Regulation of Human Sortilin Alternative Splicing by Glucagon-like Peptide-1 (GLP1) in Adipocytes
Abstract
:1. Introduction
2. Results
2.1. Alternatively Spliced Sortilin Variants in Human Adipocytes
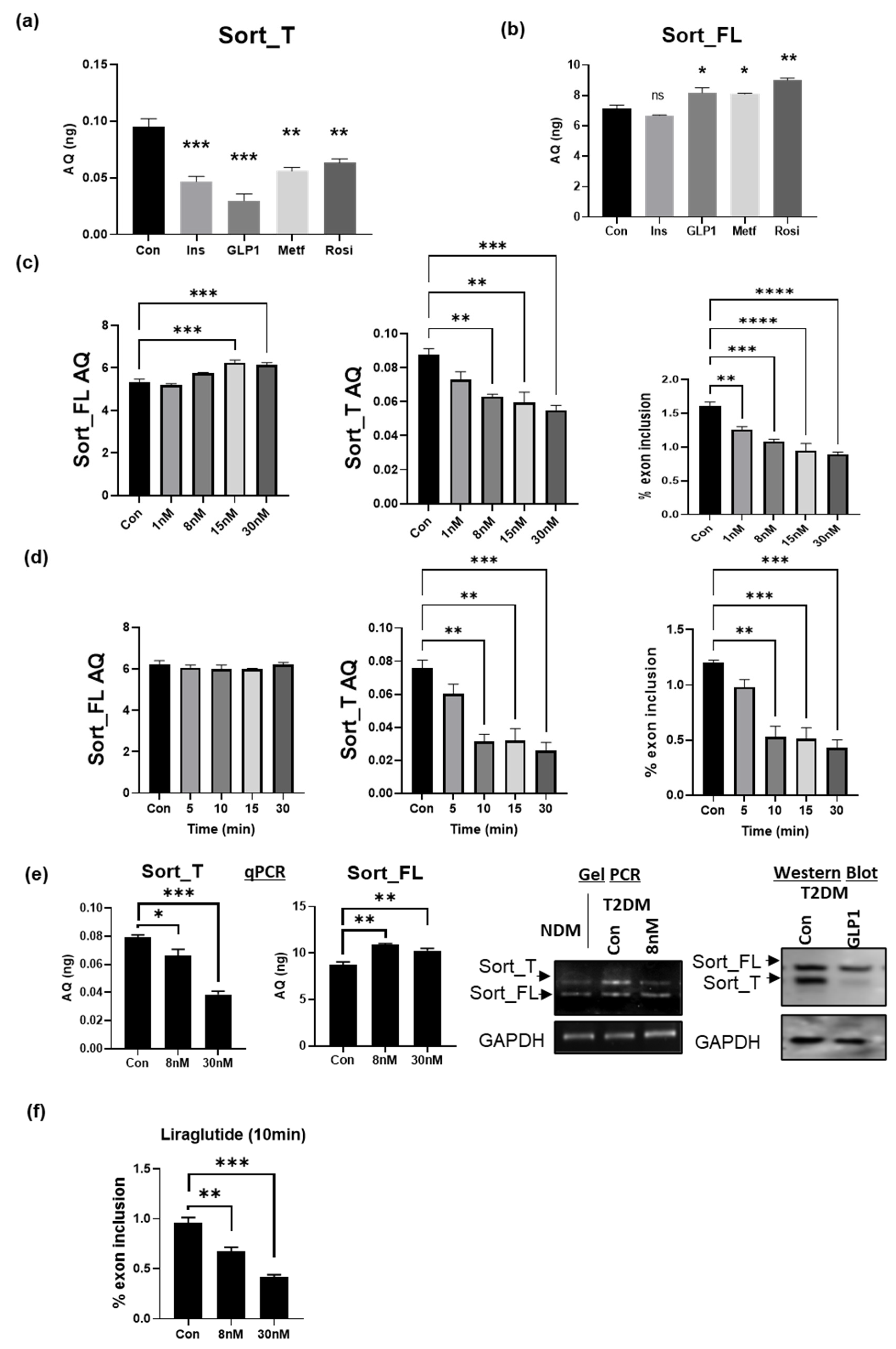
2.2. Sort_T Is Increased in Diabetic Visceral Adipose Tissue
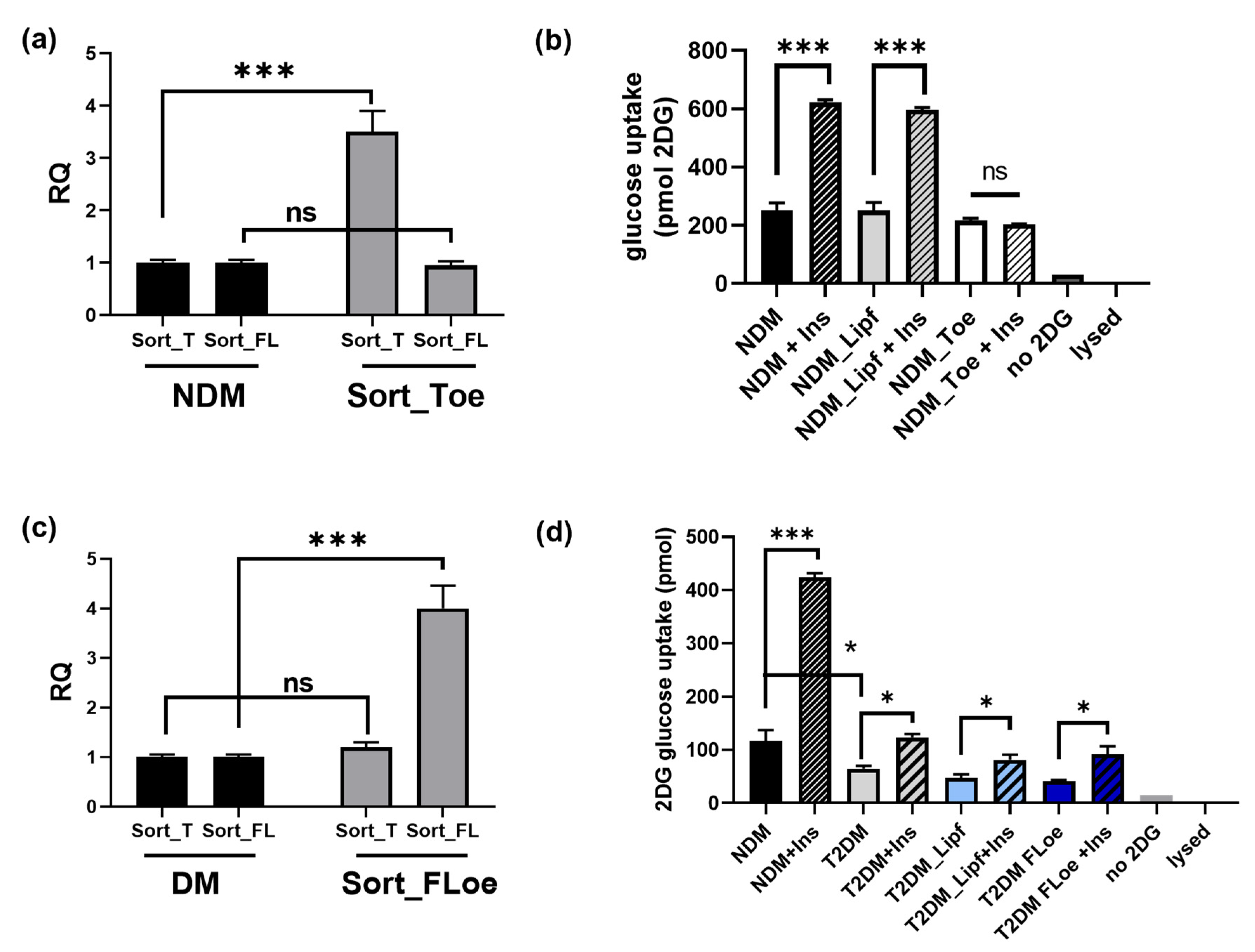
2.3. Differential Sortilin Splice Variant Expression in Nondiabetic and Diabetic Adipocytes during Adipogenesis
2.4. Hyperglycemia Increases Expression of Sort_T during Adipogenesis
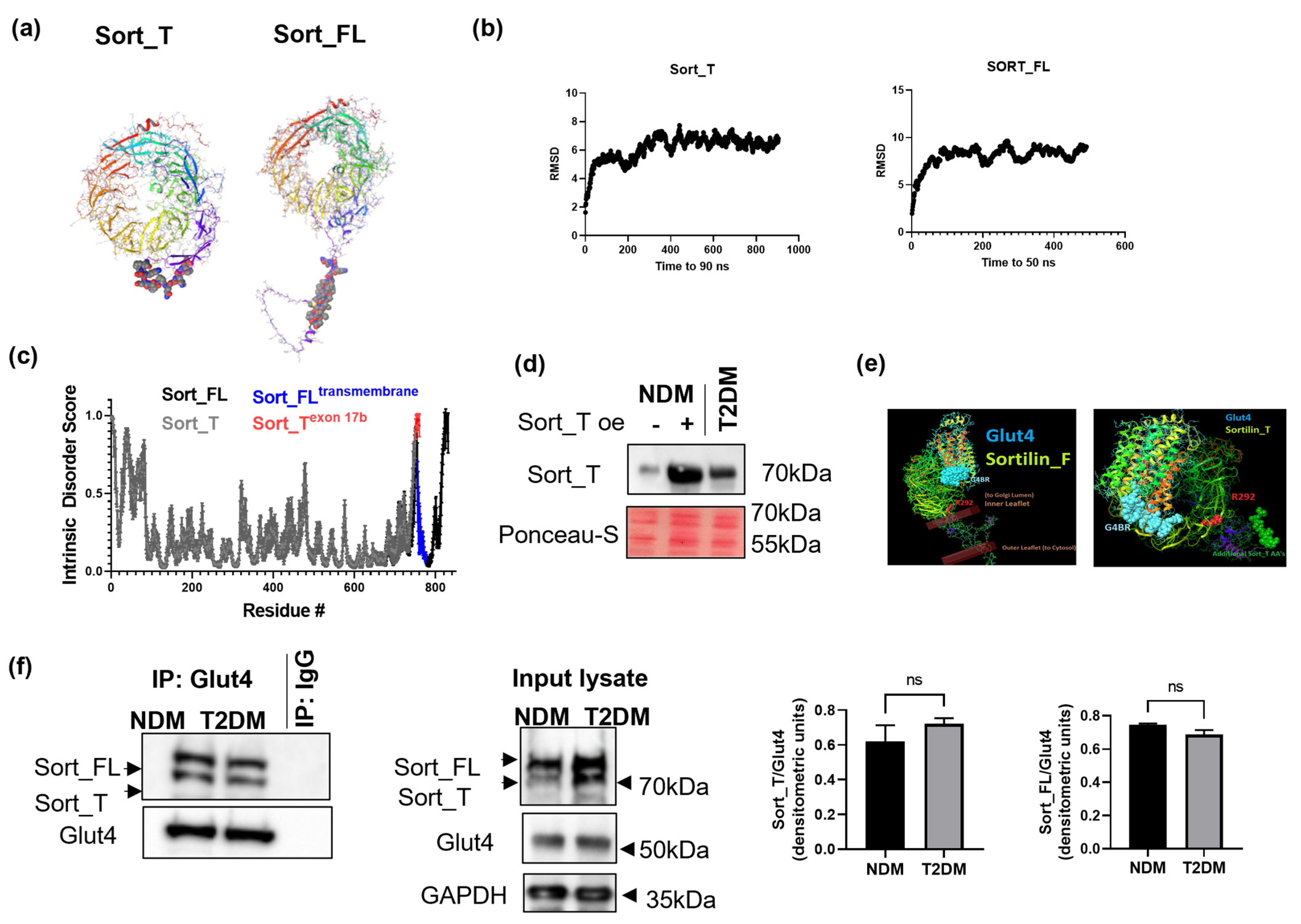
2.5. Molecular Dynamics and Intrinsic Disorder Analysis of Sortilin Splice Variants
2.6. Sort_T Is Secreted from Adipocytes
2.7. Computational Modeling Predicts Sort_T Maintains Ability to Bind Glut4
2.8. Glut4 Binds to Sort_FL and Sort_T
2.9. Effects of Sortilin Splice Variants on Insulin Stimulated Glucose Uptake
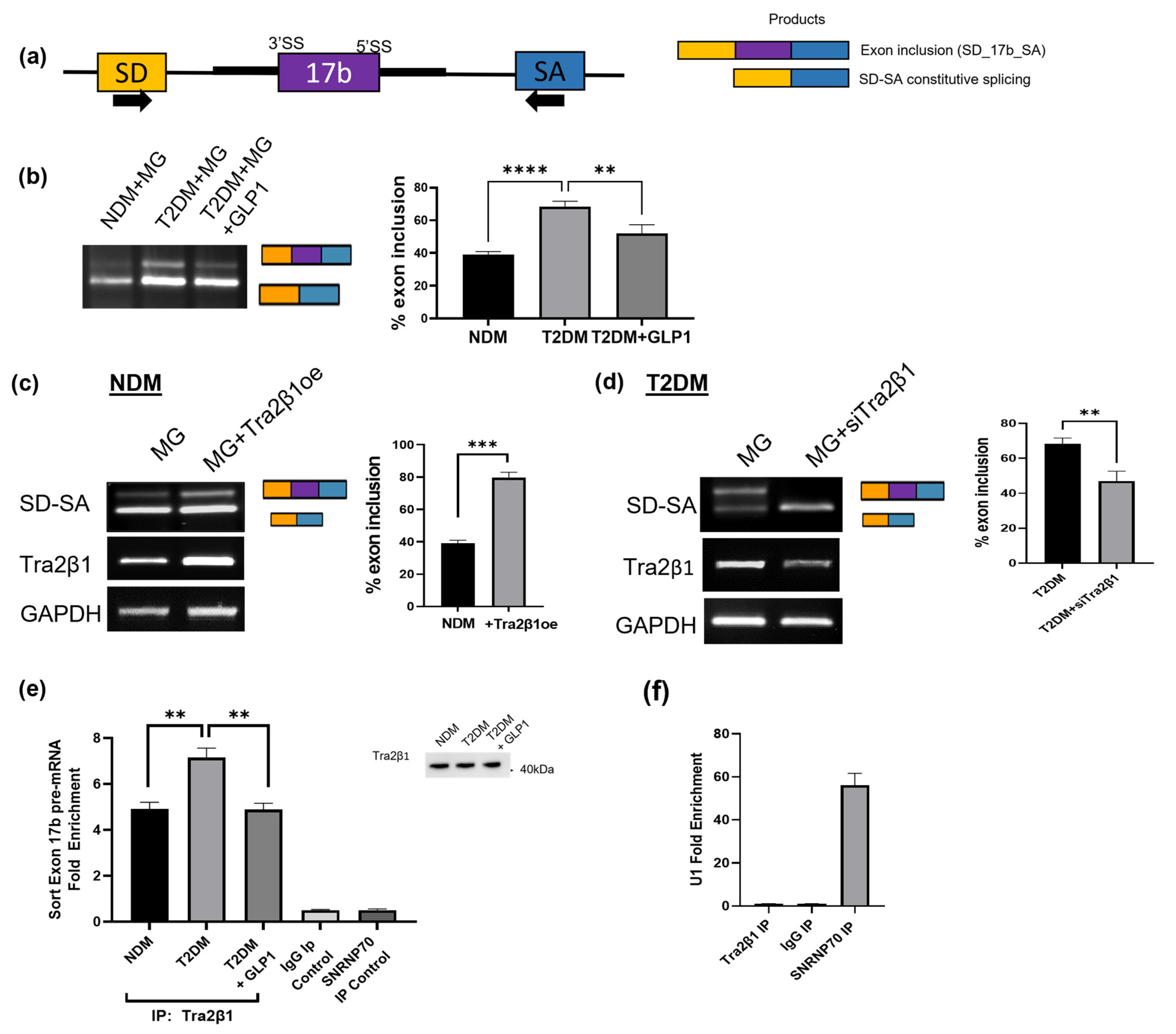
2.10. GLP1 Decreases Exon 17b Inclusion in Sortilin Pre-mRNA
2.11. GLP1R Agonist Liraglutide Decreases Exon 17b Inclusion in Sortilin Pre-mRNA
2.12. Predicted Splice Factors Regulating Sortilin Alternative Splicing in Diabetic Adipocytes
2.13. Tra2β1 Regulates Sortilin Alternative Splicing in Diabetic Adipocytes
2.14. GLP1 Decreases Inclusion of Sortilin Exon 17b in a Splicing Minigene
2.15. Tra2β1 Increases Inclusion of Sortilin Exon 17b in a Splicing Minigene
2.16. RIP Assay Shows Decreased Tra2β1 Binding to Sortilin Pre-mRNA with GLP1 Treatment
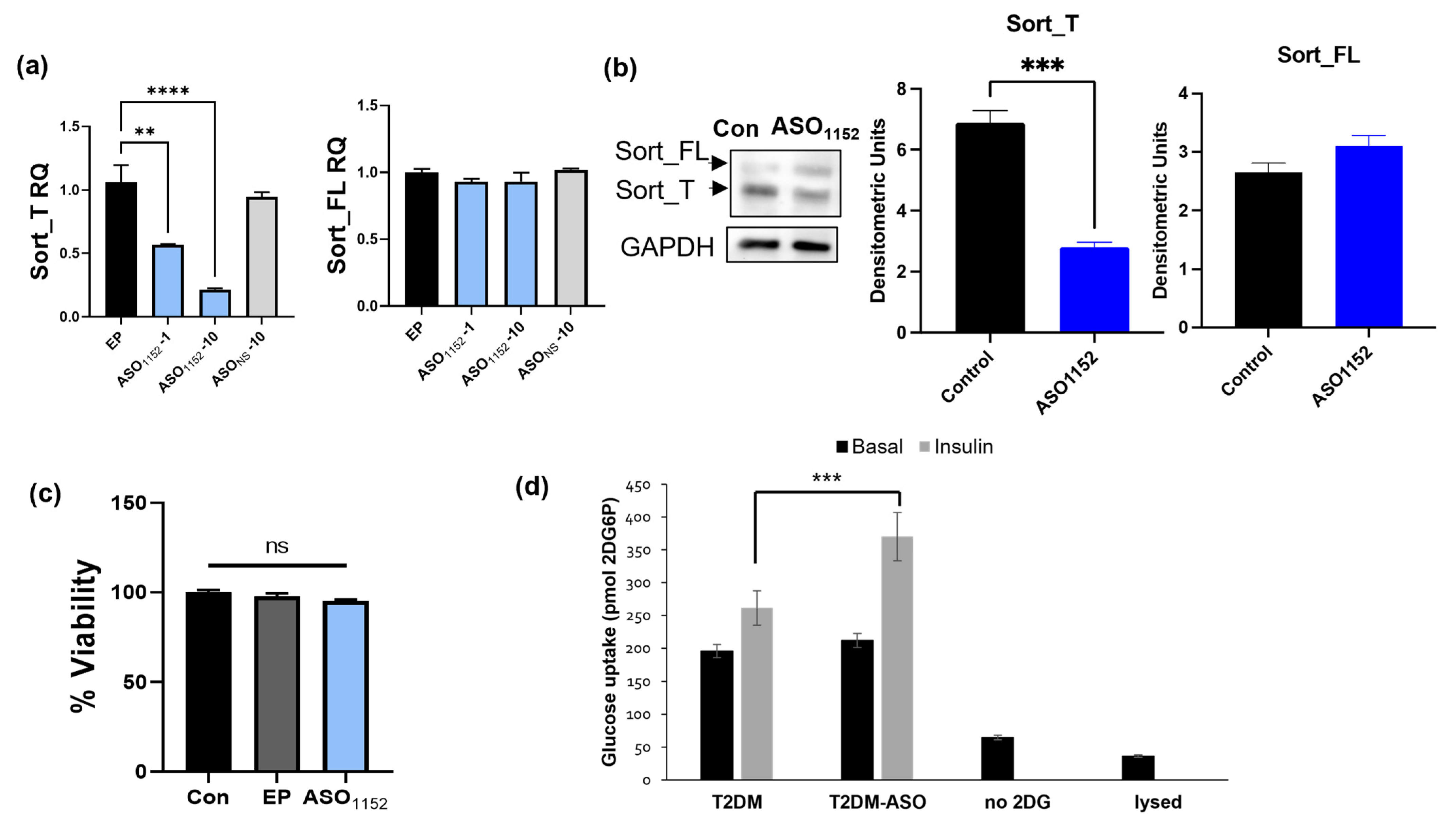
2.17. Morpholino Antisense Oligonucleotides Decrease Exon 17b Inclusion and Improve Glucose Uptake in Diabetic Adipocytes
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Human Adipose Tissue
4.3. Plasmid Transfections
4.4. Polymerase Chain Reaction
4.5. Western Blot Analysis
4.6. Coimmunoprecipitation Assay
4.7. Glucose Uptake Assay
4.8. Computational Modeling
4.9. Disorder Analysis
4.10. Cloning Human Sortilin Exon 17b-Splicing Minigene
4.11. RNA-Immunoprecipitation Assay (RIP)
4.12. Morpholino Treatment
4.13. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Center for Disease Control and Prevention. National Diabetes Statistics Report; Center for Disease Control and Prevention: Atlanta, GA, USA, 2020. [Google Scholar]
- Lin, J.; Thompson, T.J.; Cheng, Y.J.; Zhuo, X.; Zhang, P.; Gregg, E.; Rolka, D.B. Projection of the future diabetes burden in the United States through 2060. Popul. Health Metr. 2018, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Berezin, A.E.; Berezin, A.A.; Lichtenauer, M. Emerging Role of Adipocyte Dysfunction in Inducing Heart Failure among Obese Patients with Prediabetes and Known Diabetes Mellitus. Front. Cardiovasc. Med. 2020, 7, 583175. [Google Scholar] [CrossRef]
- Rea, S.; James, D.E. Moving GLUT4: The biogenesis and trafficking of GLUT4 storage vesicles. Diabetes 1997, 46, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Hashiramoto, M.; James, D.E. Characterization of insulin-responsive GLUT4 storage vesicles isolated from 3T3-L1 adipocytes. Mol. Cell Biol. 2000, 20, 416–427. [Google Scholar] [CrossRef]
- Thurmond, D.C.; Kanzaki, M.; Khan, A.H.; Pessin, J.E. Munc18c function is required for insulin-stimulated plasma membrane fusion of GLUT4 and insulin-responsive amino peptidase storage vesicles. Mol. Cell Biol. 2000, 20, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, B.J.; Griesel, B.A.; King, C.D.; Josey, M.A.; Olson, A.L. Moderate GLUT4 overexpression improves insulin sensitivity and fasting triglyceridemia in high-fat diet-fed transgenic mice. Diabetes 2013, 62, 2249–2258. [Google Scholar] [CrossRef]
- Bai, L.; Wang, Y.; Fan, J.; Chen, Y.; Ji, W.; Qu, A.; Xu, P.; James, D.E.; Xu, T. Dissecting multiple steps of GLUT4 trafficking and identifying the sites of insulin action. Cell Metab. 2007, 5, 47–57. [Google Scholar] [CrossRef]
- Jedrychowski, M.P.; Gartner, C.A.; Gygi, S.P.; Zhou, L.; Herz, J.; Kandror, K.V.; Pilch, P.F. Proteomic Analysis of GLUT4 Storage Vesicles Reveals LRP1 to Be an Important Vesicle Component and Target of Insulin Signaling. J. Biol. Chem. 2010, 285, 104–114. [Google Scholar] [CrossRef]
- Pilch, P.F. The mass action hypothesis: Formation of Glut4 storage vesicles, a tissue-specific, regulated exocytic compartment. Acta Physiol. 2008, 192, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Morris, N.J.; Ross, S.A.; Lane, W.S.; Moestrup, S.K.; Petersen, C.M.; Keller, S.R.; Lienhard, G.E. Sortilin Is the Major 110-kDa Protein in GLUT4 Vesicles From Adipocytes. J. Biol. Chem. 1998, 273, 3582–3587. [Google Scholar] [CrossRef]
- Bogan, J.S.; Kandror, K.V. Biogenesis and regulation of insulin-responsive vesicles containing GLUT4. Curr. Opin. Cell Biol. 2010, 22, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Kandror, K.V. Sortilin is essential and sufficient for the formation of Glut4 storage vesicles in 3T3-L1 adipocytes. Dev. Cell 2005, 9, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Czech, M.P. The GLUT4 Glucose Transporter. Cell Metab. 2007, 5, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kandror, K.V. The first luminal loop confers insulin responsiveness to glucose transporter 4. Mol. Biol. Cell 2012, 23, 910–917. [Google Scholar] [CrossRef]
- McCormick, P.J.; Dumaresq-Doiron, K.; Pluviose, A.-S.; Pichette, V.; Tosato, G.; Lefrancois, S. Palmitoylation Controls Recycling in Lysosomal Sorting and Trafficking. Traffic 2008, 9, 1984–1997. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Matye, D.J.; Li, T. Insulin Resistance Induces Posttranslational Hepatic Sortilin 1 Degradation in Mice. J. Biol. Chem. 2015, 290, 11526–11536. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zaarur, N.; Singh, M.; Morin, P.; Kandror, K.V. Sortilin and retromer mediate retrograde transport of Glut4 in 3T3-L1 adipocytes. Mol. Biol. Cell 2017, 28, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Seaman, M.N.J. The retromer complex—Endosomal protein recycling and beyond. J. Cell Sci. 2012, 125, 4693–4702. [Google Scholar] [CrossRef]
- Nielsen, M.S.; Madsen, P.; Christensen, E.I.; Nykjaer, A.; Gliemann, J.; Kasper, D.; Pohlmann, R.; Petersen, C.M. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001, 20, 2180–2190. [Google Scholar] [CrossRef]
- Tazi, J.; Bakkour, N.; Stamm, S. Alternative splicing and disease. Biochim. Biophys. Acta 2009, 1792, 14. [Google Scholar] [CrossRef]
- Prudencio, M.; Jansen-West, K.R.; Lee, W.C.; Gendron, T.F.; Zhang, Y.-J.; Xu, Y.-F.; Gass, J.; Stuani, C.; Stetler, C.; Rademakers, R.; et al. Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc. Natl. Acad. Sci. USA 2012, 109, 21510–21515. [Google Scholar] [CrossRef]
- Mohagheghi, F.; Prudencio, M.; Stuani, C.; Cook, C.; Jansen-West, K.; Dickson, D.W.; Petrucelli, L.; Buratti, E. TDP-43 functions within a network of hnRNP proteins to inhibit the production of a truncated human SORT1 receptor. Hum. Mol. Genet. 2016, 25, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Lui, A.; Sparks, R.; Patel, R.; Patel, N.A. Identification of Sortilin Alternatively Spliced Variants in Mouse 3T3L1 Adipocytes. Int. J. Mol. Sci. 2021, 22, 983. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Parag, S.; Patel, R.; Lui, A.; Murr, M.; Cai, J.; Patel, N.A. Stabilization of lncRNA GAS5 by a Small Molecule and Its Implications in Diabetic Adipocytes. Cell Chem. Biol. 2019, 26, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.S.; Carter, G.; El Bassit, G.; Patel, A.A.; Cooper, D.R.; Murr, M.; Patel, N.A. Adipose-derived stem cells from lean and obese humans show depot specific differences in their stem cell markers, exosome contents and senescence: Role of protein kinase C delta (PKCdelta) in adipose stem cell niche. Stem Cell Investig. 2016, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmuller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Maiorov, V.N.; Crippen, G.M. Significance of Root-Mean-Square Deviation in Comparing Three-dimensional Structures of Globular Proteins. J. Mol. Biol. 1994, 235, 625–634. [Google Scholar] [CrossRef]
- Monzon, A.M.; Zea, D.J.; Fornasari, M.S.; Saldaño, T.E.; Fernandez-Alberti, S.; Tosatto, S.C.E.; Parisi, G. Conformational diversity analysis reveals three functional mechanisms in proteins. PLoS Comput. Biol. 2017, 13, e1005398. [Google Scholar] [CrossRef] [PubMed]
- Quistgaard, E.M.; Groftehauge, M.K.; Madsen, P.; Pallesen, L.T.; Christensen, B.; Sorensen, E.S.; Nissen, P.; Petersen, C.M.; Thirup, S.S. Revisiting the structure of the Vps10 domain of human sortilin and its interaction with neurotensin. Protein Sci. 2014, 23, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Mollica, L.; Bessa, L.M.; Hanoulle, X.; Jensen, M.R.; Blackledge, M.; Schneider, R. Binding Mechanisms of Intrinsically Disordered Proteins: Theory, Simulation, and Experiment. Front. Mol. Biosci. 2016, 3, 52. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New Additions to the ClusPro Server Motivated by CAPRI. Proteins 2017, 85, 435. [Google Scholar] [CrossRef]
- Crane, J.; McGowan, B. The GLP-1 agonist, liraglutide, as a pharmacotherapy for obesity. Ther. Adv. Chronic Dis. 2016, 7, 92–107. [Google Scholar] [CrossRef]
- Fransson, L.; dos Santos, C.; Wolbert, P.; Sjöholm, Å.; Rafacho, A.; Ortsäter, H. Liraglutide counteracts obesity and glucose intolerance in a mouse model of glucocorticoid-induced metabolic syndrome. Diabetol. Metab. Syndr. 2014, 6, 3. [Google Scholar] [CrossRef]
- Sjoholm, A. Liraglutide Therapy for Type 2 Diabetes: Overcoming Unmet Needs. Pharmaceuticals 2010, 3, 764. [Google Scholar] [CrossRef]
- Cartegni, L.; Wang, J.; Zhu, Z.; Zhang, M.Q.; Krainer, A.R. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003, 31, 3568–3571. [Google Scholar] [CrossRef]
- Paz, I.; Akerman, M.; Dror, I.; Kosti, I.; Mandel-Gutfreund, Y. SFmap: A web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010, 38, W281–W285. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Ieraci, A.; Teng, H.; Dall, H.; Meng, C.-X.; Herrera, D.G.; Nykjaer, A.; Hempstead, B.L.; Lee, F.S. Sortilin Controls Intracellular Sorting of Brain-Derived Neurotrophic Factor to the Regulated Secretory Pathway. J. Neurosci. 2005, 25, 6156–6166. [Google Scholar] [CrossRef]
- Molgaard, S.; Demontis, D.; Nicholson, A.M.; Finch, N.A.; Petersen, R.C.; Petersen, C.M.; Rademakers, R.; Nykjaer, A.; Glerup, S. Soluble sortilin is present in excess and positively correlates with progranulin in CSF of aging individuals. Exp. Gerontol. 2016, 84, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.J.; Ahn, C.H.; Kim, B.R.; Kim, K.M.; Moon, J.H.; Lim, S.; Park, K.S.; Lim, C.; Jang, H.; Choi, S.H. Circulating sortilin level as a potential biomarker for coronary atherosclerosis and diabetes mellitus. Cardiovasc. Diabetol. 2017, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Trabjerg, E.; Abu-Asad, N.; Wan, Z.; Kartberg, F.; Christensen, S.; Rand, K.D. Investigating the conformational response of the Sortilin receptor upon binding endogenous peptide- and protein ligands by HDX-MS. bioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Vendrell, J.; El Bekay, R.; Peral, B.; García-Fuentes, E.; Megia, A.; Macias-Gonzalez, M.; Real, J.F.; Jimenez-Gomez, Y.; Escoté, X.; Pachón, G.; et al. Study of the Potential Association of Adipose Tissue GLP-1 Receptor with Obesity and Insulin Resistance. Endocrinology 2011, 152, 4072–4079. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.-L.; Abrahamsson, N.; Sundbom, M.; Hellström, P.M. Bariatric surgery—Time to replace with GLP-1? Scand. J. Gastroenterol. 2017, 52, 635–640. [Google Scholar] [CrossRef]
- Madsbad, S.; Holst, J.J. GLP-1 as a Mediator in the Remission of Type 2 Diabetes after Gastric Bypass and Sleeve Gastrectomy Surgery. Diabetes 2014, 63, 3172–3174. [Google Scholar] [CrossRef]
- Currie, A.; Bolckmans, R.; Askari, A.; Byrne, J.; Ahmed, A.R.; Batterham, R.L.; Mahawar, K.; Miras, A.D.; Pring, C.M.; Small, P.K.; et al. Bariatric-metabolic surgery for NHS patients with type 2 diabetes in the United Kingdom National Bariatric Surgery Registry. Diabet. Med. J. Br. Diabet. Assoc. 2023, 40, e15041. [Google Scholar] [CrossRef]
- Hollanda, A.; Lecube, A.; Rubio, M.A.; Sanchez, E.; Vilarrasa, N.; Oliva, J.G.; Fernandez-Soto, M.L.; Salas-Salvado, J.; Ballesteros-Pomar, M.D.; Ciudin, A.; et al. New Metrics to Assess Type 2 Diabetes After Bariatric Surgery: The “Time-Within-Remission Range”. J. Clin. Med. 2020, 9, 1070. [Google Scholar] [CrossRef]
- Perry, Z.H.; Netz, U.; Tzelnick, S.; Berar, O.; Atias, S.; Lantsberg, L.; Avinoach, E.; Mizrahi, S. The Effect of Bariatric Surgery upon Diabetes Mellitus: A Proof of Concept by Using the Case of the Mid-Term Effect of Lap Adjustable Gastric Banding (LAGB) on Patients with Diabetes. Metabolites 2022, 12, 1236. [Google Scholar] [CrossRef]
- Vilarrasa, N.; Goday, A.; Rubio, M.A.; Caixas, A.; Pellitero, S.; Ciudin, A.; Calanas, A.; Botella, J.I.; Breton, I.; Morales, M.J.; et al. Hyperinsulinemic Hypoglycemia after Bariatric Surgery: Diagnosis and Management Experience from a Spanish Multicenter Registry. Obes. Facts 2016, 9, 41–51. [Google Scholar] [CrossRef]
- Xu, T.Q.; Kindel, T.L. The role of weight control in the management of type 2 diabetes mellitus: Bariatric surgery. Diabetes Res. Clin. Pract. 2023, 199, 110667. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Park, M.-S.; Choung, J.-S.; Kim, S.-S.; Oh, H.-H.; Choi, C.-S.; Ha, S.-Y.; Kang, Y.; Kim, Y.; Jun, H.-S. Glucagon-like peptide-1 inhibits adipose tissue macrophage infiltration and inflammation in an obese mouse model of diabetes. Diabetologia 2012, 55, 2456–2468. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Zhang, H.; Li, S.; Tian, H. Glucagon-like peptide 1 receptor agonist ameliorates the insulin resistance function of islet β cells via the activation of PDX-1/JAK signaling transduction in C57/BL6 mice with high-fat diet-induced diabetes. Int. J. Mol. Med. 2017, 39, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, X.; Zhang, Z.; Yang, Y.; Yang, J.; Li, X.; Ning, G. GLP-1 amplifies insulin signaling by up-regulation of IRbeta, IRS-1 and Glut4 in 3T3-L1 adipocytes. Endocrine 2007, 32, 90–95. [Google Scholar] [CrossRef]
- Lee, H.M.; Joo, B.S.; Lee, C.H.; Kim, H.Y.; Ock, J.H.; Lee, Y.S. Effect of Glucagon-like Peptide-1 on the Differentiation of Adipose-derived Stem Cells into Osteoblasts and Adipocytes. J. Menopausal Med. 2015, 21, 93–103. [Google Scholar] [CrossRef] [PubMed]
- El Bekay, R.; Coín-Aragüez, L.; Fernández-García, D.; Oliva-Olivera, W.; Bernal-López, R.; Clemente-Postigo, M.; Delgado-Lista, J.; Diaz-Ruiz, A.; Guzman-Ruiz, R.; Vázquez-Martínez, R.; et al. Effects of glucagon-like peptide-1 on the differentiation and metabolism of human adipocytes. Br. J. Pharmacol. 2016, 173, 1820–1834. [Google Scholar] [CrossRef]
- Liu, H.; Zhan, Y.L.; Luo, G.J.; Zou, L.L.; Li, Y.; Lu, H.Y. Liraglutide and Insulin Have Contrary Effects on Adipogenesis of Human Adipose-Derived Stem Cells via Wnt Pathway. Diabetes Metab. Syndr. Obes. 2020, 13, 3075–3087. [Google Scholar] [CrossRef]
- Hwang, D.Y.; Cohen, J.B. Base pairing at the 5′ splice site with U1 small nuclear RNA promotes splicing of the upstream intron but may be dispensable for slicing of the downstream intron. Mol. Cell. Biol. 1996, 16, 3012–3022. [Google Scholar] [CrossRef]
- Zahler, A.M.; Roth, M.B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc. Natl. Acad. Sci. USA 1995, 92, 2642–2646. [Google Scholar] [CrossRef]
- Freund, M.; Hicks, M.J.; Konermann, C.; Otte, M.; Hertel, K.J.; Schaal, H. Extended base pair complementarity between U1 snRNA and the 5′ splice site does not inhibit splicing in higher eukaryotes, but rather increases 5′ splice site recognition. Nucleic Acids Res. 2005, 33, 5112–5119. [Google Scholar] [CrossRef]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Matsuo, N.; Ozawa, K.; Hori, O.; Higashi, T.; Nishizaki, J.; Tohnai, N.; Nagata, I.; Kawano, K.; Yutani, C.; et al. Expression of a novel RNA-splicing factor, RA301/Tra2beta, in vascular lesions and its role in smooth muscle cell proliferation. Am. J. Pathol. 2001, 158, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Takeo, K.; Kawai, T.; Nishida, K.; Masuda, K.; Teshima-Kondo, S.; Tanahashi, T.; Rokutan, K. Oxidative stress-induced alternative splicing of transformer 2β (SFRS10) and CD44 pre-mRNAs in gastric epithelial cells. Am. J. Physiol.-Cell Physiol. 2009, 297, C330–C338. [Google Scholar] [CrossRef] [PubMed]
- Kiryu-Seo, S.; Matsuo, N.; Wanaka, A.; Ogawa, S.; Tohyama, M.; Kiyama, H. A sequence-specific splicing activator, tra2beta, is up-regulated in response to nerve injury. Brain Res. Mol. Brain Res. 1998, 62, 220–223. [Google Scholar] [CrossRef]
- Pihlajamaki, J.; Lerin, C.; Itkonen, P.; Boes, T.; Floss, T.; Schroeder, J.; Dearie, F.; Crunkhorn, S.; Burak, F.; Jimenez-Chillaron, J.C.; et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011, 14, 208–218. [Google Scholar] [CrossRef]
- Li, H.; Cheng, Y.; Wu, W.; Liu, Y.; Wei, N.; Feng, X.; Xie, Z.; Feng, Y. SRSF10 regulates alternative splicing and is required for adipocyte differentiation. Mol. Cell Biol. 2014, 34, 2198–2207. [Google Scholar] [CrossRef]
- Dlamini, Z.; Mokoena, F.; Hull, R. Abnormalities in alternative splicing in diabetes: Therapeutic targets. J. Mol. Endocrinol. 2017, 59, R93–R107. [Google Scholar] [CrossRef]
- Petersen, M.C.; Shulman, G.I. Mechanisms of Insulin Action and Insulin Resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef]
- Zhao, H.; Kong, H.; Wang, B.; Wu, S.; Chen, T.; Cui, Y. RNA-Binding Proteins and Alternative Splicing Genes Are Coregulated in Human Retinal Endothelial Cells Treated with High Glucose. J. Diabetes Res. 2022, 2022, 7680513. [Google Scholar] [CrossRef]
- Renna, L.V.; Bosè, F.; Brigonzi, E.; Fossati, B.; Meola, G.; Cardani, R. Aberrant insulin receptor expression is associated with insulin resistance and skeletal muscle atrophy in myotonic dystrophies. PLoS ONE 2019, 14, e0214254. [Google Scholar] [CrossRef]
- Norgren, S.; Zierath, J.; Wedell, A.; Wallberg-Henriksson, H.; Luthman, H. Regulation of human insulin receptor RNA splicing in vivo. Proc. Natl. Acad. Sci. USA 1994, 91, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Qin, Q.; Xie, L.; Zhang, H.; Wu, F.; Stevens, R.C.; Liu, Y. GLP-1R Signaling and Functional Molecules in Incretin Therapy. Molecules 2023, 28, 751. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Mo, W.; Lan, S.; Yang, H.; Huang, Z.; Liang, X.; Li, L.; Xian, J.; Xie, X.; Qin, Y.; et al. GLP-1 RA Improves Diabetic Retinopathy by Protecting the Blood-Retinal Barrier through GLP-1R-ROCK-p-MLC Signaling Pathway. J. Diabetes Res. 2022, 2022, 1861940. [Google Scholar] [CrossRef]
- Challa, T.D.; Beaton, N.; Arnold, M.; Rudofsky, G.; Langhans, W.; Wolfrum, C. Regulation of Adipocyte Formation by GLP-1/GLP-1R Signaling. J. Biol. Chem. 2012, 287, 6421–6430. [Google Scholar] [CrossRef] [PubMed]
- Ejarque, M.; Guerrero-Perez, F.; de la Morena, N.; Casajoana, A.; Virgili, N.; Lopez-Urdiales, R.; Maymo-Masip, E.; Pujol Gebelli, J.; Garcia Ruiz de Gordejuela, A.; Perez-Maraver, M.; et al. Role of adipose tissue GLP-1R expression in metabolic improvement after bariatric surgery in patients with type 2 diabetes. Sci. Rep. 2019, 9, 6274. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, H.; Ma, X.; Zhang, Y.; Lu, S.; Wang, Y.; Zong, C.; Qin, D.; Wang, Y.; Yingfeng Yang, Y.; et al. GLP-1/GLP-1R Signaling in Regulation of Adipocyte Differentiation and Lipogenesis. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 42, 1165–1176. [Google Scholar] [CrossRef]
- Gutierrez, A.D.; Gao, Z.; Hamidi, V.; Zhu, L.; Saint Andre, K.B.; Riggs, K.; Ruscheinsky, M.; Wang, H.; Yu, Y.; Miller, C., 3rd; et al. Anti-diabetic effects of GLP1 analogs are mediated by thermogenic interleukin-6 signaling in adipocytes. Cell Rep. Med. 2022, 3, 100813. [Google Scholar] [CrossRef]
- Ferguson, D.P.; Dangott, L.J.; Lightfoot, J.T. Lessons learned from vivo-morpholinos: How to avoid vivo-morpholino toxicity. Biotechniques 2014, 56, 251–256. [Google Scholar] [CrossRef]
- Summerton, J. Morpholino antisense oligomers: The case for an RNase H-independent structural type. Biochim. Biophys. Acta (BBA)-Gene Struct. Expr. 1999, 1489, 141–158. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Havens, M.A.; Duelli, D.M.; Hastings, M.L. Targeting RNA splicing for disease therapy. Wiley Interdiscip. Rev. RNA 2013, 4, 247–266. [Google Scholar] [CrossRef] [PubMed]


| Gene | Species | Forward and Reverse Primer Sequences (5′-3′) |
|---|---|---|
| β-Actin | Human | CTCTTCCAGCCTTCCTTCCT AGCACTGTGTTGGCGTACAG |
| Sortilin exon 16–20 | Human | AAGGGCCACGACCTGGAGTT CAATCTCGAGCTATTCCAAGAGGTCCTCATCTG |
| Sort_T | Human | TTGTCCTAGAATCCAGCTCTGC TCTGGGTGGGAAATAGGGAGT |
| Sort_FL | Human | TGGGGTAAATCCAGTTCGAG GACTTGGAATTCTGTTTTTCCGGAC |
| GAPDH | Human | GATCATCAGCAATGCCTCCT TGTGGTCATGAGTCCTTCCA |
| TRA2B | Human | TGTCTACTCGCAGGCGTCATGT AGACACATCGGCAATGGGACCA |
| SD-SA | N/A | GTGAACTGCACTGTGACAAGC CACCTGAGGAGTGAATTGGTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lui, A.; Patel, R.S.; Krause-Hauch, M.; Sparks, R.P.; Patel, N.A. Regulation of Human Sortilin Alternative Splicing by Glucagon-like Peptide-1 (GLP1) in Adipocytes. Int. J. Mol. Sci. 2023, 24, 14324. https://doi.org/10.3390/ijms241814324
Lui A, Patel RS, Krause-Hauch M, Sparks RP, Patel NA. Regulation of Human Sortilin Alternative Splicing by Glucagon-like Peptide-1 (GLP1) in Adipocytes. International Journal of Molecular Sciences. 2023; 24(18):14324. https://doi.org/10.3390/ijms241814324
Chicago/Turabian StyleLui, Ashley, Rekha S. Patel, Meredith Krause-Hauch, Robert P. Sparks, and Niketa A. Patel. 2023. "Regulation of Human Sortilin Alternative Splicing by Glucagon-like Peptide-1 (GLP1) in Adipocytes" International Journal of Molecular Sciences 24, no. 18: 14324. https://doi.org/10.3390/ijms241814324
APA StyleLui, A., Patel, R. S., Krause-Hauch, M., Sparks, R. P., & Patel, N. A. (2023). Regulation of Human Sortilin Alternative Splicing by Glucagon-like Peptide-1 (GLP1) in Adipocytes. International Journal of Molecular Sciences, 24(18), 14324. https://doi.org/10.3390/ijms241814324





