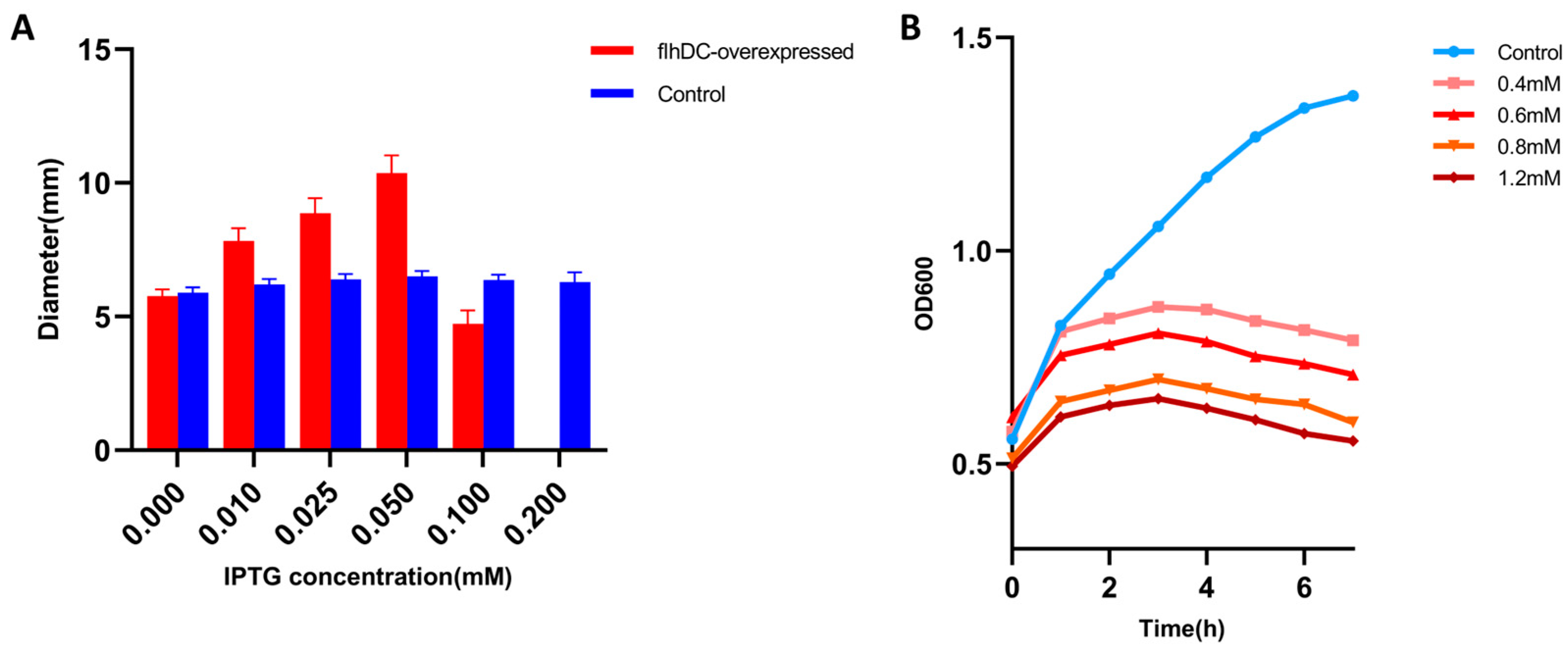
2.1. Overexpression of flhDC Can Be Lethal to Host Hacteria
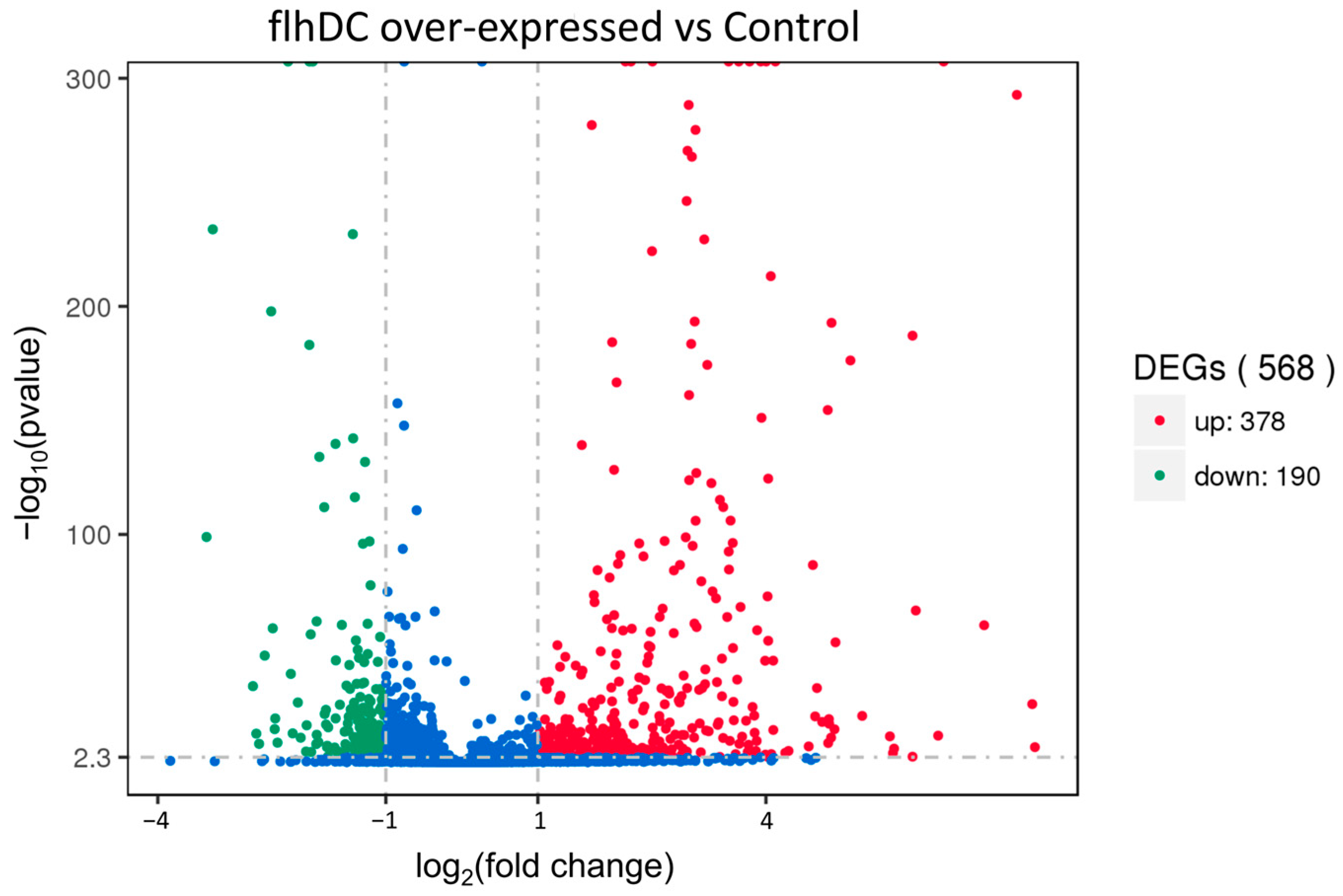
2.2. Analysis of Differentially Expressed Genes
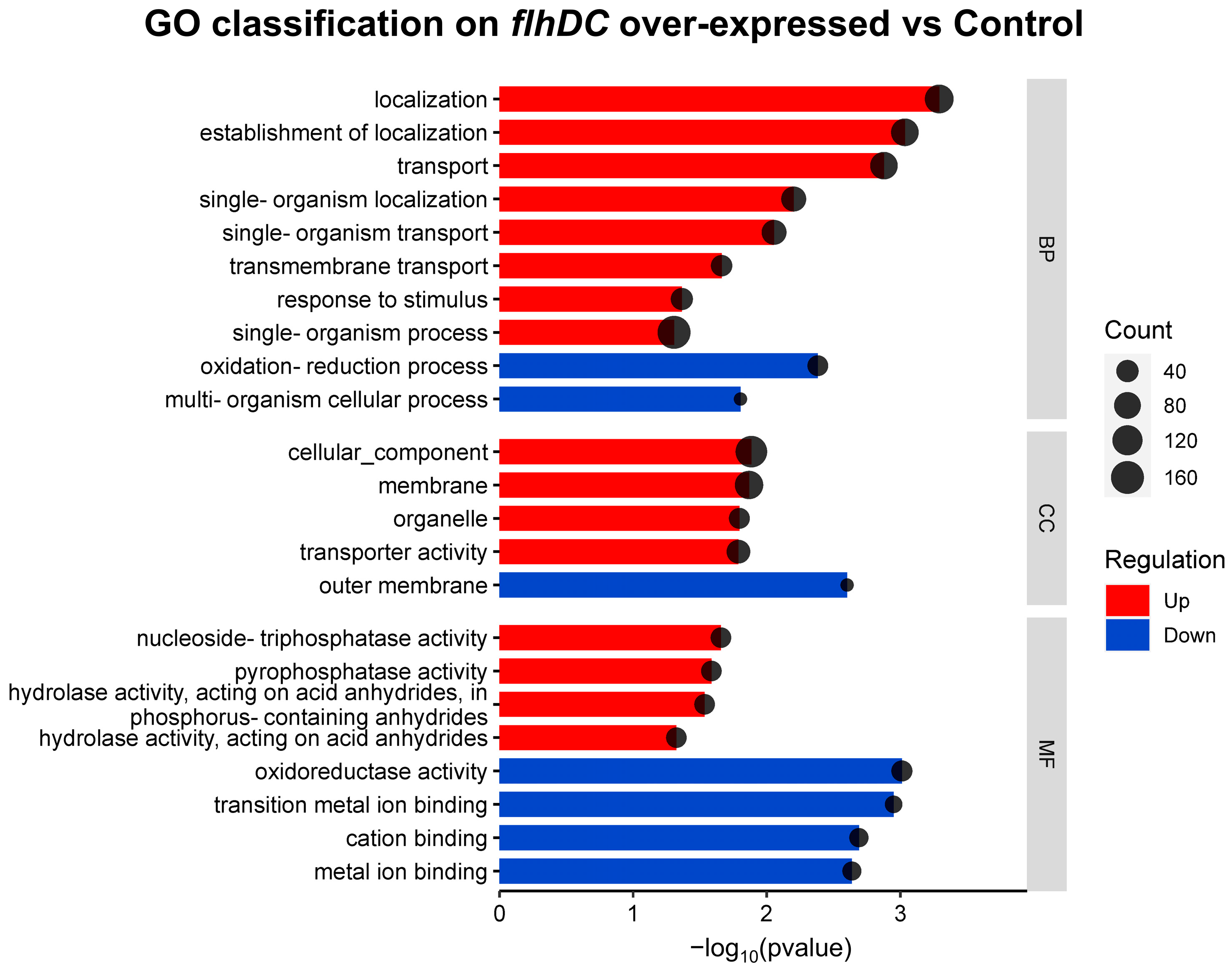
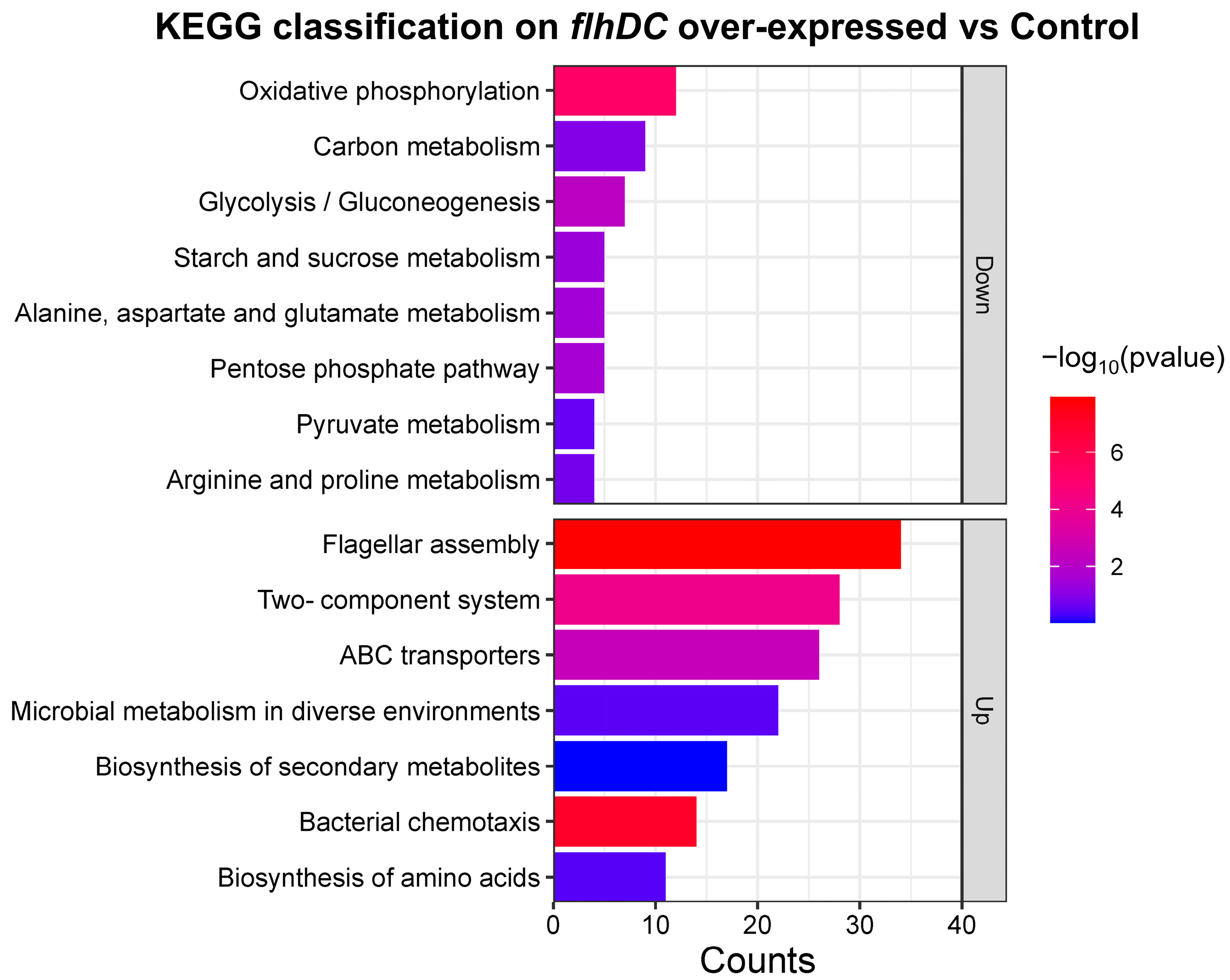
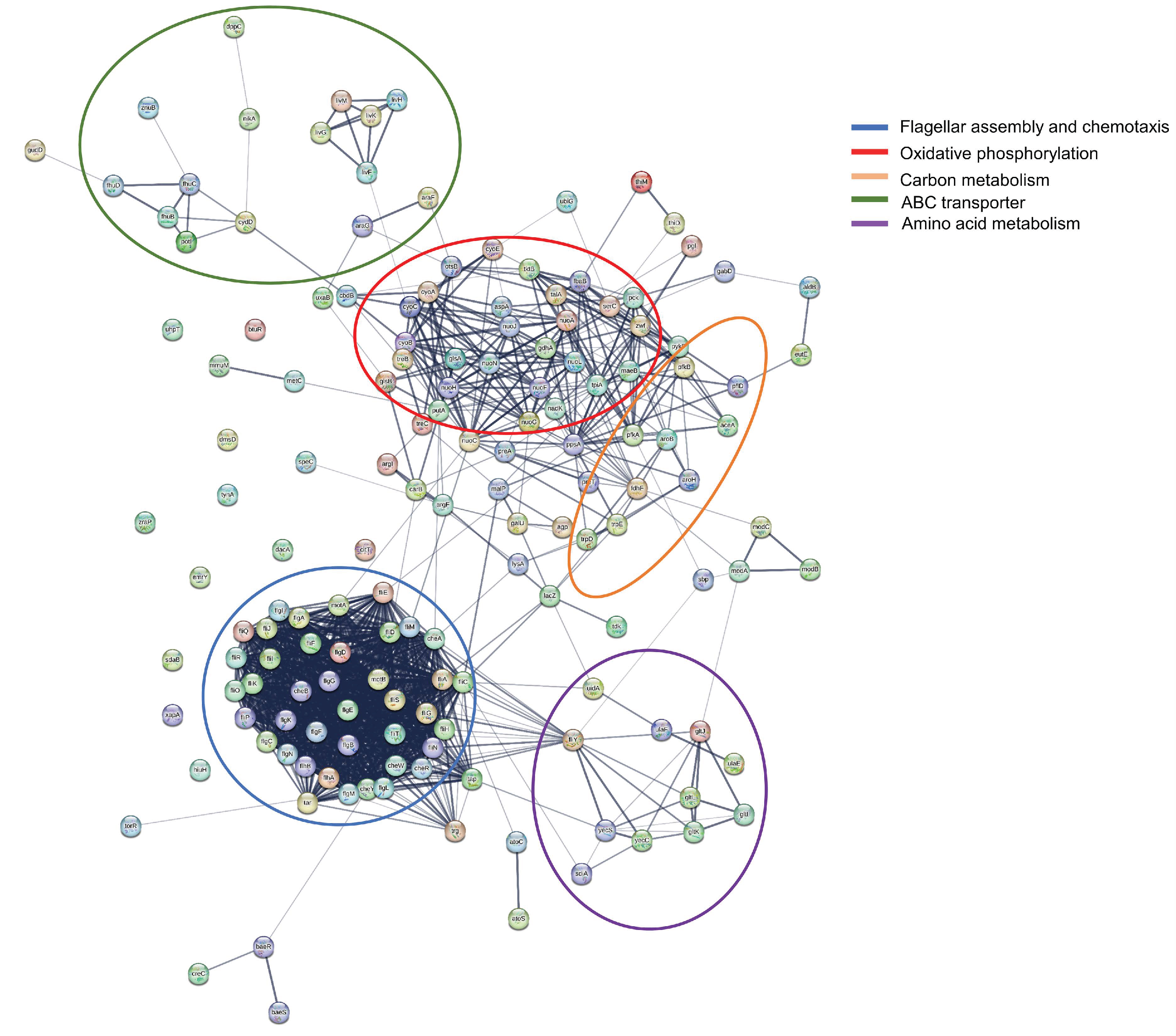
2.3. Functional Enrichment Analysis of DEGs
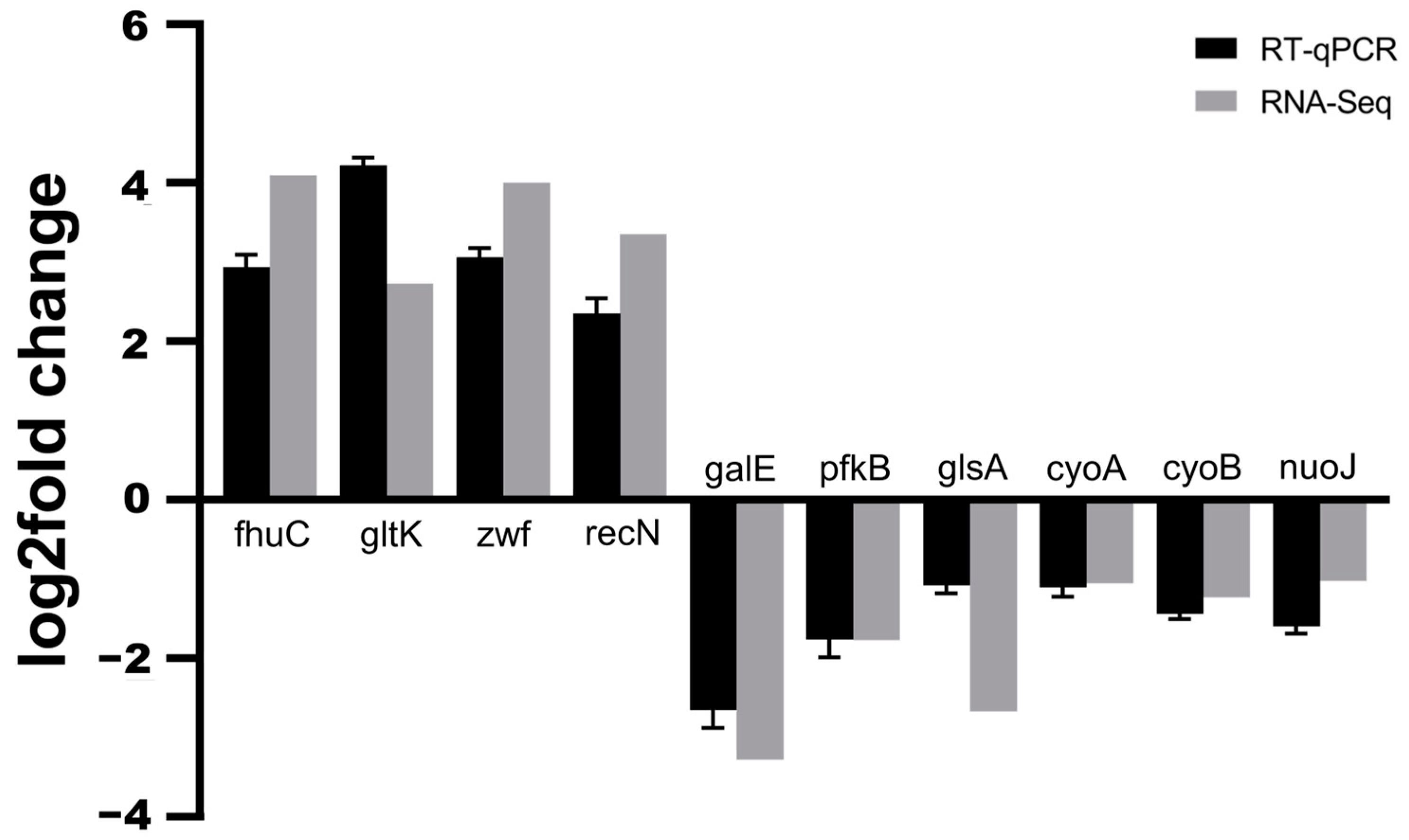
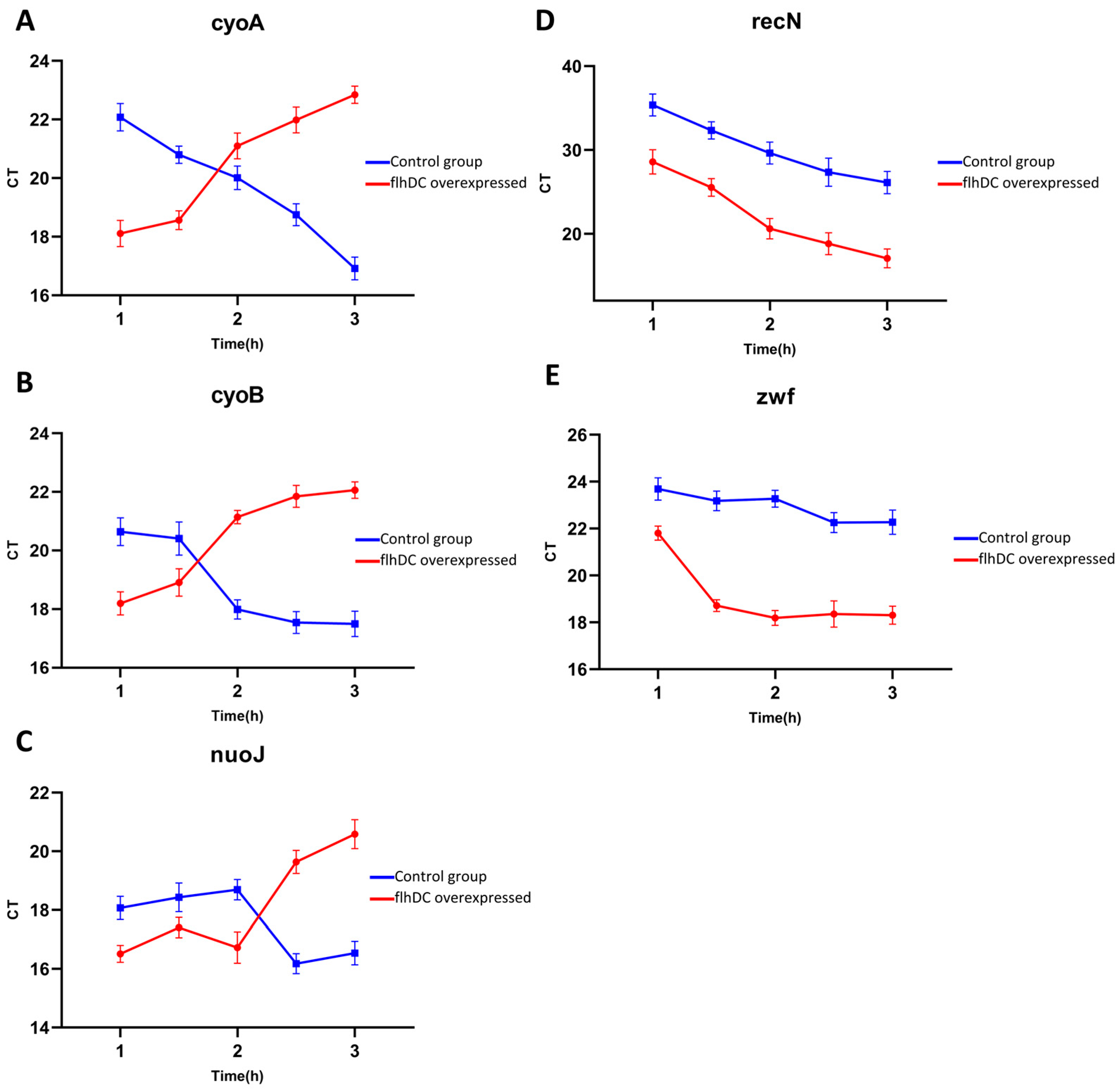
2.4. Quantitative Real-Time PCR Validation
2.5. Excessive Expression of the Oxidative Phosphorylation Pathway Is the Primary Cause of Bacterial Eell Death
2.6. Excessive Oxidative Phosphorylation Leads to High ROS Production
2.7. Cellular Response to ROS Damage
3. Discussion
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Growth of flhDC-E. coli Recombinant Bacteria on Soft Agar and LB Liquid
4.3. RNA Extraction, Library Preparation, and Sequencing
4.4. Bioinformatic Data Analysis
4.5. Differential Expression Genes (DEGs) Analysis and Annotation
4.6. Functional and Protein-Protein Interaction (PPI) Analysis of DEGs
4.7. Reverse Transcription and Quantitative Real-Time RT-PCR (qRT-PCR)
4.8. ROS Detection by Flow Cytometry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Minamino, T.; Macnab, R.M. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 1999, 181, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Zhou, M.; Zhu, L.; Zhu, G. Flagella and bacterial pathogenicity. J. Basic Microbiol. 2013, 53, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Macnab, R.M. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 1992, 26, 131–158. [Google Scholar] [CrossRef] [PubMed]
- Chevance, F.F.V.; Hughes, K.T. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 2008, 6, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Merino, S.; Shaw, J.G.; Tomás, J.M. Bacterial lateral flagella: An inducible flagella system. FEMS Microbiol. Lett. 2006, 263, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Okumura, M.; Kobayashi, N.; Nakamura, D.; Iwakura, M.; Aizawa, S.I. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol. Microbiol. 1997, 24, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.L.; Brun-Zinkernagel, A.M.; Simecka, J.W.; Prüß, B.M.; Babitzke, P.; Romeo, T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 2001, 40, 245–256. [Google Scholar] [CrossRef]
- Deng, W.; Marshall, N.C.; Rowland, J.L.; McCoy, J.M.; Worrall, L.J.; Santos, A.S.; Strynadka, N.C.; Finlay, B.B. Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 2017, 15, 323–337. [Google Scholar] [CrossRef]
- Macnab, R.M. How bacteria assemble flagella. Annu. Rev. Microbiol. 2003, 57, 77–100. [Google Scholar] [CrossRef]
- Ohnishi, K.; Kutsukake, K.; Suzuki, H.; Lino, T. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: An antisigma factor inhibits the activity of the flagellum-specific sigma factor, sigma F. Mol. Microbiol. 1992, 6, 3149–3157. [Google Scholar] [CrossRef]
- Kundu, T.K.; Kusano, S.; Ishihama, A. Promoter selectivity of Escherichia coli RNA polymerase sigmaF holoenzyme involved in transcription of flagellar and chemotaxis genes. J. Bacteriol. 1997, 179, 4264–4269. [Google Scholar] [CrossRef] [PubMed]
- Stafford, G.P.; Ogi, T.; Hughes, C. Binding and transcriptional activation of non-flagellar genes by the Escherichia coli flagellar master regulator FlhD2C2. Microbiology 2005, 151, 1779–1788. [Google Scholar] [CrossRef] [PubMed]
- Prüβ, B.M.; Campbell, J.W.; Van Dyk, T.K.; Zhu, C.; Kogan, Y.; Matsumura, P. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 2003, 185, 534–543. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.; Tremblay, J.; Déziel, E. Swarming motility: A multicellular behaviour conferring antimicrobial resistance. Environ. Microbiol. 2009, 11, 126–136. [Google Scholar] [CrossRef]
- Stabryla, L.M.; Johnston, K.A.; Diemler, N.A.; Cooper, V.S.; Millstone, J.E.; Haig, S.J.; Gilbertson, L.M. Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions. Nat. Nanotechnol. 2021, 16, 996–1003. [Google Scholar] [CrossRef]
- Butler, M.T.; Wang, Q.; Harshey, R.M. Cell density and mobility protect swarming bacteria against antibiotics. Proc. Natl. Acad. Sci. USA 2010, 107, 3776–3781. [Google Scholar] [CrossRef]
- Gustavsson, N.; Diez, A.; Nyström, T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 2002, 43, 107–117. [Google Scholar] [CrossRef]
- Zhang, Z.; Kukita, C.; Humayun, M.Z.; Saier, M.H. Environment-directed activation of the Escherichia coli flhDC operon by transposons. Microbiology 2017, 163, 554–569. [Google Scholar] [CrossRef]
- Ko, M.; Park, C. H-NS-Dependent Regulation of Flagellar Synthesis Is Mediated by a LysR Family Protein. J. Bacteriol. 2000, 182, 4670–4672. [Google Scholar] [CrossRef]
- Lehti, T.A.; Bauchart, P.; Dobrindt, U.; Korhonen, T.K.; Westerlund-Wikström, B. The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC. Microbiology 2012, 158, 1444–1455. [Google Scholar] [CrossRef]
- Francez-Charlot, A.; Laugel, B.; Van Gemert, A.; Dubarry, N.; Wiorowski, F.; Castanié-Cornet, M.P.; Gutierrez, C.; Cam, K. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli: Regulation of flhDC by RcsB. Mol. Microbiol. 2004, 49, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Lehti, T.A.; Heikkinen, J.; Korhonen, T.K.; Westerlund-Wikström, B. The Response Regulator RcsB Activates Expression of Mat Fimbriae in Meningitic Escherichia coli. J. Bacteriol. 2012, 194, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Pesavento, C.; Hengge, R. The global repressor FliZ antagonizes gene expression by σS-containing RNA polymerase due to overlapping DNA binding specificity. Nucleic Acids Res. 2012, 40, 4783–4793. [Google Scholar] [CrossRef] [PubMed]
- Fahrner, K.A.; Berg, H.C. Mutations That Stimulate flhDC Expression in Escherichia coli K-12. J. Bacteriol. 2015, 197, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Torres, A.G.; Kaper, J.B. Quorum sensing Escherichia coli regulators B and C (QseBC): A novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 2002, 43, 809–821. Available online: https://pubmed.ncbi.nlm.nih.gov/11929534/ (accessed on 12 October 2022). [CrossRef]
- Yamada, H.; Muramatsu, S.; Mizuno, T. An Escherichia coli protein that preferentially binds to sharply curved DNA. J. Biochem. 1990, 108, 420–425. [Google Scholar] [CrossRef]
- Pérez-Martín, J.; Rojo, F.; de Lorenzo, V. Promoters responsive to DNA bending: A common theme in prokaryotic gene expression. Microbiol. Rev. 1994, 58, 268–290. [Google Scholar] [CrossRef]
- Lehnen, D.; Blumer, C.; Polen, T.; Wackwitz, B.; Wendisch, V.F.; Unden, G. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 2002, 45, 521–532. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, M.; Burgess, R.R. Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 2010, 38, 1273–1283. [Google Scholar] [CrossRef]
- Djordjevic, M. Redefining Escherichia coli σ(70) promoter elements: -15 motif as a complement of the -10 motif. J. Bacteriol. 2011, 193, 6305–6314. [Google Scholar] [CrossRef]
- Fitzgerald, D.M.; Bonocora, R.P.; Wade, J.T. Comprehensive Mapping of the Escherichia coli Flagellar Regulatory Network. PLoS Genet. 2014, 10, e1004649. [Google Scholar] [CrossRef]
- De Lay, N.; Gottesman, S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012, 86, 524–538. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Strakhova, T.S.; Smagin, V.A.; Ravin, N.V. Tightly regulated, high-level expression from controlled copy number vectors based on the replicon of temperate phage N15. Gene 2007, 395, 15–21. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Yang, S.; Lian, G. ROS and diseases: Role in metabolism and energy supply. Mol. Cell. Biochem. 2020, 467, 1–12. [Google Scholar] [CrossRef]
- Gupta, N.; Verma, K.; Nalla, S.; Kulshreshtha, A.; Lall, R.; Prasad, S. Free Radicals as a Double-Edged Sword: The Cancer Preventive and Therapeutic Roles of Curcumin. Molecules 2020, 25, 5390. [Google Scholar] [CrossRef]
- Angelova, P.R.; Dinkova-Kostova, A.T.; Abramov, A.Y. Assessment of ROS Production in the Mitochondria of Live Cells. Methods Mol. Biol. Clifton NJ 2021, 2202, 33–42. [Google Scholar]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Cardenas, P.P.; Gándara, C.; Alonso, J.C. DNA double strand break end-processing and RecA induce RecN expression levels in Bacillus subtilis. DNA Repair 2014, 14, 1–8. [Google Scholar] [CrossRef]
- Walsh, C. Where will new antibiotics come from? Nat. Rev. Microbiol. 2003, 1, 65–70. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A Common Mechanism of Cellular Death Induced by Bactericidal Antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Camon, E. The Gene Ontology Annotation (GOA) Database: Sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 2004, 32, D262–D266. [Google Scholar] [CrossRef]
- Zhou, B.B.; Elledge, S.J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–439. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, M.R.G.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Altenhöfer, S.; Radermacher, K.A.; Kleikers, P.W.M.; Wingler, K.; Schmidt, H.H.H.W. Evolution of NADPH Oxidase Inhibitors: Selectivity and Mechanisms for Target Engagement. Antioxid. Redox Signal. 2015, 23, 406–427. [Google Scholar] [CrossRef]
- Lin, Y.-H. MicroRNA Networks Modulate Oxidative Stress in Cancer. Int. J. Mol. Sci. 2019, 20, 4497. [Google Scholar] [CrossRef]
- Checa, J.; Aran, J.M. Reactive Oxygen Species: Drivers of Physiological and Pathological Processes. J. Inflamm. Res. 2020, 13, 1057–1073. [Google Scholar] [CrossRef]
- Alonso, J.C.; Cardenas, P.P.; Sanchez, H.; Hejna, J.; Suzuki, Y.; Takeyasu, K. Early steps of double-strand break repair in Bacillus subtilis. DNA Repair 2013, 12, 162–176. [Google Scholar] [CrossRef]
- Nicolas, P.; Mäder, U.; Dervyn, E.; Rochat, T.; Leduc, A.; Pigeonneau, N.; Bidnenko, E.; Marchadier, E.; Hoebeke, M.; Aymerich, S.; et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 2012, 335, 1103–1106. [Google Scholar] [CrossRef]
- Ighodaro, O.M. Molecular pathways associated with oxidative stress in diabetes mellitus. Biomed. Pharmacother. 2018, 108, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Chaput, V.; Martin, A.; Lejay, L. Redox metabolism: The hidden player in carbon and nitrogen signaling? J. Exp. Bot. 2020, 71, 3816–3826. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Wamelink, M.M.; Kowald, A.; Gerisch, B.; Heeren, G.; Struys, E.A.; Klipp, E.; Jakobs, C.; Breitenbach, M.; Lehrach, H.; et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6, 10. [Google Scholar] [CrossRef]
- Anastasiou, D.; Poulogiannis, G.; Asara, J.M.; Boxer, M.B.; Jiang, J.K.; Shen, M.; Bellinger, G.; Sasaki, A.T.; Locasale, J.W.; Auld, D.S.; et al. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses. Science 2011, 334, 1278–1283. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Wamelink, M.M.; Latkolik, S.; Jansen, E.E.; Lehrach, H.; Jakobs, C. Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat. Biotechnol. 2009, 27, 604–605. [Google Scholar] [CrossRef]
- Grant, C.M. Metabolic reconfiguration is a regulated response to oxidative stress. J. Biol. 2008, 7, 1. [Google Scholar] [CrossRef]

| Sample | flhDC Over-Expressed | Control | |
|---|---|---|---|
| Parameter | |||
| Total raw reads | 16,354,716 | 15,013,008 | |
| Total clean reads | 15,980,318 | 14,770,190 | |
| Mapped reads | 13,683,571 | 14,396,239 | |
| Mapped rate (%) | 85.63% | 97.47% | |
| FPKM 0−1 | 383 (8.51%) | 471 (10.47%) | |
| FPKM 1−3 | 221 (4.91%) | 324 (7.20%) | |
| FPKM 3−15 | 777 (17.27%) | 773 (17.19%) | |
| FPKM 15−60 | 1135 (25.23%) | 1018 (22.63%) | |
| FPKM > 60 | 1982 (44.06%) | 1912 (42.51%) | |
| Genes’ Name | log2fold Change | Description |
|---|---|---|
| recN | 3.3559 | DNA repair protein |
| recJ | 2.0536 | single-stranded-DNA-specific exonuclease |
| held | 1.1701 | DNA helicase IV |
| torR | 2.1721 | DNA-binding response regulator |
| sbmC | 4.8629 | DNA gyrase inhibitor |
| rsgA | 1.7846 | Small ribosomal subunit biogenesis GTPase |
| zwf | 4.0027 | Glucose-6-phosphate dehydrogenase |
| pfkB | −1.7747 | 6-phosphofructokinase |
| pykF | −1.0239 | pyruvate kinase |
| ppsA | 1.1415 | phosphoenolpyruvate synthase |
| pck | −1.0299 | phosphoenolpyruvate carboxykinase |
| fbaB | −1.2847 | fructose-diphosphate aldolase |
| maeB | 1.7846 | malic enzyme |
| Genes’ Name | Sequence (5′-3′) | Length | |
|---|---|---|---|
| cyoA | Forward | GTTCACTGATACTGACGGCAT | 121 bp |
| Reverse | TTCGGGCTGTACTTAGCATC | ||
| cyoB | Forward | TGGCCTGATCACTTACTTCGG | 138 bp |
| Reverse | ATAATGGCGTCAGCAAAACCAC | ||
| nuoJ | Forward | GCCATCCTCGGTGTTAACGAT | 114 bp |
| Reverse | CAGCATAGAAGCCAGTTCCAC | ||
| fhuC | Forward | TCTTGATGCCCAACCGCTGGA | 138 bp |
| Reverse | CCATGCCACGGGTAACGACCA | ||
| gltK | Forward | GGTTTGCCAAAGCCTACGTT | 111 bp |
| Reverse | GCGATAATCCCAGCACGTT | ||
| glsA | Forward | GTGGATCAGGCTTACACCCAA | 140 bp |
| Reverse | TCACCCGCACTATAGACGTTG | ||
| zwf | Forward | CCAGGTTTACCGTATCGACCAC | 112 bp |
| Reverse | ATCAATGGTGCGATTGTCCC | ||
| galE | Forward | AAGGCATTCCGAATAACCTGA | 100 bp |
| Reverse | CCATCTTCGGTCGGATAATCGTT | ||
| pfkB | Forward | ATGAAAATGTCCCCGTCGCTAC | 198 bp |
| Reverse | GCAGGCTTCCGCTTATGACCA | ||
| recN | Forward | GCCTATTGCCAAAGTCGCATC | 124 bp |
| Reverse | CTGTTGGACCGCTAATCCCT | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, G.; Yu, Z.; Li, Q.; Zhang, Y.; Wang, M.; Liu, Y.; Liu, J.; Liu, L.; Yu, X. Mechanism of Escherichia coli Lethality Caused by Overexpression of flhDC, the Flagellar Master Regulator Genes, as Revealed by Transcriptome Analysis. Int. J. Mol. Sci. 2023, 24, 14058. https://doi.org/10.3390/ijms241814058
Sun G, Yu Z, Li Q, Zhang Y, Wang M, Liu Y, Liu J, Liu L, Yu X. Mechanism of Escherichia coli Lethality Caused by Overexpression of flhDC, the Flagellar Master Regulator Genes, as Revealed by Transcriptome Analysis. International Journal of Molecular Sciences. 2023; 24(18):14058. https://doi.org/10.3390/ijms241814058
Chicago/Turabian StyleSun, Guanglu, Zihao Yu, Qianwen Li, Yuanxing Zhang, Mingxiao Wang, Yunhui Liu, Jinze Liu, Lei Liu, and Xuping Yu. 2023. "Mechanism of Escherichia coli Lethality Caused by Overexpression of flhDC, the Flagellar Master Regulator Genes, as Revealed by Transcriptome Analysis" International Journal of Molecular Sciences 24, no. 18: 14058. https://doi.org/10.3390/ijms241814058
APA StyleSun, G., Yu, Z., Li, Q., Zhang, Y., Wang, M., Liu, Y., Liu, J., Liu, L., & Yu, X. (2023). Mechanism of Escherichia coli Lethality Caused by Overexpression of flhDC, the Flagellar Master Regulator Genes, as Revealed by Transcriptome Analysis. International Journal of Molecular Sciences, 24(18), 14058. https://doi.org/10.3390/ijms241814058




