Comparison of Transcriptome between Tolerant and Susceptible Rice Cultivar Reveals Positive and Negative Regulators of Response to Rhizoctonia solani in Rice
Abstract
1. Introduction
2. Results
2.1. Screening of Rice Varieties for Sheath Blight Resistance
2.2. RNA-Seq Data and Alignment Analysis
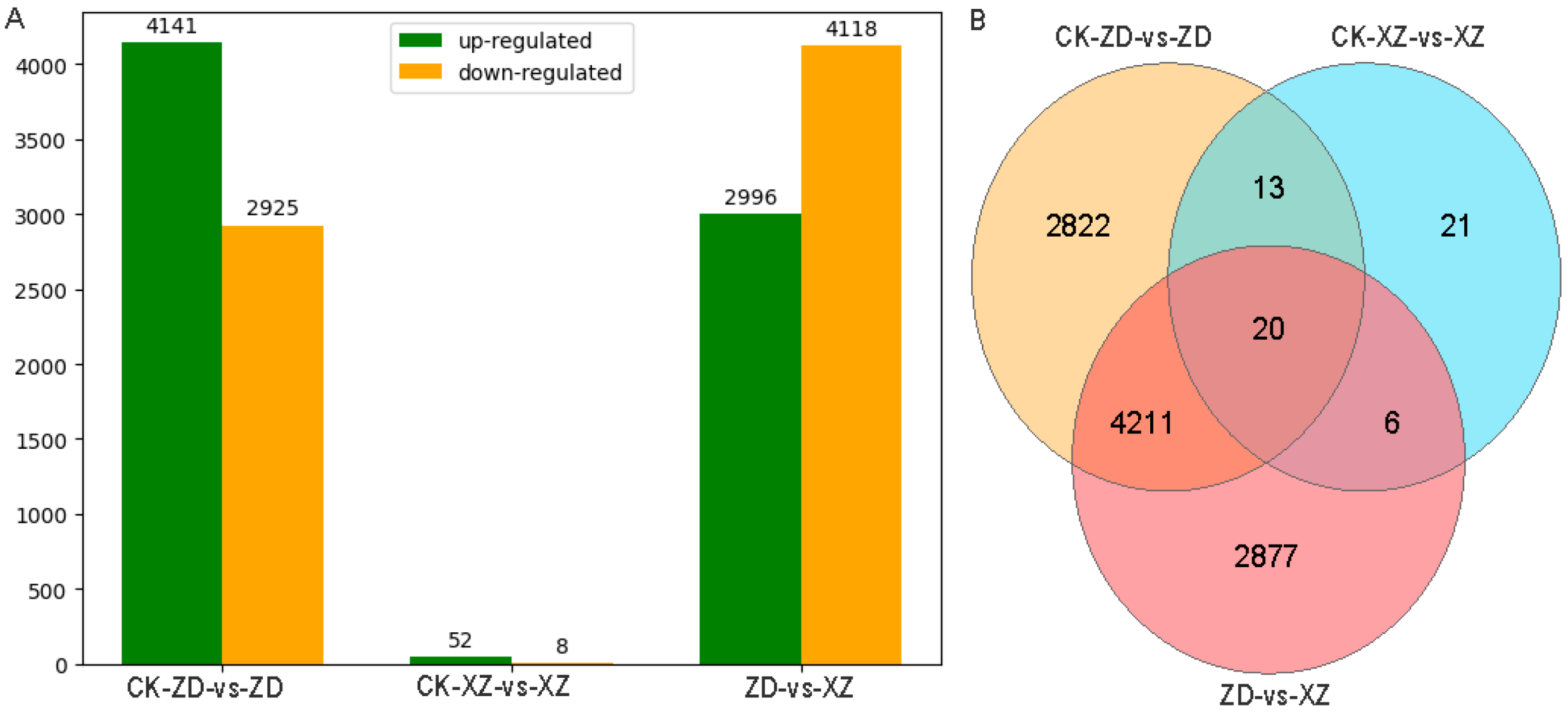
2.3. Differential Expression Gene Analysis of Tolerant and Susceptible Cultivar Response to R. solani
2.4. GO Enrichment Analysis of DEG Response to R. solani in Tolerant Cultivar ZD
2.5. KEGG Enrichment Analysis of DEG Response to R. solani in Tolerant Cultivar ZD
2.6. Analysis of DEGs of Transcription Factors Response to R. solani in Tolerant Cultivar ZD
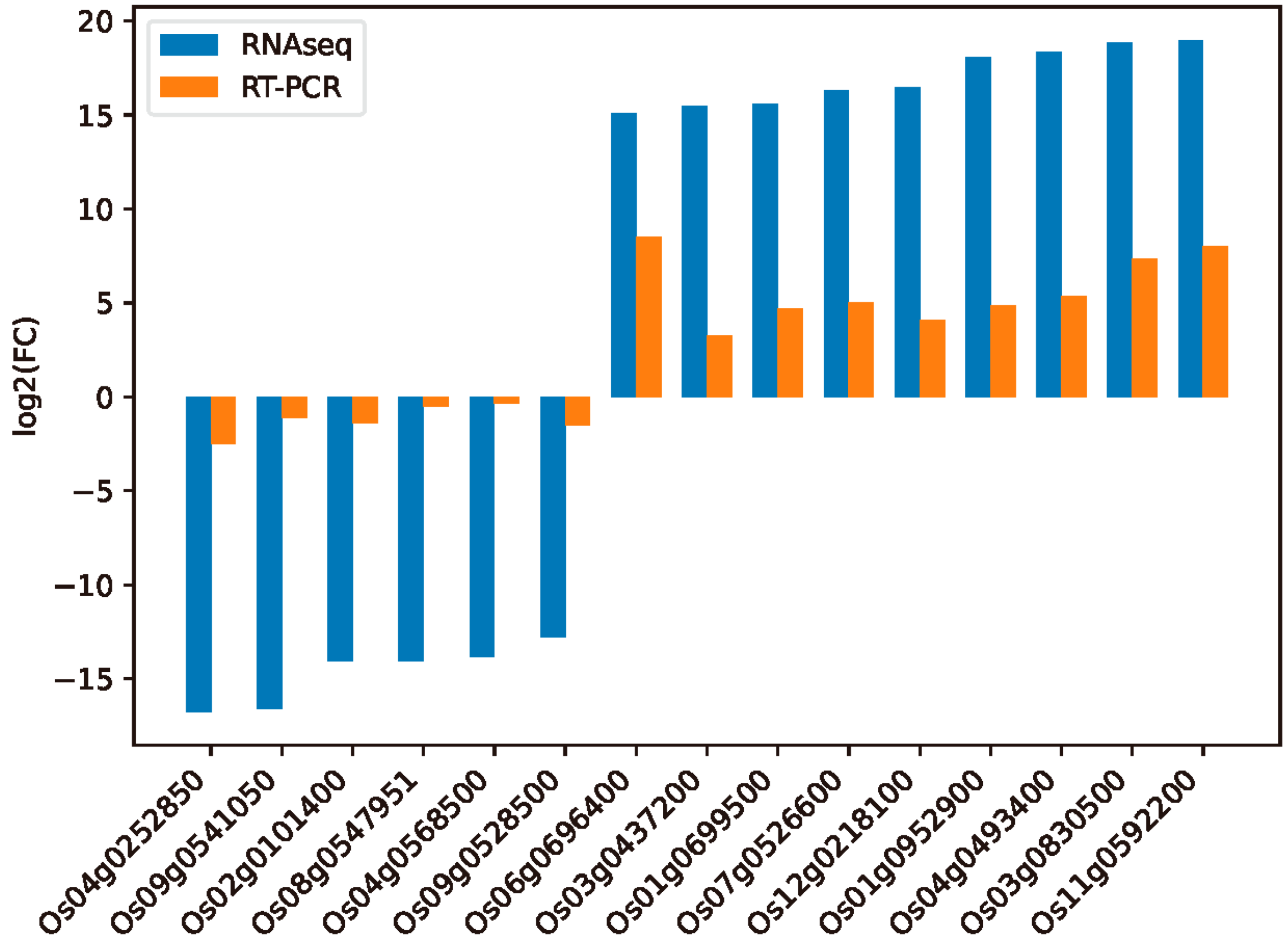
2.7. Validation of Differentially Expressed Genes through qRT-PCR
3. Discussion
3.1. Activation of Cell Surface Pattern-Recognition Receptors
3.2. Activation of Ca2+ Signaling Pathway in Response to R. solani
3.3. Activation of MAPK Cascade
3.4. Positive and Negative Regulators in Plant Hormone Signal Transduction
3.5. Activation and Inhibition of Transcription Factors
3.6. Chloroplast and Inhibition of Photosynthesis in Plant Immunity
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Pathogen Inoculation and Disease Scoring
4.3. Sample Collection, RNA Extraction, and RNA Sequencing
4.4. Differentially Expressed Genes Analysis and qPCR Validation
4.5. Functional Enrichment Analysis
4.6. Transcription Factors Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H.; et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.K.; Cunningham, A.A.; Patel, N.G.; Morales, F.J.; Epstein, P.R.; Daszak, P. Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 2004, 19, 535–544. [Google Scholar] [CrossRef]
- Shi, W.; Zhao, S.-L.; Liu, K.; Sun, Y.-B.; Ni, Z.-B.; Zhang, G.-Y.; Tang, H.-S.; Zhu, J.-W.; Wan, B.-J.; Sun, H.-Q.; et al. Comparison of leaf transcriptome in response to Rhizoctonia solani infection between resistant and susceptible rice cultivars. BMC Genom. 2020, 21, 245. [Google Scholar] [CrossRef] [PubMed]
- Taguchi-Shiobara, F.; Ozaki, H.; Sato, H.; Maeda, H.; Kojima, Y.; Ebitani, T.; Yano, M. Mapping and validation of QTLs for rice sheath blight resistance. Breed. Sci. 2013, 63, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.-J.; Wang, A.-R.; Shi, Y.-J.; Wang, L.-Q.; Liu, W.-D.; Wang, Z.-H.; Lu, G.-D. Identification of defense-related genes in rice responding to challenge by Rhizoctonia solani. Theor. Appl. Genet. 2008, 116, 501–516. [Google Scholar] [CrossRef]
- Brooks, S.A. Sensitivity to a phytotoxin from Rhizoctonia solani correlates with sheath blight susceptibility in rice. Phytopathology 2007, 97, 1207–1212. [Google Scholar] [CrossRef]
- Lin, R.; He, L.; He, J.; Qin, P.; Wang, Y.; Deng, Q.; Yang, X.; Li, S.; Wang, S.; Wang, W.; et al. Comprehensive analysis of microRNA-Seq and target mRNAs of rice sheath blight pathogen provides new insights into pathogenic regulatory mechanisms. DNA Res. 2016, 23, 415–425. [Google Scholar] [CrossRef]
- Liu, G.; Jia, Y.; McClung, A.; Oard, J.H.; Lee, F.N.; Correll, J.C.; Zeng, Y.; Dong, J.; Ji, Z.; Liang, Y.; et al. Confirming QTLs and finding additional loci responsible for resistance to rice sheath blight disease. Plant Dis. 2013, 97, 113–117. [Google Scholar] [CrossRef]
- Wang, A.; Shu, X.; Jing, X.; Jiao, C.; Chen, L.; Zhang, J.; Ma, L.; Jiang, Y.; Yamamoto, N.; Li, S.; et al. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 2021, 19, 1553–1566. [Google Scholar] [CrossRef]
- Peng, X.; Hu, Y.; Tang, X.; Zhou, P.; Deng, X.; Wang, H.; Guo, Z. Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 2012, 236, 1485–1498. [Google Scholar] [CrossRef]
- Peng, X.; Wang, H.; Jang, J.-C.; Xiao, T.; He, H.; Jiang, D.; Tang, X. OsWRKY80-OsWRKY4 Module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice 2016, 9, 63. [Google Scholar] [CrossRef]
- Tonnessen, B.W.; Manosalva, P.; Lang, J.M.; Baraoidan, M.; Bordeos, A.; Mauleon, R.; Oard, J.; Hulbert, S.; Leung, H.; Leach, J.E. Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 2015, 87, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Y.; Zhang, L.; He, Z.; Huang, B.; Chen, C.; Zhang, Q.; Zuo, S. Amino acid substitutions in a polygalacturonase inhibiting protein (OsPGIP2) increases sheath blight resistance in rice. Rice 2019, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, L.; Fu, C.; Wang, L.; Liu, H.; Cheng, Y.; Li, S.; Deng, Q.; Wang, S.; Zhu, J.; et al. Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection inresistant and susceptible rice varieties. Front. Plant Sci. 2017, 8, 1422. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Gu, X.; Ding, J.; Yao, L.; Gao, X.; Zhang, M.; Meng, Q.; Wei, S.; Fu, J. Gene expression analysis of resistant and susceptible rice cultivars to sheath blight after inoculation with Rhizoctonia solani. BMC Genom. 2022, 23, 278. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.-N.; Liu, R.; Li, T.; Li, M.; Liu, X.-Y.; Wang, W.-J.; Yu, Y.-K.; Cao, J.; Tan, X.-L. Physiological and comparative transcriptome analyses reveal the mechanisms underlying waterlogging tolerance in a rapeseed anthocyanin-more mutant. Biotechnol. Biofuels Bioprod. 2022, 15, 55. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ding, P.; Jones, J.D.G. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef]
- Naithani, S.; Dikeman, D.; Garg, P.; Al-Bader, N.; Jaiswal, P. Beyond gene ontology (GO): Using biocuration approach to improve the gene nomenclature and functional annotation of rice S-domain kinase subfamily. PeerJ 2021, 9, e11052. [Google Scholar] [CrossRef]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef]
- Kouzai, Y.; Kimura, M.; Watanabe, M.; Kusunoki, K.; Osaka, D.; Suzuki, T.; Matsui, H.; Yamamoto, M.; Ichinose, Y.; Toyoda, K.; et al. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018, 217, 771–783. [Google Scholar] [CrossRef]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.-F.; Wang, M.; Wang, J.; Liu, B.; Wang, H.-B. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y.; Nakagawa, H.; Nishizawa, Y.; Kamakura, T.; Mori, M. Broad-spectrum disease resistance conferred by the overexpression of rice RLCK BSR1 results from an enhanced immune response to multiple MAMPs. Int. J. Mol. Sci. 2019, 20, 5523. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhao, L.; Qi, F.; Htwe, N.M.P.S.; Li, Q.; Zhang, D.; Lin, F.; Shang-Guan, K.; Liang, Y. The receptor-like cytoplasmic kinase RIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol. Plant 2021, 14, 1652–1667. [Google Scholar] [CrossRef]
- Li, Z.; Ao, Y.; Feng, D.; Liu, J.; Wang, J.; Wang, H.-B.; Liu, B. OsRLCK 57, OsRLCK107 and OsRLCK118 positively regulate chitin- and PGN-induced immunity in Rice. Rice 2017, 10, 6. [Google Scholar] [CrossRef] [PubMed]
- Delteil, A.; Gobbato, E.; Cayrol, B.; Estevan, J.; Michel-Romiti, C.; Dievart, A.; Kroj, T.; Morel, J.-B. Several wall-associated kinases participate positively and negatively in basal defense against rice blast fungus. BMC Plant Biol. 2016, 16, 17. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Li, L.; Meng, L.; Singh, J.; Jiang, N.; Deng, X.-W.; He, Z.-H.; Lemaux, P.G. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005, 139, 1107–1124. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 2001, re22. [Google Scholar] [CrossRef]
- Wang, J.; Qu, B.; Dou, S.; Li, L.; Yin, D.; Pang, Z.; Zhou, Z.; Tian, M.; Liu, G.; Xie, Q.; et al. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015, 15, 49. [Google Scholar] [CrossRef]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510.e5. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020, 225, 87–104. [Google Scholar] [CrossRef]
- Yadav, M.; Pandey, J.; Chakraborty, A.; Hassan, I.; Kundu, J.K.; Roy, A.; Singh, I.K.; Singh, A. A comprehensive analysis of calmodulin-like proteins of glycine max indicates their role in calcium signaling and plant defense against insect attack. Front. Plant Sci. 2022, 13, 817950. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Pottosin, I.I.; Shabala, S.S.; Palmgren, M.G.; Shabala, S. calcium efflux systems in stress signaling and adaptation in plants. Front. Plant Sci. 2011, 2, 85. [Google Scholar] [CrossRef] [PubMed]
- Bhar, A.; Chakraborty, A.; Roy, A. The captivating role of calcium in plant-microbe interaction. Front. Plant Sci. 2023, 14, 1138252. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Mohanta, N.; Mohanta, Y.K.; Parida, P.; Bae, H. Genome-wide identification of Calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 2015, 15, 189. [Google Scholar] [CrossRef] [PubMed]
- Luan, S.; Kudla, J.; Rodriguez-Concepcion, M.; Yalovsky, S.; Gruissem, W. Calmodulins and calcineurin b–like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 2002, 14 (Suppl. S1), S389–S400. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wu, Z.; Tian, H.; Li, X.; Zhang, Y. Arabidopsis CALMODULIN-BINDING PROTEIN 60b plays dual roles in plant immunity. Plant Commun. 2021, 2, 100213. [Google Scholar] [CrossRef]
- Tuteja, N.; Mahajan, S. Calcium signaling network in plants: An overview. Plant Signal. Behav. 2007, 2, 79–85. [Google Scholar] [CrossRef]
- Vadassery, J.; Scholz, S.S.; Mithöfer, A. Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion. Plant Signal. Behav. 2012, 7, 1277–1280. [Google Scholar] [CrossRef]
- Zhu, X.; Robe, E.; Jomat, L.; Aldon, D.; Mazars, C.; Galaud, J.-P. CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol. 2017, 58, 307–319. [Google Scholar] [CrossRef][Green Version]
- Zhu, K.; Fan, P.; Liu, H.; Tan, P.; Ma, W.; Mo, Z.; Zhao, J.; Chu, G.; Peng, F. Insight into the CBL and CIPK gene families in pecan (Carya illinoinensis): Identification, evolution and expression patterns in drought response. BMC Plant Biol. 2022, 22, 221. [Google Scholar] [CrossRef]
- Yu, C.; Ke, Y.; Qin, J.; Huang, Y.; Zhao, Y.; Liu, Y.; Wei, H.; Liu, G.; Lian, B.; Chen, Y.; et al. Genome-wide identification of calcineurin B-like protein-interacting protein kinase gene family reveals members participating in abiotic stress in the ornamental woody plant Lagerstroemia indica. Front. Plant Sci. 2022, 13, 942217. [Google Scholar] [CrossRef]
- Kurusu, T.; Hamada, J.; Hamada, H.; Hanamata, S.; Kuchitsu, K. Roles of calcineurin B-like protein-interacting protein kinases in innate immunity in rice. Plant Signal. Behav. 2010, 5, 1045–1047. [Google Scholar] [CrossRef] [PubMed]
- Freymark, G.; Diehl, T.; Miklis, M.; Romeis, T.; Panstruga, R.; Bredow, M.; Monaghan, J.; Kusch, S.; Moscou, M.J.; Lauter, N.; et al. Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol. Plant-Microbe Interact. 2007, 20, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-L.; Huang, H.-J. ROS and CDPK-like kinase-mediated activation of MAP kinase in rice roots exposed to lead. Chemosphere 2008, 71, 1377–1385. [Google Scholar] [CrossRef]
- Zhang, M.; Su, J.; Zhang, Y.; Xu, J.; Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 2018, 45, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, Z.; Li, J.; Xie, W.; Qin, X.; Kang, Y.; Zhang, Q.; Li, X.; Xiao, J.; Ma, H.; et al. Two VQ Proteins are substrates of the OsMPKK6-OsMPK4 cascade in rice defense against bacterial blight. Rice 2021, 14, 39. [Google Scholar] [CrossRef]
- Suarez Rodriguez, M.C.; Petersen, M.; Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [CrossRef]
- Yang, Z.; Ma, H.; Hong, H.; Yao, W.; Xie, W.; Xiao, J.; Li, X.; Wang, S. Transcriptome-based analysis of mitogen-activated protein kinase cascades in the rice response to Xanthomonas oryzae infection. Rice 2015, 8, 4. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Yuan, M. Update on the roles of rice MAPK cascades. Int. J. Mol. Sci. 2021, 22, 1679. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Zhang, C.; Zhu, P.; Dai, H.; Yu, N.; He, Z.; Xu, L.; Wang, E. OsCERK1-mediated chitin perception and immune signaling requires receptor-like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant 2017, 10, 619–633. [Google Scholar] [CrossRef]
- Ueno, Y.; Yoshida, R.; Kishi-Kaboshi, M.; Matsushita, A.; Jiang, C.-J.; Goto, S.; Takahashi, A.; Hirochika, H.; Takatsuji, H. Abiotic stresses antagonize the rice defence pathway through the tyrosine-dephosphorylation of OsMPK6. PLoS Pathog. 2015, 11, e1005231. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, H.; Yuan, B.; Li, X.; Xu, C.; Wang, S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011, 34, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Chern, M.; Bartley, L.E.; Han, M.; Jung, K.-H.; Lee, I.; Walia, H.; Richter, T.; Xu, X.; Cao, P.; et al. Towards establishment of a rice stress response interactome. PLoS Genet. 2011, 7, e1002020. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Liu, Q.; Cao, Y.; Zhang, Y.; Chen, D.; Lou, X.; Cheng, S.; Cao, L. The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice. Front. Plant Sci. 2019, 10, 752. [Google Scholar] [CrossRef]
- Nguyen, H.T.; To, H.T.M.; Lebrun, M.; Bellafiore, S.; Champion, A. Jasmonates-The master regulator of rice development, adaptation and defense. Plants 2019, 8, 339. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed]
- Tezuka, D.; Kawamata, A.; Kato, H.; Saburi, W.; Mori, H.; Imai, R. The rice ethylene response factor OsERF83 positively regulates disease resistance to Magnaporthe oryzae. Plant Physiol. Biochem. 2019, 135, 263–271. [Google Scholar] [CrossRef]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping ethylene response: The role of EIN3/EIL1 transcription factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef]
- Huang, P.-Y.; Catinot, J.; Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 2016, 67, 1231–1241. [Google Scholar] [CrossRef]
- Kunkel, B.N.; Johnson, J.M. Auxin plays multiple roles during plant–pathogen interactions. Cold Spring Harb. Perspect. Biol. 2021, 13, a040022. [Google Scholar] [CrossRef]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhu, Z.; Chern, M.; Yin, J.; Yang, C.; Ran, L.; Cheng, M.; He, M.; Wang, K.; Wang, J.; et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 2017, 170, 114–126.e15. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Deng, Y.; Liang, D.; Tang, J.; Liu, J.; Yan, B.; Yin, X.; Lin, H.; Chen, F.; Yang, D.; et al. RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol. Cell 2019, 74, 996–1009.e7. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Xu, F.; Kapos, P.; Cheng, Y.T.; Li, M.; Zhang, Y.; Li, X. NLR-Associating transcription factor bHLH84 and its paralogs function redundantly in plant immunity. PLoS Pathog. 2014, 10, e1004312. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, T. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genom. 2017, 18, 118. [Google Scholar] [CrossRef]
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef]
- Qiu, D.; Xiao, J.; Xie, W.; Cheng, H.; Li, X.; Wang, S. Exploring transcriptional signalling mediated by OsWRKY13, a potential regulator of multiple physiological processes in rice. BMC Plant Biol. 2009, 9, 74. [Google Scholar] [CrossRef]
- Lv, G.; Han, R.; Shi, J.; Chen, K.; Liu, G.; Yu, Q.; Yang, C.; Jiang, J. Genome-wide identification of the TIFY family reveals JAZ subfamily function in response to hormone treatment in Betula platyphylla. BMC Plant Biol. 2023, 23, 143. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, F.; Wang, H.M.; Xue, D.X.; Liu, Y.-G.; Zhang, Q.Y. A rice mTERF protein V14 sustains photosynthesis establishment and temperature acclimation in early seedling leaves. BMC Plant Biol. 2021, 21, 406. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Gao, Y.; Song, S.; Hassani, D.; Lu, J. Identification, characterization and functional analysis of grape (Vitis vinifera L.) mitochondrial transcription termination factor (mTERF) genes in responding to biotic stress and exogenous phytohormone. BMC Genom. 2021, 22, 136. [Google Scholar] [CrossRef] [PubMed]
- Kuźniak, E.; Kopczewski, T. The chloroplast reactive oxygen species-redox system in plant immunity and disease. Front. Plant Sci. 2020, 11, 572686. [Google Scholar] [CrossRef] [PubMed]
- Shapiguzov, A.; Vainonen, J.P.; Wrzaczek, M.; Kangasjärvi, J. ROS-talk—How the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012, 3, 292. [Google Scholar] [CrossRef]
- Brunkard, J.O.; Runkel, A.M.; Zambryski, P.C. Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. USA 2015, 112, 10044–10049. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Yao, J. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 2018, 19, 3900. [Google Scholar] [CrossRef]
- Grimmer, M.K.; Foulkes, J.; Paveley, N.D. Foliar pathogenesis and plant water relations: A review. J. Exp. Bot. 2012, 63, 4321–4331. [Google Scholar] [CrossRef]
- Yadav, S.; Anuradha, G.; Kumar, R.R.; Vemireddy, L.R.; Sudhakar, R.; Donempudi, K.; Venkata, D.; Jabeen, F.; Narasimhan, Y.K.; Marathi, B.; et al. Identification of QTLs and possible candidate genes conferring sheath blight resistance in rice (Oryza sativa L.). SpringerPlus 2015, 4, 175. [Google Scholar] [CrossRef]
- IRRI, International Rice Research Institute. Standard Evaluation System for Rice; IRRI, International Rice Research Institute: Los Baños, Philippines, 2002. [Google Scholar]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef]
- Jung, K.-H.; Cao, P.; Seo, Y.-S.; Dardick, C.; Ronald, P.C. The Rice Kinase Phylogenomics Database: A guide for systematic analysis of the rice kinase super-family. Trends Plant Sci. 2010, 15, 595–599. [Google Scholar] [CrossRef]

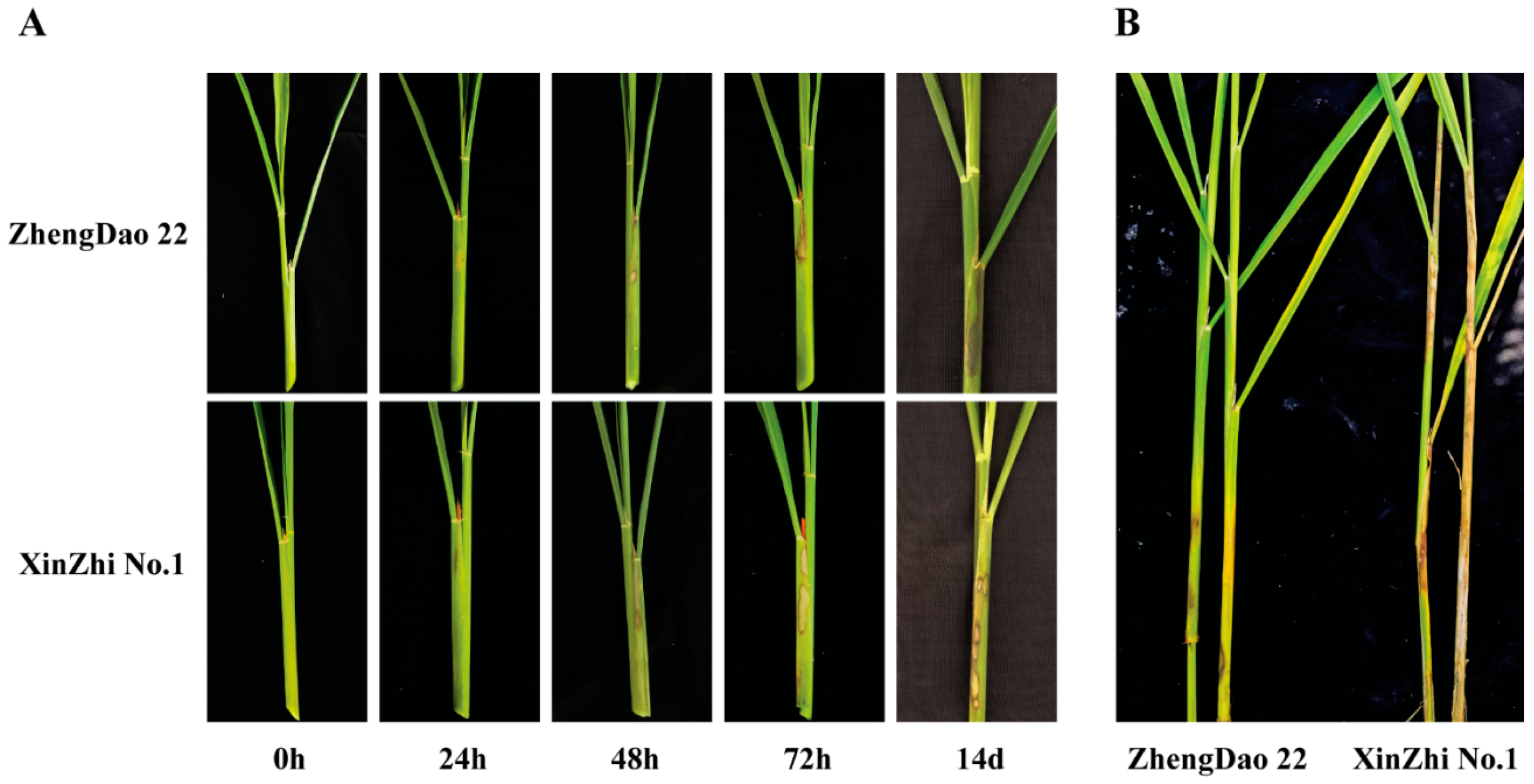



| No. of Sample | Cultivar | Relative Lesion Height (%) | Reaction * |
|---|---|---|---|
| 1 | JingHua 1832 | 56.0 | S |
| 2 | XinZhi No.1 | 82.9 | HS |
| 3 | XinDao 575 | 44.1 | MS |
| 4 | XinFeng 21 | 50.0 | S |
| 5 | XinLiang 501 | 52.4 | S |
| 6 | ZhengDao 201 | 59.5 | S |
| 7 | YiDao 178 | 67.2 | HS |
| 8 | XinKeDao 37 | 52.1 | S |
| 9 | YueNongDao No. 1 | 38.2 | MS |
| 10 | ShengDao 735 | 34.3 | MS |
| 11 | Xu72985 | 69.4 | HS |
| 12 | YuanDao26 | 52.9 | S |
| 13 | ZhengDao 22 | 14.5 | MR |
| 14 | YuDao 24 | 63.2 | S |
| 15 | XinKeDao 42 | 26.5 | MR |
| 16 | LianDao819 | 48.8 | S |
| 17 | XinLiang 320 | 29.8 | MR |
| 18 | YiDao 675 | 38.2 | MS |
| 19 | JingGuang17 | 47.4 | S |
| 20 | LQ202 | 55.3 | S |
| 21 | ZhengDao25 | 39.4 | MS |
| 22 | XinDao No.18 | 51.4 | S |
| Combination | Up-Regulated | Down-Regulated | All DEGs |
|---|---|---|---|
| CK-ZD-vs-ZD | 4141 | 2925 | 7066 |
| CK-XZ-vs-XZ | 52 | 8 | 60 |
| ZD-vs-XZ | 2996 | 4118 | 7114 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Yan, S.; Li, Y.; Li, G.; Sun, S.; Li, J.; Cui, Z.; Huo, J.; Sun, Y.; Wang, X.; et al. Comparison of Transcriptome between Tolerant and Susceptible Rice Cultivar Reveals Positive and Negative Regulators of Response to Rhizoctonia solani in Rice. Int. J. Mol. Sci. 2023, 24, 14310. https://doi.org/10.3390/ijms241814310
Yang X, Yan S, Li Y, Li G, Sun S, Li J, Cui Z, Huo J, Sun Y, Wang X, et al. Comparison of Transcriptome between Tolerant and Susceptible Rice Cultivar Reveals Positive and Negative Regulators of Response to Rhizoctonia solani in Rice. International Journal of Molecular Sciences. 2023; 24(18):14310. https://doi.org/10.3390/ijms241814310
Chicago/Turabian StyleYang, Xiurong, Shuangyong Yan, Yuejiao Li, Guangsheng Li, Shuqin Sun, Junling Li, Zhongqiu Cui, Jianfei Huo, Yue Sun, Xiaojing Wang, and et al. 2023. "Comparison of Transcriptome between Tolerant and Susceptible Rice Cultivar Reveals Positive and Negative Regulators of Response to Rhizoctonia solani in Rice" International Journal of Molecular Sciences 24, no. 18: 14310. https://doi.org/10.3390/ijms241814310
APA StyleYang, X., Yan, S., Li, Y., Li, G., Sun, S., Li, J., Cui, Z., Huo, J., Sun, Y., Wang, X., & Liu, F. (2023). Comparison of Transcriptome between Tolerant and Susceptible Rice Cultivar Reveals Positive and Negative Regulators of Response to Rhizoctonia solani in Rice. International Journal of Molecular Sciences, 24(18), 14310. https://doi.org/10.3390/ijms241814310






