Abstract
Even though male breast cancer (MBC) risk encompasses both genetic and environmental aetiologies, the primary risk factor is a germline pathogenic variant (PV) or likely pathogenic variant (LPV) in BRCA2, BRCA1 and/or PALB2 genes. To identify new potential MBC-specific predisposition genes, we sequenced a panel of 585 carcinogenesis genes in an MBC cohort without BRCA1/BRCA2/PALB2 PV/LPV. We identified 14 genes carrying rare PVs/LPVs in the MBC population versus noncancer non-Finnish European men, predominantly coding for DNA repair and maintenance of genomic stability proteins. We identified for the first time PVs/LPVs in PRCC (pre-mRNA processing), HOXA9 (transcription regulation), RECQL4 and WRN (maintenance of genomic stability) as well as in genes involved in other cellular processes. To study the specificity of this MBC PV/LPV profile, we examined whether variants in the same genes could be detected in a female breast cancer (FBC) cohort without BRCA1/BRCA2/PALB2 PV/LPV. Only 5/109 women (4.6%) carried a PV/LPV versus 18/85 men (21.2%) on these genes. FBC did not carry any PV/LPV on 11 of these genes. Although 5.9% of the MBC cohort carried PVs/LPVs in PALLD and ERCC2, neither of these genes were altered in our FBC cohort. Our data suggest that in addition to BRCA1/BRCA2/PALB2, other genes involved in DNA repair/maintenance or genomic stability as well as cell adhesion may form a specific MBC PV/LPV signature.
1. Introduction
Male breast cancer (MBC), a rare cancer and a poorly understood malignancy, represents less than 1% of all cancers in men and all breast cancers in Western countries [1], but its incidence is increasing worldwide. A man’s lifetime risk of breast cancer is approximately 1:1000, compared to 1:8 for a woman [2]. On average, MBC cases are diagnosed at an older age than women (67 versus 62 years). The breast cancer prognosis, corrected for age and tumour stage, is similar for men and women, but men are often diagnosed later in life and therefore at a more advanced stage of disease, which is reflected in their lower overall survival rate [3,4]. MBC is often compared to postmenopausal breast cancer in women due to the relatively older age at onset and the high prevalence of estrogen receptor (ER) positivity. Nevertheless, several lines of evidence suggest that MBC tumours differ from the female equivalent. Although genomic approaches are not as developed in MBC as they are in female breast cancer (FBC), several studies have identified sex-specific differences in mutated genes [5] as well as copy-number variations [6] or epigenetic mutations [7]. Given the rarity of the disease, few MBC clinical trials have been conducted to date. The mainstay of MBC treatments, therefore, follows the same recommendations as FBC treatments and includes hormone-, chemo- and radiotherapy as well as targeted therapies such as PARP inhibitors for germline BRCA1/BRCA2-mutation carriers. However, the differences between MBC and FBC have led the American Association of Clinical Oncology (ASCO) to recently publish guidelines providing practice recommendations for the specific management of MBC [2].
MBC risk factors encompass both genetic or environmental factors: a personal and/or family history of cancer [8]; ethnicity [9]; a hormonal imbalance [10] such as Klinefelter syndrome [11]; obesity; and exposure to specific toxic substances such as solvents, exhaust gases, petrol and ionizing radiation [12]. Having a first-degree relative with breast cancer increases the risk of MBC (relative risk (RR) = 1.92); the risk increases even more with an affected sister (RR = 2.25), with a dramatic increase observed when the family history includes both an affected mother and an affected sister (RR = 9.73) [11]. To date, the only clearly identified risk factor for MBC is the presence of a germline pathogenic or likely pathogenic variant (PV or LPV, respectively) in the BRCA2 or BRCA1 genes. Several studies have investigated the involvement of other genes in the risk of developing MBC, with populations ranging from 6 to 767 patients and panels ranging from 16 to 94 genes, predominantly restricted to genes previously implicated in FBC [13,14,15,16] or even at the exome scale (4600 genes), albeit only including 6 patients [17]. These studies have demonstrated the presence of PVs associated with the risk of MBC in the PALB2 (OR = 6.6–17.3), CHEK2 (OR = 3.7), ATM and RAD51D (OR = 8.6–10.2) genes (see review [18]). A recent study identified three novel MBC susceptibility loci, two mapping to 6q25.1 and one to 11q13.3, which were also associated with FBC risk [19].
In France, all MBC cases are offered genetic counselling and germline genetic testing of cancer predisposition genes. Genetic testing is currently performed on a consensus panel of predisposition genes, initially BRCA1/BRCA2/PALB2 genes, and now an HBOC (hereditary breast and ovarian cancer) panel including BRCA1, BRCA2, PALB2, TP53, CDH1, PTEN, RAD51C, RAD51D, MLH1, MSH2, MSH6, PMS2 and EPCAM genes. However, these genetic panel analyses only reveal the presence of a PV in approximately 15–20% of high-risk breast and/or ovarian cancer families, and there are currently no specific MBC risk panels. To identify genes potentially involved in MBC risk in an autosomic dominant model, we focused our attention on rare germline variants presented in an MBC population (that tested negative for BRCA1/BRCA2/PALB2 genes) by investigating a panel of 585 genes. We then compared the germline mutation profile of these genes detected in MBC to that of a population with FBC.
2. Results
2.1. Patient Characteristics
All patients diagnosed with MBC but testing negative for PV/LPV on BRCA1/BRCA2/PALB2 genes and referred for an oncogenetics consultation in our institute over four years were included in this study (85 patients). Patients in our cohort were diagnosed with MBC at a median age of 66 years (ranging from 23 to 88 years of age), which is similar to the median age classically described in different works [20,21] (Table 1). MBCs were predominantly invasive ductal carcinoma (80.7%), HR-positive (98.8%) and HER2-negative (96.3%) tumours that were treated by hormone therapy (90.1%). The majority of patients were Caucasian (98.8%), with 1.2% from other ethnic groups. Our cohort presented with 27.1% having first-degree relatives with a history of breast and/or ovarian cancer and 8.2% with an affected second-degree relative. Twenty-five patients were diagnosed with a second cancer: bilateral breast (n = 5), prostate (n = 12) and colon, urinary tract or lung cancers (n = 8). More than half of our MBC patients (58.7%) had a BMI greater than 25, and 15.9% had a BMI greater than 30; furthermore, 35.2% of them had smoked tobacco at some stage during their lives.

Table 1.
Patient characteristics. Data are presented as n (%, with percentages excluding any missing data) or median (range).
2.2. Germline Pathogenic and Likely Pathogenic Variants
The sequencing of our 585-gene panel generated a mean of 6295 variants per patient. Figure 1 describes the filters used to select the variants. Briefly, we selected rare coding variants with a population minor allele frequency from GnomAD (MAF) < 1% and a CADD_phred score > 20 [22,23], and we excluded variants previously detected >300 times from our sequencing databases to avoid introducing recurrent artefact-related variants. The same filters were applied to the GnomAD database v2.1.1 restricted to male noncancer non-Finnish Europeans (NFE). We then selected the variants preferentially detected in our MBC population (adjusted p-value < 0.05), thus identifying 281 variants. The American College of Medical Genetics and Genomics (ACMG) criteria [24] were manually applied to classify 218 variants of unknown significance (VUS; Supplementary Tables S1 and S2) and 21 PVs/LPVs preferentially detected in our MBC population compared to the male NFE (Table 2). Some of these PV/LPV carriers also carried one or several ACMG-classified VUS (up to six different VUS per patient; Supplementary Table S2), including for patient #40 a PALB2 VUS. According to ACMG recommendations concerning the nonuse of VUS in clinical decision, we restricted our present study to PVs or LPVs. Five of the PV/LPV-carrying genes identified had already been associated with an increased risk of FBC in the case of BARD1 (BRCA1-associated Ring Domain 1) and MRE11 (MRE11 homolog, double-strand break repair nuclease); colon cancer in the case of MUTYH (mutY DNA glycosylase) when both copies of the gene are mutated; ovarian cancer in the case of RAD51C (RAD51 paralog C); or pancreatic cancer and melanoma in the case of CDKN2A (Cyclin Dependent Kinase Inhibitor 2A). Men carrying these PVs/LPVs did not report any familial history of colon, pancreatic or ovarian cancer, although we did not have a family history for patient #32, who was adopted (Table 2).
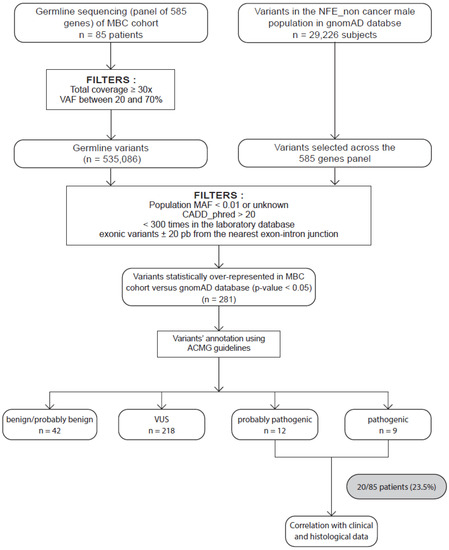
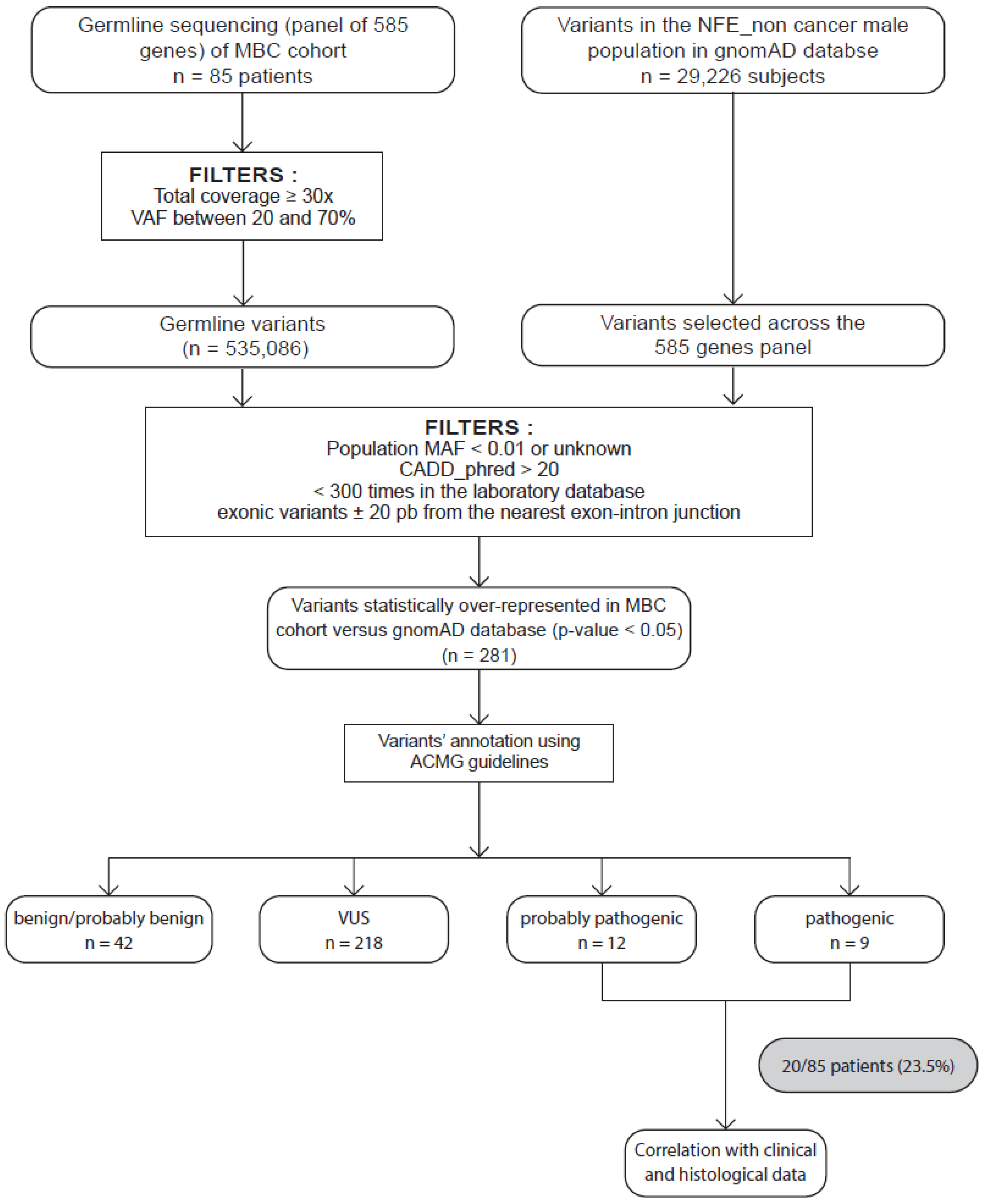
Figure 1.
Strategy to identify candidate MBC susceptibility genes. Eighty-five patients with male breast cancer (MBC) were included. Germline variants of a panel of 585 genes were sequenced and aligned to the human genome hg19 before variant calling and annotation. All germline variants were identified and filtered as described in the Materials and Methods section. We obtained a total of 535086 variants. In parallel, we collected and applied the same filters on variants in the GnomAD database restricted to male noncancer NFE and selected across our 585-gene panel. Statistical tests were performed on 495 variants. We then selected the variants preferentially detected in our MBC population (adjusted p-value < 0.05), identifying 281 variants. These variants were then classified as PV, LPV, or variant of uncertain significance (VUS) and likely benign (LBV) or benign (BV) according to the ACMG criteria. We obtained 12 LPVs and 9 PVs. We also performed correlations with clinical and histological characteristics of patients.

Table 2.
Molecular characteristics of PV/LPV germline variants in MBC patients. VAF, variant allele frequency; p-value and ACMG criteria for each variant; MAF, population minor allele frequency from GnomAD.
2.3. MBC Patients with Germline Variants
A number of MBC patients who had at least one PV/LPV had a personal history of cancer: prostate carcinoma for patients #42 (diagnosed at 72 years), #56 (at 68 years) and #72 (at 68 years), melanoma for patient #41, lymphoma for patient #18 and gallbladder carcinoma for patient #32. Only three MBC patients with a PV/LPV had a first-degree relative with breast or ovarian cancer history: patient #41 was identified with a RECQL4 PV/LPV and diagnosed with MBC at 77 years of age (his sister was diagnosed with breast cancer at 35 and his brother with pancreatic cancer at 79), patient #83 had a PALLD PV/LPV and was diagnosed with MBC at 59 years of age (his mother was diagnosed with breast cancer at 55) and patient #14 was an ERCC2 PV/LPV carrier and was diagnosed with MBC at 77 years of age (his mother was diagnosed with ovarian cancer at the age of 88).
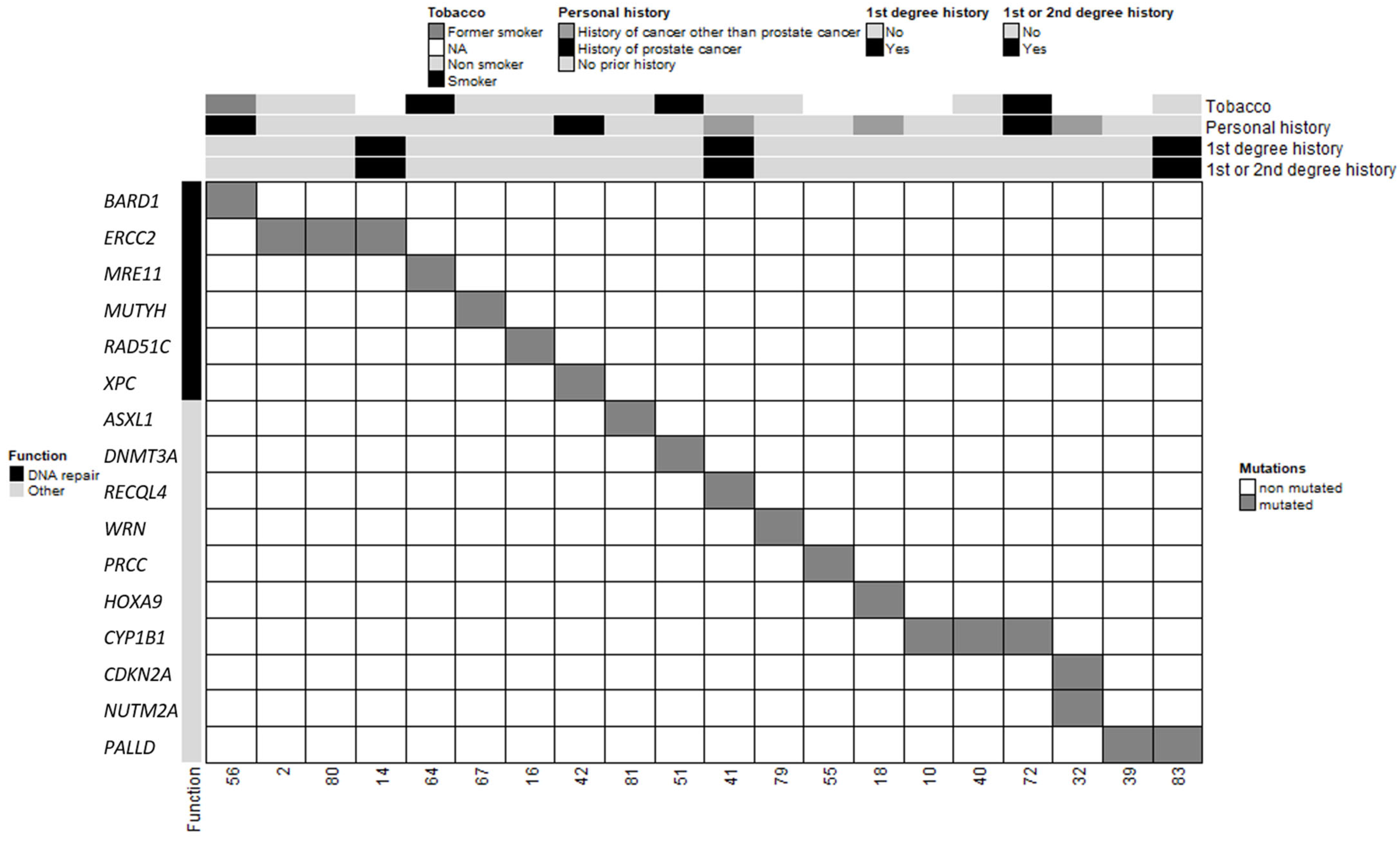
In total, 20 patients in our cohort carried at least one PV or LPV in 1 of the 585 genes screened, and 3 patients carried 2 variants (Table 2 and Figure 2). Among the three patients with two distinct PVs/LPVs, patient #32 was notable because his two variants mapped to two distinct genes: CDKN2A and NUTM2A. Two other MBC patients (patients #2 and #80) carried the same two PVs on the ERCC2 gene: c.1381C>G and c.2150C>G. The literature describes multiple individuals in whom these two ERCC2 variants occur in cis and as part of a complex allele of compound heterozygous genotypes with other pathogenic alleles and features of xeroderma pigmentosum (XPD; OMIM #278730) and trichothiodystrophy (TTD1; OMIM #601675) [25,26,27]. Our two MBC patients carrying these two ERCC2 variants did not present any of the characteristics of these diseases. Patient #2 only reported a case of a brain tumour in a paternal aunt diagnosed between 40 and 50 years of age, and patient #80 did not report a family history of cancer.
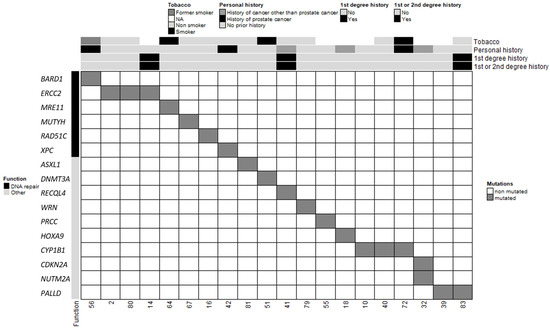
Figure 2.
Clinical and molecular characteristics of MBC patients and PVs/LPVs. Each column corresponds to a patient (patient ID specified). The upper section shows the patient’s clinical characteristics (smoking status, personal history of cancer, first-degree relatives with breast and/or ovarian cancer and first- and second-degree relatives with breast and/or ovarian cancer).
The other PVs/LPVs detected in our MBC cohort mapped to 13 distinct genes (Figure 2). However, because DNMT3A and ASXL1 often contain somatic mutations in a variety of adult hematologic malignancies [28], we chose to exclude these variants from subsequent analyses. The remaining 11 genes predominantly encode proteins involved in DNA repair (MUTYH, MRE11, XPC, RAD51C and BARD1), maintenance of genomic stability (RECQL4, WRN), pre-mRNA processing (PRCC) and transcription regulation (HOXA9). These results suggest that the risk of MBC may be mainly associated with DNA repair and maintenance of genomic integrity. Five patients presented with PVs/LPVs in genes that control other cellular pathways such as the PALLD gene, which encodes the f-actin organising protein palladin (patients #39 and #83), and the CYP1B1 gene that encodes the cytochrome P450 family 1 subfamily B member 1 (patients #10, #40 and #72). Segregation of a PALLD PV (c.2176C>T, p.Pro726Ser) was first identified in members of a high-risk pancreatic cancer family [29]. More recently, another PALLD PV (c.154G>A, p.Asp52Asn) was reported in another high-risk pancreatic cancer family [30]. However, patients #39 and #83 did not have a family history of pancreatic cancer. CYP1B1 encodes for a key estrogen metabolism enzyme, which metabolises polycyclic aromatic hydrocarbons and 17beta-estradiol [31]. We did not have any information relating to the exposure of patients #10, #40 and #72 to hydrocarbons. The preferential expression of CYP1B1 PVs/LPVs in our MBC population when compared to NFE males suggests that a constitutive defect in CYP1B1-controlled metabolism may increase the risk of MBC in men exposed to these hydrocarbon molecules during their lives.
2.4. Clinical and Family Characteristics of MBC PV/LPV Patients
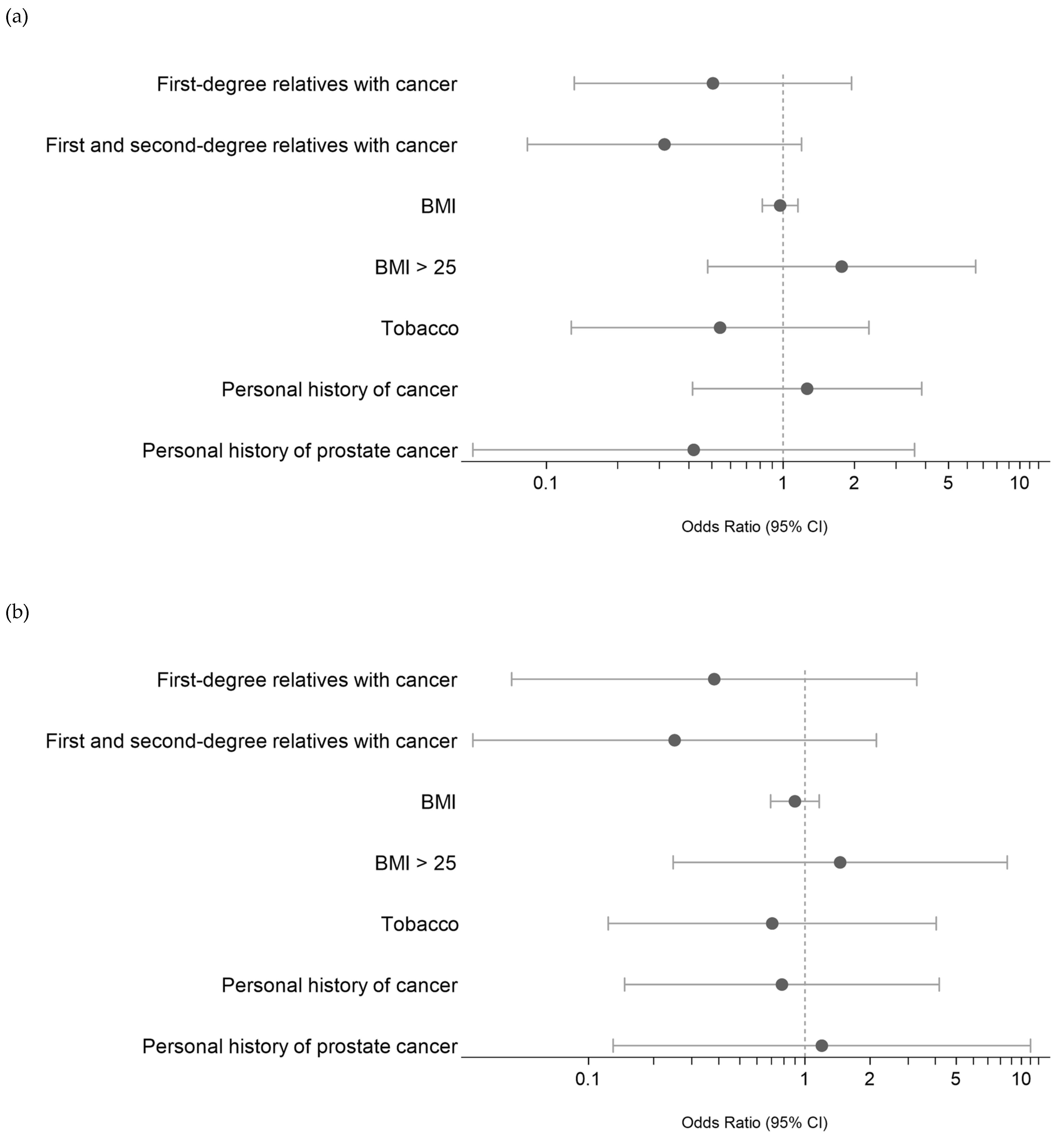
We then attempted to identify any potential associations between these PVs/LPVs (DNMT3A and ASXL1 excluded) and the clinical characteristics of MBC patients, namely, the presence of a first- or second-degree relative with cancer, personal history of cancer, BMI and smoking. As shown in Figure 3A, we did not find any significant association. A subanalysis of the eight MBC patients with PVs/LPVs in DNA repair pathway genes (Figure 3B) also found no significant association.
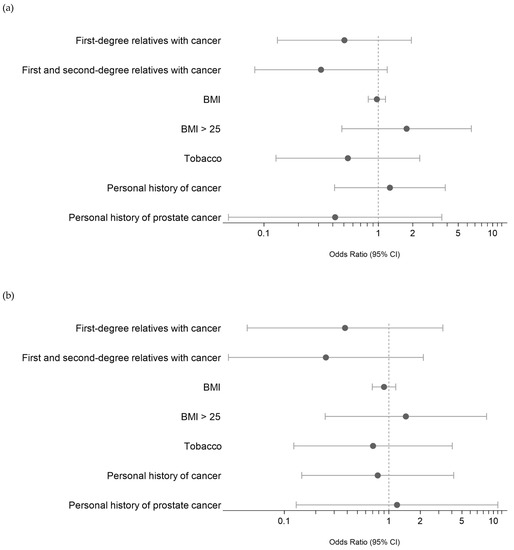
Figure 3.
(a) Risk evaluation (ODD) of PV/LPV germline variants in MBC based on clinical characteristics. References of categorical values: first- or first- and second-degree relatives with cancer: presence of cancer; personal history of cancer: presence; smoking status = former and current. (b) Risk evaluation of PV/LPV germline DNA damage variants in MBC based on clinical characteristics. References of categorical values: first- or first- and second-degree relatives with cancer: presence of cancer; personal history of cancer: presence; smoking status = former and current.
2.5. Study of the 14 MBC PV/LPV-Carrying Genes in a Population of High-risk Breast Cancer Women
To investigate whether MBC and FBC risk might be at least in part due to different genes, we then examined their mutability in a population of high-risk breast cancer women without BRCA1/BRCA2/PALB2 PV/LPV detected on the same 585-gene panel analysis. These women (n = 109) had families that fulfilled the criteria admitted for high-risk breast/ovarian cancer families according to the national recommendations; all of them were addressed to the oncogenetics medical consultation of our institute. We examined whether the fourteen MBC PVs/LPVs were also present in high-risk breast cancer women. Similar to our MBC cohort, we selected women diagnosed with invasive intraductal carcinoma (95.4%), HR-positive and HER2-negative tumours (92.7%) and screened the whole coding region of the 14 previously identified genes for any PVs/LPVs. As shown in Table 3, only 5/109 women (4.6%) were identified as having a PV/LPV in these fourteen genes versus 18/85 men (21.2%). Among these genes, 11 did not contain any PVs/LPVs in the FBC population. These genes that were not mutated in the FBC cohort also included genes involved in DNA repair functions (MUTYH, RECQL4, BARD1, ERCC2 and XPC) and other cellular functions (CDKN2A, HOXA9, NUTM2A, PALLD, PRCC and WRN). Three MBC patients presented one PV/LPV in the ERCC2 gene and two MBC patients in the PALLD gene, but none presented in our FBC population. These results show that the PV/LPV profiles identified in our MBC cohort are not identical to our FBC population.

Table 3.
Comparing the frequencies of PV/LPV-carrying genes in the male and female breast cancer cohorts. The fourteen genes (DNT3A and ASXL1 genes excluded) were examined in a population of 109 women with breast cancer and not carrying PV/LPV on BRCA1, BRCA2 and PALB2 genes.
3. Discussion
Our work identified germline PVs/LPVs from a panel of 585 genes in a population of 85 MBC BRCA1/BRCA2/PALB2-negative patients. We detected PVs/LPVs in fourteen genes preferentially mutated in our MBC population when compared to a male noncancer NFE population (DNMT3A and ASXL1 excluded). Previous works have tended to identify genes involved in MBC risk based on either more or less restricted panels of genes on large MBC populations or exome analysis of small MBC populations. Bucalo et al. recently screened 1349 MBC cases with a 50-gene panel and reported the involvement of ATM and PALB2 genes in the risk of MBC [16]. Another study examining 94 genes in 102 Greek MBC patients identified PVs in 6 genes: BRCA2, ATM, BRCA1, CHEK2, PMS2 and FANCL [15]. In a cohort of Italian MBC (523 patients) analysed with a panel of 50 cancer-associated genes, PALB2 and RAD51D gene variants were significantly associated with MBC risk [21]. Whole exome sequencing performed on six MBC cases revealed germline variants in BRCA2, MSH5, DCC, ERBB3, NOTCH3, DIAPH1 and DNAH11, but no statistical tests were performed in this study [17]. Our current study showed that our MBC cohort presents a distinct mutation profile when compared to a male noncancer NFE population by preferentially carrying at least one PV/LPV variant in the CDKN2A, HOXA9, NUTM2A, PALLD, PRCC, RECQL4, WRN, CYP1B1, BARD1, ERCC2, MRE11, MUTYH, RAD51C or XPC genes. Our results are consistent with a previous study identifying BARD1 and MUTYH mutations in MBCs [21]. BARD1 (BRCA1-associated Ring Domain 1), which encodes a nuclear partner of BRCA1 interacting with the N-terminal region of BRCA1, has been identified as a moderate penetrance predisposition gene in FBC [32]. The high allelic frequency of the more common MUTYH variants in the general population may explain the concordant MUTYH results in our study and the Rizzolo et al. study (c.1105delC in our study (MAF = 0.01% in NFE population); c.494A > G (MAF = 0.25% in NFE population) and c.679C > T (MAF = 0.005% in NFE population) in Rizzolo et al.’s work) [21].
Consistent with what has been described in the literature, our study confirms that PVs/LPVs in MBC patients predominantly map to key DNA repair genes (ERCC2, MRE11, XPC, RAD51C and BARD1) or to genes that safeguard genomic stability (RECQL4, WRN). The DNA repair pathways controlled by these genes involve nucleotide excision repair pathways (ERCC2, XPC), homologous recombination (HR) repair pathways (BARD1, RAD51C) and nonhomologous DNA end joining (NHEJ) repair pathways (MRE11). Our data are in line with previously published data [21] which suggest that, as for other cancer predisposition syndromes, some MBCs may be favoured by a constitutive defect in at least one DNA repair pathway. Our work demonstrated that genes involved in pre-mRNA processing (PRCC) and transcription regulation (HOXA9) are also preferentially mutated in our MBC cohort. A PRCC (which codes for the Proline Rich Mitotic Checkpoint Control Factor) and TFE3 (transcription factor E3) gene translocation was identified in papillary renal carcinoma in 1996 [33]. HOXA9, a member of the homeobox gene family that controls gene expression during early development, plays a crucial role in haematopoiesis. Indeed, HOXA9 dysregulation is necessary for leukemic transformation [34]. RECQL4 codes for a DNA helicase of the Rec family, which unwinds double-stranded DNA into single-stranded DNA and is a key factor involved in maintaining genomic stability. Homozygous or compound heterozygous germline mutations in the RECQL4 gene are linked to the recessive syndromes Rapadilino syndrome (RAPADILINO syndrome; OMIM #266280) and Baller–Gerold syndrome (BGS; OMIM #218600). Recent work investigating the cancer risk in a cohort of 123 individuals with heterozygous germline RECQL4 mutations showed that the prevalence of cancer was not increased in these patients but that patients with type II Rothmund–Thomson syndrome with biallelic REC mutations (including RECQL4) were specifically at increased risk of developing lymphomas and osteosarcomas [35]. Another member of the Rec family, WRN, is associated with Werner syndrome (WS; OMIM #277700) and increases the risk of thyroid carcinoma, melanoma, breast cancer and meningioma as well as soft tissue and bone sarcomas [36]. None of our MBC patients presented with the characteristic phenotypes of these different syndromes. To the best of our knowledge, no germline PVs have to date been reported in the PRCC, HOXA9, RECQL4 or WRN genes in MBC patients. These results suggest that in addition to the DNA repair pathways typically reported in cancer predisposition syndromes, pathways controlling genomic stability, in particular heterozygous PVs in genes of family members of the Rec helicases, may also be involved in the risk of some MBCs.
Our study also identified PVs/LPVs in four other genes involved in cellular functions independent of DNA repair or genomic integrity functions (CDKN2A, NUTM2A, PALLD and CYP1B1). CDKN2A is a well-established melanoma and pancreatic cancer predisposition gene. Given that the family history of patient #32, who had a germline PV in the CDKN2A gene, is unknown, a familial predisposition to these two cancers cannot be ruled out. CYP1B1 (cytochrome P450 1B1), a member of the cytochrome P450 family, is involved in the metabolism of estrogens and a variety of xenobiotics, such as polycyclic aromatic hydrocarbons, which can be oxidized to active carcinogenic products by CYP1B1. Homozygous or compound heterozygous germline mutations in the CYP1B1 gene are linked to the recessive syndrome primary congenital glaucoma (GLC3A; OMIM #231300). CYP1B1 polymorphisms have also been found to increase the risk of prostate and breast cancer [37,38]. Three patients in our MBC cohort carried one of the three different PVs/LPVs, two of which were predicted to encode a truncated, potentially inactive CYP1B1 protein. Several studies have, in turn, shown significant associations between MBC risk and estrogen imbalance [10] as well as exposure of men to polycyclic aromatic hydrocarbons [39]. It may be reasonable to extrapolate that the presence of CYP1B1 gene PVs/LPVs that affect both estrogen and hydrocarbon metabolism might increase the risk of developing MBC, at least in men exposed to hydrocarbons. Interestingly, our study of 85 MBCs did not identify any associations between MBC and a patient or family history of cancer. This may be consistent with MBC predisposition being due to a combination of genetic variants affecting specific metabolic genes and genes involved in the clearance of environmental hydrocarbons.
We also examined whether the PVs/LPVs in the 14 genes of the 18 MBC patients were present at similar frequencies in a cohort of 109 high-risk breast/ovarian cancer women without BRCA1/BRCA2/PALB2 PV/LPV. It was notable that two genes, PALLD and ERCC2, presented PVs/LPVs in 5.9% of our MBC population (respectively, 2.4 and 3.5%) but none in our 109 FBC patients. The PALLD gene encodes palladin, a protein that regulates actin cytoskeleton organisation as well as cell adhesion and migration/invasion [40]. Germline mutations in PALLD have been reported in high-risk pancreatic cancer families [29,30,41]. To date, only one study has reported PALLD germline variants in a woman diagnosed with breast cancer at the age of 38 from a high-risk breast/ovarian cancer Chinese family (a total of 389 patients investigated) [42], but the involvement of this variant in breast cancer risk remains to be clearly established. To the best of our knowledge, we are the first study to demonstrate that germline PVs/LPVs mapping to a gene involved in cellular invasion, PALLD, are represented at an increased prevalence in an MBC cohort compared with a male noncancer NFE and an FBC population. The second gene, ERCC2, encoding an adenosine triphosphate (ATP)-dependent DNA helicase, is frequently mutated in families with increased nasopharyngeal carcinoma risk [43]. A polymorphism in this gene (p.Asp312Asn) is associated with bladder, oesophageal, and gastric cancers but not with breast or prostate cancer [44]. Taken together, we show that 11 out of the 14 genes tested in our MBC cohort (n = 85) contained PVs/LPVs but that these 11 genes were all wild type in the FBC cohort (n = 109). This suggests that in addition to the well-established high-risk breast cancer genes previously described in FBC and MBC, other genes involved in cell adhesion and DNA repair may form part of a specific MBC risk signature.
To conclude, we have identified germline PVs/LPVs in genes not previously associated with MBC risk, which may suggest a potential specific role of at least some of these genes in male breast cancer. However, our present results do not allow us to firmly associate these genes with MBC risk. To further explore the involvement of these genes in MBC risk, larger MBC population studies will be needed along with FBC cohorts to firmly establish the presence of a specific MBC profile risk. In the meantime, identifying variants in these MBC-specific genes in the clinic may be important for better patient care and for guiding personalized therapy, as evidenced by the success of PARP inhibitors in patients with BRCA1/BRCA2-mutated cancers or other defects in homologous recombination DNA repair mechanisms including those reported here.
4. Materials and Methods
4.1. Patient Cohort
This is a retrospective study of inherited defects in cancer genes in all patients diagnosed with MBC, followed by the oncogenetics department at the IUCT-Oncopole, Toulouse (France), between 2017 and 2020. This study included 85 consecutive MBC patients who tested negative for pathogenic BRCA1/BRCA2/PALB2 gene variants. Clinical data, tumour characteristics and molecular results were retrospectively reviewed for all MBC patients. Informed consent for using information and biological samples was obtained from all participants. For comparison with FBC and for descriptive purposes only, we selected a population of 109 FBC (without MBC family history) from the last ones referred to our laboratory to perform the same genetic analysis. All these women were BRCA1/BRCA2/PALB2-negative and shared the same tumour characteristics as the male population.
4.2. NGS Analysis Panel of 585 Genes
DNA was extracted from whole blood with a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany), captured using a KAPA library amplification kit (Roche; list of genes available on Supplementary Table S3) and sequenced on an Illumina NextSeq500 sequencer (Illumina, San Diego, CA, USA) according to manufacturers’ instructions. Germline variants were then aligned to the human genome hg19 and called using HaplotypeCaller from GATK 3.3.0 [45] and VarScan 2.3.7 [46]. Only high-confidence variants (total sequencing depth ≥ 30 and variant allele frequency (VAF) between 20% (to avoid any sequencing artefact) and 70% (to avoid homozygous variants)) were reported in this study. These variants have been confirmed by a review of BAM files with Integrative Genomics Viewer (IGV) [47].
4.3. Germline Data Analysis
The sequencing data were analysed for the presence of single nucleotide variants and small insertions and deletions (coding variants or variants which were less than 20 bp away from the nearest exon–intron junction). All variants with a frequency of less than 1% in the public GnomAD v2.1.1 database were investigated and then manually classified according to the published ACMG recommendations [24]. Classification criteria included information from curated databases, computer predictions of the effects of mutations on protein function and data from the medical literature. PVs/LPVs including nonsense, frameshift, splice site and missense variants were included. For missense variants, we only selected those with published evidence supporting a pathogenic or likely pathogenic function in the ClinVar [48] or Uniprot databases [49]. Details of the data analysis and interpretation are provided in Figure 1.
4.4. Statistical Analysis
Categorical variables are expressed as frequencies and percentages and continuous variables as medians and ranges. Comparisons of individual variant frequencies between MBC and a control male NFE population from the GnomAD database were performed using the Fisher exact test with a Benjamini–Hochberg procedure for multiple testing. Associations between clinical characteristics and the presence of LPV or PV were assessed using a logistic regression model. Odds ratios (ORs) were estimated with a 95% confidence interval (95% CI). All statistical tests were two-sided, and p-values < 0.05 were considered statistically significant. Statistical analyses were conducted using R (v4.0.2) and STATA software (v16) (Stata Corporation, College Station, TX, USA).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms241814348/s1.
Author Contributions
Conceptualization, C.T., A.A.S., J.P., J.G. and P.V.P.; methodology, J.P., J.G., N.M., B.C. and T.F.; formal analysis, J.P., J.G., N.M., B.C. and T.F.; investigation, A.A.S., D.T., E.P.-L. and N.L.; resources, P.V.P., L.G., V.F., A.S., M.M., E.C. and G.C.; data curation, A.A.S. and N.L.; writing—original draft preparation, C.T. and A.A.S.; writing—review and editing, C.T., A.A.S., F.T. and L.G.; visualization, A.A.S.; supervision, C.T. and A.A.S.; project administration, C.T.; funding acquisition, C.T. and A.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Ligue Contre le Cancer (comités 31 and 65).
Institutional Review Board Statement
This study has been registered on the Health Data Hub (number: F20201005100612; MR004).
Informed Consent Statement
Informed consent was obtained from all subjects involved in this study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to the terms of patients’ inform consent.
Acknowledgments
The authors thank the Oncopole Claudius Regaud’s oncogenetics laboratory staff for their contributions: L. Loncle, A. Da Cruz, I. Gitlaw, C. Villarzel, L. Gourdain and F. Fouque.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cottenet, J.; Dabakuyo-Yonli, T.S.; Mariet, A.; Roussot, A.; Arveux, P.; Quantin, C. Prevalence of Patients Hospitalised for Male Breast Cancer in France Using the French Nationwide Hospital Administrative Database. Eur. J. Cancer Care 2019, 28, e13117. [Google Scholar] [CrossRef]
- Hassett, M.J.; Somerfield, M.R.; Baker, E.R.; Cardoso, F.; Kansal, K.J.; Kwait, D.C.; Plichta, J.K.; Ricker, C.; Roshal, A.; Ruddy, K.J.; et al. Management of Male Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 1849–1863. [Google Scholar] [CrossRef]
- Korde, L.A.; Zujewski, J.A.; Kamin, L.; Giordano, S.; Domchek, S.; Anderson, W.F.; Bartlett, J.M.S.; Gelmon, K.; Nahleh, Z.; Bergh, J.; et al. Multidisciplinary Meeting on Male Breast Cancer: Summary and Research Recommendations. J. Clin. Oncol. 2010, 28, 2114–2122. [Google Scholar] [CrossRef]
- Giordano, S.H.; Cohen, D.S.; Buzdar, A.U.; Perkins, G.; Hortobagyi, G.N. Breast Carcinoma in Men: A Population-Based Study. Cancer 2004, 101, 51–57. [Google Scholar] [CrossRef]
- Johansson, I.; Ringnér, M.; Hedenfalk, I. The Landscape of Candidate Driver Genes Differs between Male and Female Breast Cancer. PLoS ONE 2013, 8, e78299. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Nilsson, C.; Berglund, P.; Strand, C.; Jönsson, G.; Staaf, J.; Ringnér, M.; Nevanlinna, H.; Barkardottir, R.B.; Borg, A.; et al. High-Resolution Genomic Profiling of Male Breast Cancer Reveals Differences Hidden behind the Similarities with Female Breast Cancer. Breast Cancer Res. Treat. 2011, 129, 747–760. [Google Scholar] [CrossRef] [PubMed]
- André, S.; Nunes, P.S.; Silva, F.; Henrique, R.; Félix, A.; Jerónimo, C. Analysis of Epigenetic Alterations in Homologous Recombination DNA Repair Genes in Male Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2715. [Google Scholar] [CrossRef] [PubMed]
- Brinton, L.A.; Richesson, D.A.; Gierach, G.L.; Lacey, J.V.; Park, Y.; Hollenbeck, A.R.; Schatzkin, A. Prospective Evaluation of Risk Factors for Male Breast Cancer. J. Natl. Cancer Inst. 2008, 100, 1477–1481. [Google Scholar] [CrossRef]
- Anderson, W.F.; Althuis, M.D.; Brinton, L.A.; Devesa, S.S. Is Male Breast Cancer Similar or Different than Female Breast Cancer? Breast Cancer Res. Treat. 2004, 83, 77–86. [Google Scholar] [CrossRef]
- Weiss, J.R.; Moysich, K.B.; Swede, H. Epidemiology of Male Breast Cancer. Cancer Epidemiol. Biomarkers Prev. 2005, 14, 20–26. [Google Scholar] [CrossRef]
- Brinton, L.A.; Cook, M.B.; McCormack, V.; Johnson, K.C.; Olsson, H.; Casagrande, J.T.; Cooke, R.; Falk, R.T.; Gapstur, S.M.; Gaudet, M.M.; et al. Anthropometric and Hormonal Risk Factors for Male Breast Cancer: Male Breast Cancer Pooling Project Results. J. Natl. Cancer Inst. 2014, 106, djt465. [Google Scholar] [CrossRef] [PubMed]
- Abdelwahab Yousef, A.J. Male Breast Cancer: Epidemiology and Risk Factors. Semin. Oncol. 2017, 44, 267–272. [Google Scholar] [CrossRef]
- Pritzlaff, M.; Summerour, P.; McFarland, R.; Li, S.; Reineke, P.; Dolinsky, J.S.; Goldgar, D.E.; Shimelis, H.; Couch, F.J.; Chao, E.C.; et al. Male Breast Cancer in a Multi-Gene Panel Testing Cohort: Insights and Unexpected Results. Breast Cancer Res. Treat. 2017, 161, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Tedaldi, G.; Tebaldi, M.; Zampiga, V.; Cangini, I.; Pirini, F.; Ferracci, E.; Danesi, R.; Arcangeli, V.; Ravegnani, M.; Martinelli, G.; et al. Male Breast Cancer: Results of the Application of Multigene Panel Testing to an Italian Cohort of Patients. Diagnostics 2020, 10, 269. [Google Scholar] [CrossRef] [PubMed]
- Fostira, F.; Saloustros, E.; Apostolou, P.; Vagena, A.; Kalfakakou, D.; Mauri, D.; Tryfonopoulos, D.; Georgoulias, V.; Yannoukakos, D.; Fountzilas, G.; et al. Germline Deleterious Mutations in Genes Other than BRCA2 Are Infrequent in Male Breast Cancer. Breast Cancer Res. Treat. 2018, 169, 105–113. [Google Scholar] [CrossRef]
- Bucalo, A.; Conti, G.; Valentini, V.; Capalbo, C.; Bruselles, A.; Tartaglia, M.; Bonanni, B.; Calistri, D.; Coppa, A.; Cortesi, L.; et al. Male Breast Cancer Risk Associated with Pathogenic Variants in Genes Other than BRCA1/2: An Italian Case-Control Study. Eur. J. Cancer 2023, 188, 183–191. [Google Scholar] [CrossRef]
- Ben Kridis-Rejeb, W.; Ben Ayed-Guerfali, D.; Ammous-Boukhris, N.; Ayadi, W.; Kifagi, C.; Charfi, S.; Saguem, I.; Sellami-Boudawara, T.; Daoud, J.; Khanfir, A.; et al. Identification of Novel Candidate Genes by Exome Sequencing in Tunisian Familial Male Breast Cancer Patients. Mol. Biol. Rep. 2020, 47, 6507–6516. [Google Scholar] [CrossRef]
- Campos, F.A.B.; Rouleau, E.; Torrezan, G.T.; Carraro, D.M.; Casali da Rocha, J.C.; Mantovani, H.K.; da Silva, L.R.; de Osório, C.A.B.T.; Moraes Sanches, S.; Caputo, S.M.; et al. Genetic Landscape of Male Breast Cancer. Cancers 2021, 13, 3535. [Google Scholar] [CrossRef]
- Maguire, S.; Perraki, E.; Tomczyk, K.; Jones, M.E.; Fletcher, O.; Pugh, M.; Winter, T.; Thompson, K.; Cooke, R.; kConFab Consortium; et al. Common Susceptibility Loci for Male Breast Cancer. J. Natl. Cancer Inst. 2021, 113, 453–461. [Google Scholar] [CrossRef]
- Cardoso, F.; Bartlett, J.M.S.; Slaets, L.; van Deurzen, C.H.M.; van Leeuwen-Stok, E.; Porter, P.; Linderholm, B.; Hedenfalk, I.; Schröder, C.; Martens, J.; et al. Characterization of Male Breast Cancer: Results of the EORTC 10085/TBCRC/BIG/NABCG International Male Breast Cancer Program. Ann. Oncol. 2018, 29, 405–417. [Google Scholar] [CrossRef]
- Rizzolo, P.; Zelli, V.; Silvestri, V.; Valentini, V.; Zanna, I.; Bianchi, S.; Masala, G.; Spinelli, A.M.; Tibiletti, M.G.; Russo, A.; et al. Insight into Genetic Susceptibility to Male Breast Cancer by Multigene Panel Testing: Results from a Multicenter Study in Italy. Int. J. Cancer 2019, 145, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A General Framework for Estimating the Relative Pathogenicity of Human Genetic Variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.; Danks, D.M.; Salazar, E.P.; Cleaver, J.E.; Weber, C.A. DNA Repair Characteristics and Mutations in the ERCC2 DNA Repair and Transcription Gene in a Trichothiodystrophy Patient. Hum. Mutat. 1997, 9, 519–525. [Google Scholar] [CrossRef]
- Fujimoto, M.; Leech, S.N.; Theron, T.; Mori, M.; Fawcett, H.; Botta, E.; Nozaki, Y.; Yamagata, T.; Moriwaki, S.-I.; Stefanini, M.; et al. Two New XPD Patients Compound Heterozygous for the Same Mutation Demonstrate Diverse Clinical Features. J. Investig. Dermatol. 2005, 125, 86–92. [Google Scholar] [CrossRef]
- Horibata, K.; Kono, S.; Ishigami, C.; Zhang, X.; Aizawa, M.; Kako, Y.; Ishii, T.; Kosaki, R.; Saijo, M.; Tanaka, K. Constructive Rescue of TFIIH Instability by an Alternative Isoform of XPD Derived from a Mutated XPD Allele in Mild but Not Severe XP-D/CS. J. Hum. Genet. 2015, 60, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Brunet, T.; Berutti, R.; Dill, V.; Hecker, J.S.; Choukair, D.; Andres, S.; Deschauer, M.; Diehl-Schmid, J.; Krenn, M.; Eckstein, G.; et al. Clonal Hematopoiesis as a Pitfall in Germline Variant Interpretation in the Context of Mendelian Disorders. Hum. Mol. Genet. 2022, 31, 2386–2395. [Google Scholar] [CrossRef]
- Pogue-Geile, K.L.; Chen, R.; Bronner, M.P.; Crnogorac-Jurcevic, T.; Moyes, K.W.; Dowen, S.; Otey, C.A.; Crispin, D.A.; George, R.D.; Whitcomb, D.C.; et al. Palladin Mutation Causes Familial Pancreatic Cancer and Suggests a New Cancer Mechanism. PLoS Med. 2006, 3, e516. [Google Scholar] [CrossRef]
- Liotta, L.; Lange, S.; Maurer, H.C.; Olive, K.P.; Braren, R.; Pfarr, N.; Burger, S.; Muckenhuber, A.; Jesinghaus, M.; Steiger, K.; et al. PALLD Mutation in a European Family Conveys a Stromal Predisposition for Familial Pancreatic Cancer. JCI Insight 2021, 6, e141532. [Google Scholar] [CrossRef]
- Gamboa-Loira, B.; López-Carrillo, L.; Mar-Sánchez, Y.; Stern, D.; Cebrián, M.E. Epidemiologic Evidence of Exposure to Polycyclic Aromatic Hydrocarbons and Breast Cancer: A Systematic Review and Meta-Analysis. Chemosphere 2022, 290, 133237. [Google Scholar] [CrossRef] [PubMed]
- Rofes, P.; Del Valle, J.; Torres-Esquius, S.; Feliubadaló, L.; Stradella, A.; Moreno-Cabrera, J.M.; López-Doriga, A.; Munté, E.; De Cid, R.; Campos, O.; et al. BARD1 Pathogenic Variants Are Associated with Triple-Negative Breast Cancer in a Spanish Hereditary Breast and Ovarian Cancer Cohort. Genes 2021, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Weterman, M.A.; Wilbrink, M.; Geurts van Kessel, A. Fusion of the Transcription Factor TFE3 Gene to a Novel Gene, PRCC, in t(X;1)(P11;Q21)-Positive Papillary Renal Cell Carcinomas. Proc. Natl. Acad. Sci. USA 1996, 93, 15294–15298. [Google Scholar] [CrossRef] [PubMed]
- Aryal, S.; Zhang, Y.; Wren, S.; Li, C.; Lu, R. Molecular Regulators of HOXA9 in Acute Myeloid Leukemia. FEBS J. 2021, 290, 321–339. [Google Scholar] [CrossRef]
- Martin-Giacalone, B.A.; Rideau, T.-T.; Scheurer, M.E.; Lupo, P.J.; Wang, L.L. Cancer Risk among RECQL4 Heterozygotes. Cancer Genet. 2022, 262–263, 107–110. [Google Scholar] [CrossRef]
- Lauper, J.M.; Krause, A.; Vaughan, T.L.; Monnat, R.J. Spectrum and Risk of Neoplasia in Werner Syndrome: A Systematic Review. PLoS ONE 2013, 8, e59709. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sasaki, M.; Kaneuchi, M.; Shiina, H.; Igawa, M.; Dahiya, R. Polymorphisms of the CYP1B1 Gene Have Higher Risk for Prostate Cancer. Biochem. Biophys. Res. Commun. 2002, 296, 820–826. [Google Scholar] [CrossRef]
- Liu, J.-Y.; Yang, Y.; Liu, Z.-Z.; Xie, J.-J.; Du, Y.-P.; Wang, W. Association between the CYP1B1 Polymorphisms and Risk of Cancer: A Meta-Analysis. Mol. Genet. Genomics 2015, 290, 739–765. [Google Scholar] [CrossRef]
- Palli, D.; Masala, G.; Mariani-Costantini, R.; Zanna, I.; Saieva, C.; Sera, F.; Decarli, A.; Ottini, L. A Gene-Environment Interaction between Occupation and BRCA1/BRCA2 Mutations in Male Breast Cancer? Eur. J. Cancer 2004, 40, 2474–2479. [Google Scholar] [CrossRef]
- Najm, P.; El-Sibai, M. Palladin Regulation of the Actin Structures Needed for Cancer Invasion. Cell Adhes. Migr. 2014, 8, 29–35. [Google Scholar] [CrossRef]
- Landi, S. Genetic Predisposition and Environmental Risk Factors to Pancreatic Cancer: A Review of the Literature. Mutat. Res. 2009, 681, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Lang, G.-T.; Shi, J.-X.; Huang, L.; Cao, A.-Y.; Zhang, C.-H.; Song, C.-G.; Zhuang, Z.-G.; Hu, X.; Huang, W.; Shao, Z.-M. Multiple Cancer Susceptible Genes Sequencing in BRCA-Negative Breast Cancer with High Hereditary Risk. Ann. Transl. Med. 2020, 8, 1417. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-M.; He, Y.-Q.; Xue, W.-Q.; Zhang, J.-B.; Xia, Y.-F.; Deng, C.-M.; Zhang, W.-L.; Xiao, R.-W.; Liao, Y.; Yang, D.-W.; et al. Whole-Exome Sequencing Study of Familial Nasopharyngeal Carcinoma and Its Implication for Identifying High-Risk Individuals. J. Natl. Cancer Inst. 2022, 114, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Pu, J.; Wen, Q.; Huang, Q.; Zhang, Q.; Huang, B.; Huang, S.; Lan, A.; Zhang, Y.; Li, J.; et al. Association between the ERCC2 Asp312Asn Polymorphism and Risk of Cancer. Oncotarget 2017, 8, 48488–48506. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic Mutation and Copy Number Alteration Discovery in Cancer by Exome Sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Wenger, A.M.; Zehir, A.; Mesirov, J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017, 77, e31–e34. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving Access to Variant Interpretations and Supporting Evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- UniProt Consortium UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).