Is HOXA5 a Novel Prognostic Biomarker for Uterine Corpus Endometrioid Adenocarcinoma?
Abstract
:1. Introduction
2. Results
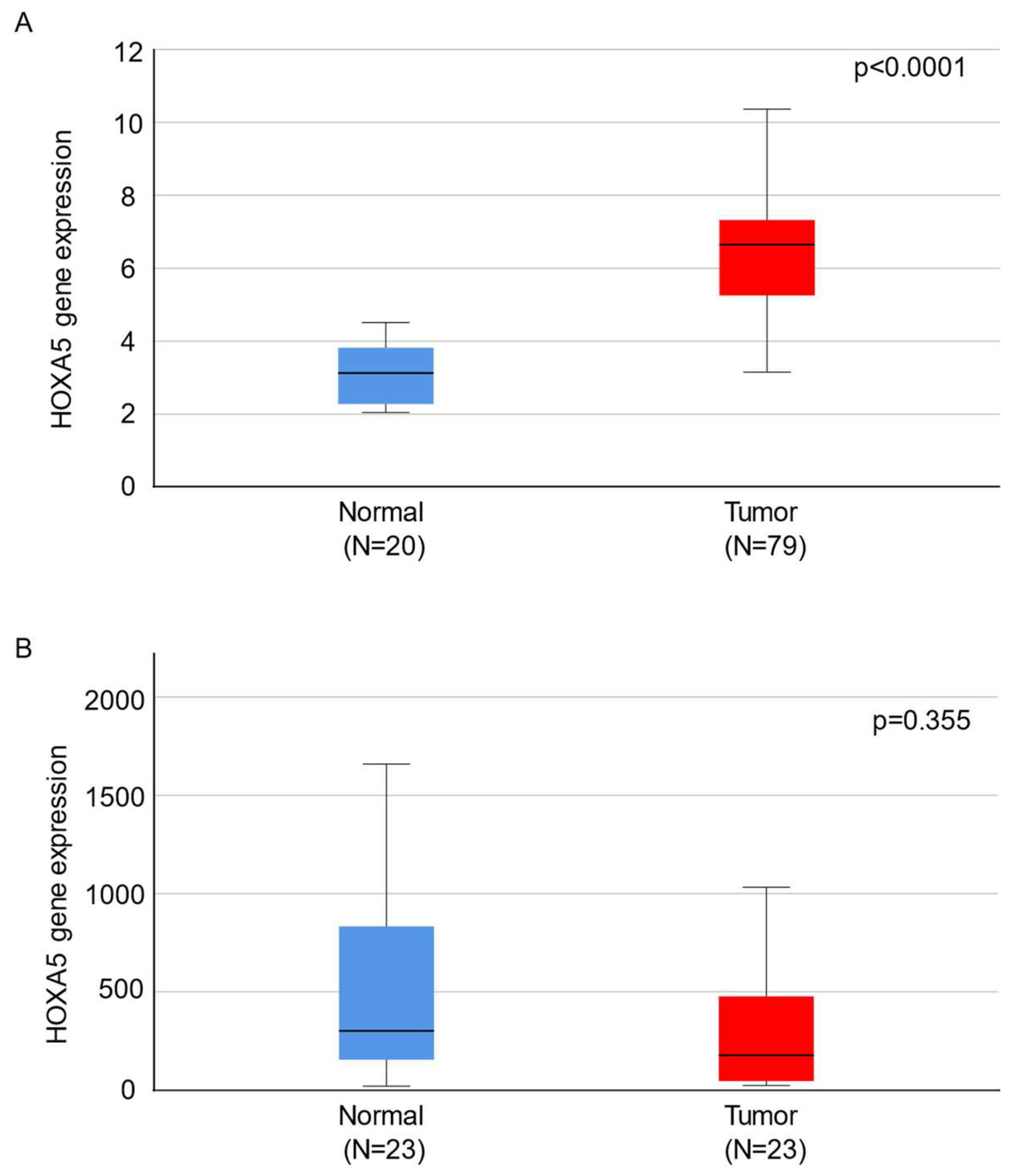
2.1. HOXA5 Is Overexpressed in Endometrial Cancer Tissues
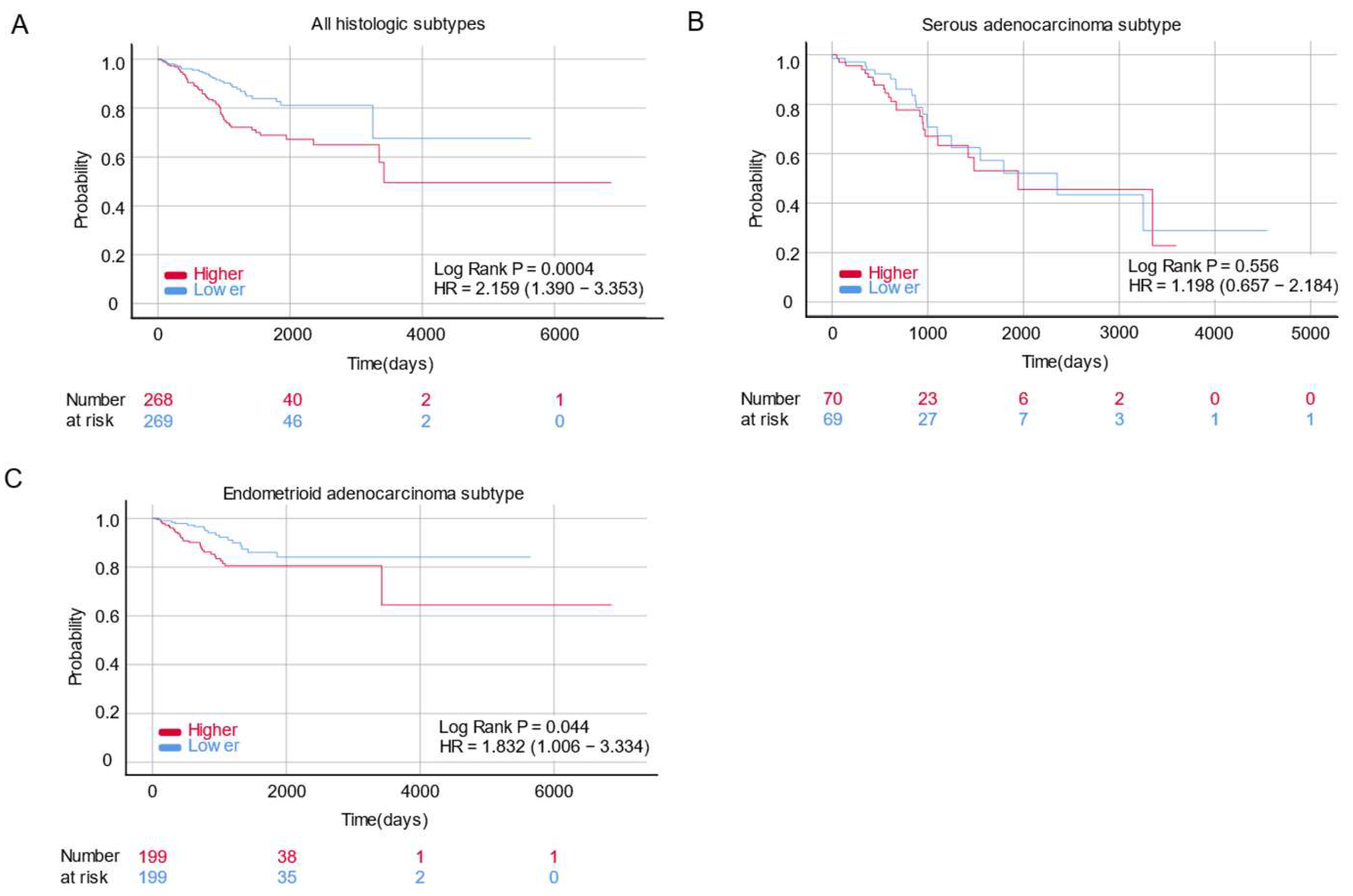
2.2. HOXA5 Is Associated with Poor Survival in Patients with Endometrial Cancer
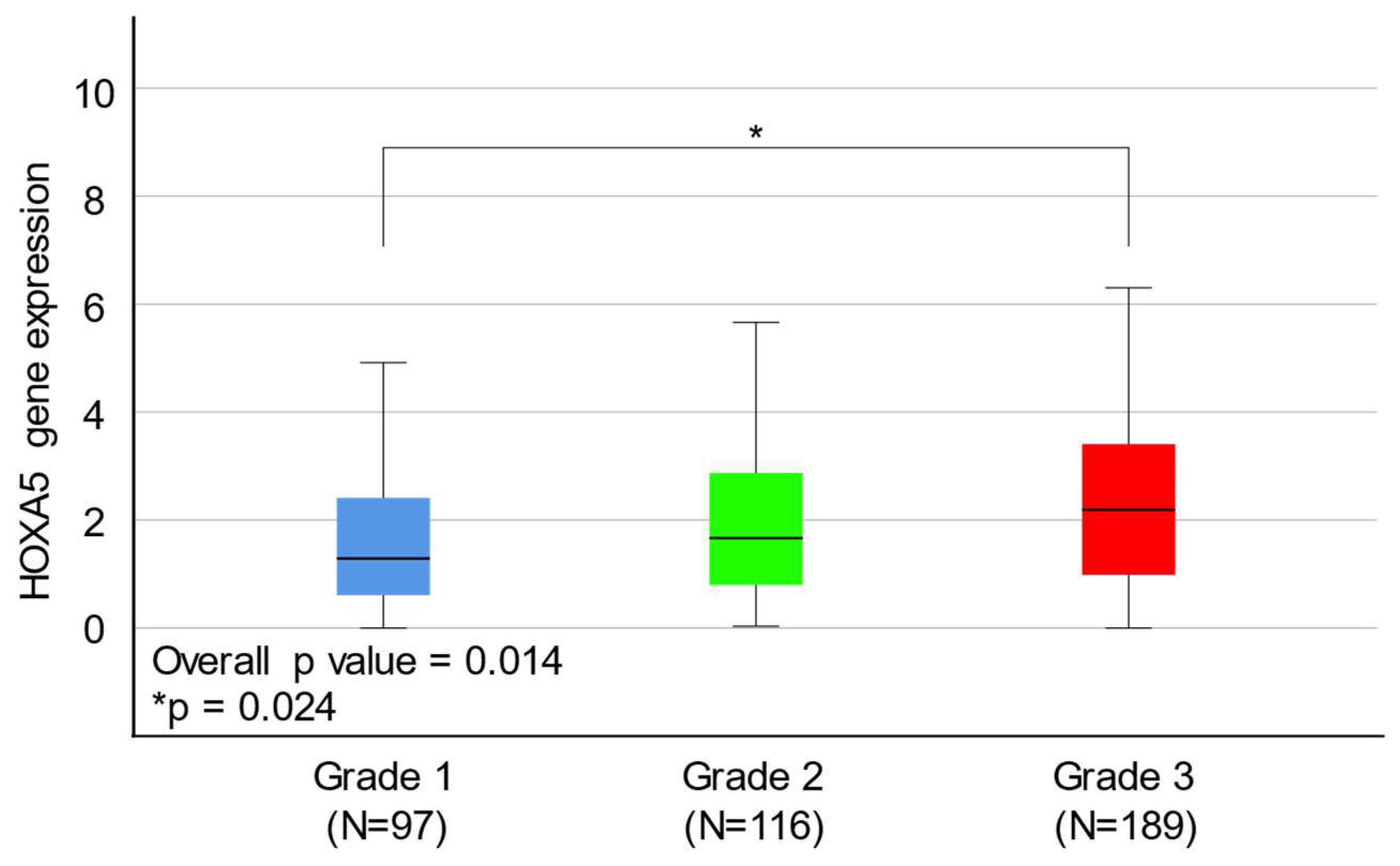
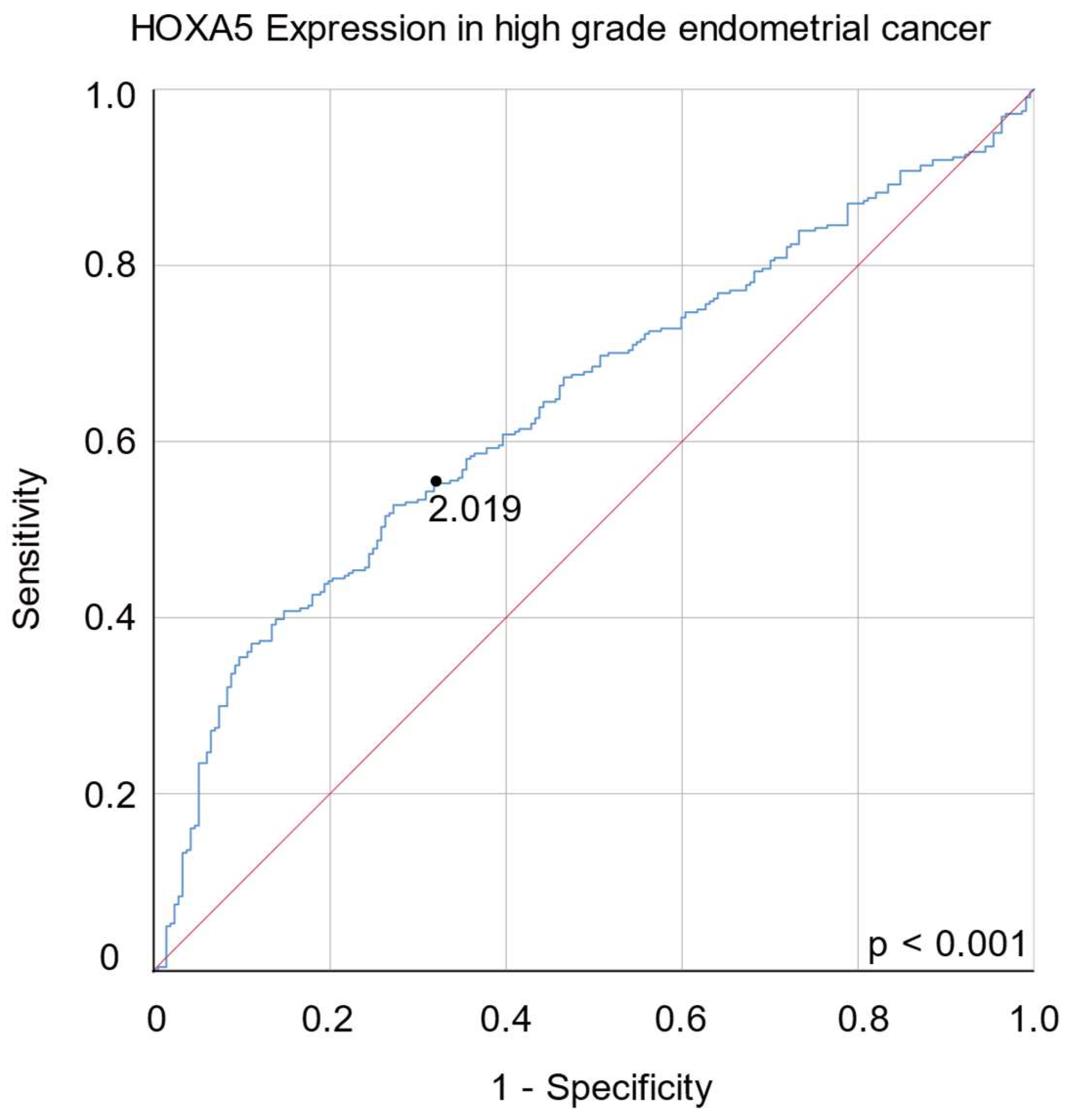
2.3. HOXA5 Overexpression Is Associated with a Higher Histological Grade of Endometrial Cancer
2.4. HOXA5 Is Overexpressed in the High-Copy-Number Endometrial Carcinoma Group
3. Discussion
4. Materials and Methods
4.1. Acquisition of Microarray Datasets
4.2. Data Normalization
4.3. Acquisition and Analysis of Clinical Data
4.4. Subgrouping According to Molecular Classification
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Wang, K.L.; Chen, M.Y.; Yu, M.H.; Wu, C.H.; Ke, Y.M.; Chen, Y.J.; Chang, Y.Y.; Hsu, K.F.; Yen, M.S. Risk factor analysis of coexisting endometrial carcinoma in patients with endometrial hyperplasia: A retrospective observational study of Taiwanese Gynecologic Oncology Group. J. Gynecol. Oncol. 2013, 24, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Hwang, S.O.; Lee, B.; Kim, K.; Kim, Y.B.; Park, S.H.; Choi, H.Y. Risk factors of progression to endometrial cancer in women with endometrial hyperplasia: A retrospective cohort study. PLoS ONE 2020, 15, e0243064. [Google Scholar] [CrossRef] [PubMed]
- Amant, F.; Moerman, P.; Neven, P.; Timmerman, D.; Van Limbergen, E.; Vergote, I. Endometrial cancer. Lancet 2005, 366, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Schmeler, K.M.; Soliman, P.T.; Sun, C.C.; Slomovitz, B.M.; Gershenson, D.M.; Lu, K.H. Endometrial cancer in young, normal-weight women. Gynecol. Oncol. 2005, 99, 388–392. [Google Scholar] [CrossRef]
- Setiawan, V.W.; Yang, H.P.; Pike, M.C.; McCann, S.E.; Yu, H.; Xiang, Y.B.; Wolk, A.; Wentzensen, N.; Weiss, N.S.; Webb, P.M.; et al. Type I and II endometrial cancers: Have they different risk factors? J. Clin. Oncol. 2013, 31, 2607–2618. [Google Scholar] [CrossRef]
- Clarke, M.A.; Long, B.J.; Del Mar Morillo, A.; Arbyn, M.; Bakkum-Gamez, J.N.; Wentzensen, N. Association of endometrial cancer risk with postmenopausal bleeding in women: A systematic review and meta-analysis. JAMA Intern. Med. 2018, 178, 1210–1222. [Google Scholar] [CrossRef]
- Dowdy, S.C. Improving oncologic outcomes for women with endometrial cancer: Realigning our sights. Gynecol. Oncol. 2014, 133, 370–374. [Google Scholar] [CrossRef]
- Tejerizo-García, A.; Jiménez-López, J.S.; Muñoz-González, J.L.; Bartolomé-Sotillos, S.; Marqueta-Marqués, L.; López-González, G.; Gómez, J.F. Overall survival and disease-free survival in endometrial cancer: Prognostic factors in 276 patients. Onco Targets Ther. 2013, 9, 1305–1313. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, D.Y.; Kim, T.J.; Kim, J.W.; Kim, J.H.; Kim, Y.M.; Kim, Y.T.; Bae, D.S.; Nam, J.H. Hormonal therapy for women with stage IA endometrial cancer of all grades. Obstet. Gynecol. 2013, 122, 7–14. [Google Scholar] [CrossRef]
- Singh, N.; Hirschowitz, L.; Zaino, R.; Alvarado-Cabrero, I.; Duggan, M.A.; Ali-Fehmi, R.; Euscher, E.; Hecht, J.L.; Horn, L.C.; Ioffe, O.; et al. Pathologic prognostic factors in endometrial carcinoma (other than tumor type and grade). Int. J. Gynecol. Pathol. 2019, 38 (Suppl. 1), S93–S113. [Google Scholar] [CrossRef] [PubMed]
- Yarandi, F.; Shirali, E.; Akhavan, S.; Nili, F.; Ramhormozian, S. The impact of lymphovascular space invasion on survival in early stage low-grade endometrioid endometrial cancer. Eur. J. Med. Res. 2023, 28, 118. [Google Scholar] [CrossRef] [PubMed]
- Guerra, E.; Matias-Guiu, X. Relevance of pathologic features in risk stratification for early-stage endometrial cancer. J. Gynecol. Oncol. 2021, 32, e67. [Google Scholar] [CrossRef] [PubMed]
- Huvila, J.; Orte, K.; Vainio, P.; Mettälä, T.; Joutsiniemi, T.; Hietanen, S. Molecular subtype diagnosis of endometrial carcinoma: Comparison of the next-generation sequencing panel and Proactive Molecular Risk Classifier for Endometrial Cancer classifier. Hum. Pathol. 2021, 111, 98–109. [Google Scholar] [CrossRef]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jürgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Yang, W.; Lum, A.; Senz, J.; Boyd, N.; Pike, J.; Anglesio, M.; Kwon, J.S.; et al. Confirmation of ProMisE: A simple, genomics-based clinical classifier for endometrial cancer. Cancer 2017, 123, 802–813. [Google Scholar] [CrossRef]
- Yen, T.T.; Wang, T.L.; Fader, A.N.; Shih, I.M.; Gaillard, S. Molecular classification and emerging targeted therapy in endometrial cancer. Int. J. Gynecol. Pathol. 2020, 39, 26–35. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Zakany, J.; Duboule, D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007, 17, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Boucherat, O.; Montaron, S.; Bérubé-Simard, F.A.; Aubin, J.; Philippidou, P.; Wellik, D.M.; Dasen, J.S.; Jeannotte, L. Partial functional redundancy between Hoxa5 and Hoxb5 paralog genes during lung morphogenesis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L817–L830. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Mo, H.; Zhang, H.; Dai, Z.; Wang, Z.; Qu, C.; Liu, F.; Zhang, L.; Luo, P.; Zhang, J.; et al. HOXA5: A crucial transcriptional factor in cancer and a potential therapeutic target. Biomed. Pharmacother. 2022, 155, 113800. [Google Scholar] [CrossRef] [PubMed]
- Ordóñez-Morán, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 counteracts stem cell traits by inhibiting Wnt signaling in colorectal cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Hussain, I.; Deb, P.; Chini, A.; Obaid, M.; Bhan, A.; Ansari, K.I.; Mishra, B.P.; Bobzean, S.A.; Udden, S.M.N.; Alluri, P.G.; et al. HOXA5 expression is elevated in breast cancer and is transcriptionally regulated by estradiol. Front. Genet. 2020, 11, 592436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.L.; Nie, F.Q.; Sun, M.; Xia, R.; Xie, M.; Lu, K.H.; Li, W. HOXA5 indicates poor prognosis and suppresses cell proliferation by regulating p21 expression in non small cell lung cancer. Tumour Biol. 2015, 36, 3521–3531. [Google Scholar] [CrossRef]
- Peng, X.; Zha, L.; Chen, A.; Wang, Z. HOXA5 is a tumor suppressor gene that is decreased in gastric cancer. Oncol. Rep. 2018, 40, 1317–1329. [Google Scholar] [CrossRef]
- Pei, L.; Wang, Z.Q.; Shen, J.; Yang, Y.Z.; Tian, J.; He, X.; Lin, J.; Hou, Q.Y.; Mo, W.F.; Zhao, H.L.; et al. Expression and clinical significance of HOXA5, E-cadherin, and β-catenin in cervical squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2018, 11, 3091–3096. [Google Scholar]
- Ma, H.M.; Cui, N.; Zheng, P.S. HOXA5 inhibits the proliferation and neoplasia of cervical cancer cells via downregulating the activity of the Wnt/β-catenin pathway and transactivating TP53. Cell Death Dis. 2020, 11, 420. [Google Scholar] [CrossRef]
- Dziobek, K.; Oplawski, M.; Zmarzły, N.; Gabarek, B.O.; Kiełbasiński, R.; Kiełbasiński, K.; Kieszkowski, P.; Talkowski, K.; Boroń, D. Assessment of expression of Homeobox A5 in endometrial cancer on the mRNA and protein level. Curr. Pharm. Biotechnol. 2020, 21, 635–641. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Okayama, A.; Fujita, M.; Enomoto, T.; Tanizawa, O.; Ueshima, H. A case-control study on risk factors for uterine endometrial cancer in Japan. Jpn. J. Cancer Res. 1994, 85, 346–350. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Homepage. Available online: http://cancergenome.nih.gov/abouttcga (accessed on 7 March 2023).
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.; Sukumar, S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 2010, 10, 361–371. [Google Scholar] [CrossRef]
- Mallo, M.; Alonso, C.R. The regulation of Hox gene expression during animal development. Development 2013, 140, 3951–3963. [Google Scholar] [CrossRef]
- Holland, P.W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Taylor, H.S. The role of HOX genes in the development and function of the female reproductive tract. Semin. Reprod. Med. 2000, 18, 81–89. [Google Scholar] [CrossRef]
- Quinonez, S.C.; Innis, J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014, 111, 4–15. [Google Scholar] [CrossRef]
- Grier, D.G.; Thompson, A.; Kwasniewska, A.; McGonigle, G.J.; Halliday, H.L.; Lappin, T.R. The pathophysiology of HOX genes and their role in cancer. J. Pathol. 2005, 205, 154–171. [Google Scholar] [CrossRef]
- Holland, P.W.; Booth, H.A.; Bruford, E.A. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007, 5, 47. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zeng, T.; Li, W.; Wu, H.; Sun, C.; Yang, F.; Yang, M.; Fu, Z.; Yin, Y. Long non-coding RNA SNHG1 activates HOXA1 expression via sponging miR-193a-5p in breast cancer progression. Aging 2020, 12, 10223–10234. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Dong, C.; Cui, J.; Wang, Y.; Hong, X. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J. Exp. Clin. Cancer Res. 2018, 37, 265. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Zhu, X.; Han, Y.; Song, C.; Liu, C.; Lu, S.; Zhang, M.; Yu, F.; Peng, Z.; Zhou, C. Elevated HOXA1 expression correlates with accelerated tumor cell proliferation and poor prognosis in gastric cancer partly via cyclin D1. J. Exp. Clin. Cancer Res. 2016, 35, 15. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, R.; An, X.; Li, Z.; Fang, C.; Pan, B.; Chen, W.; Xu, G.; Han, W. LncRNA HOXA-AS3 confers cisplatin resistance by interacting with HOXA3 in non-small-cell lung carcinoma cells. Oncogenesis 2019, 8, 60. [Google Scholar] [CrossRef]
- Jiang, L.; Wu, Z.; Meng, X.; Chu, X.; Huang, H.; Xu, C. LncRNA HOXA-AS2 facilitates tumorigenesis and progression of papillary thyroid cancer by modulating the miR-15a-5p/HOXA3 axis. Hum. Gene Ther. 2019, 30, 618–631. [Google Scholar] [CrossRef]
- Lin, J.; Zhu, H.; Hong, L.; Tang, W.; Wang, J.; Hu, H.; Wu, X.; Chen, Y.; Liu, G.; Yang, Q.; et al. Coexpression of HOXA6 and PBX2 promotes metastasis in gastric cancer. Aging 2021, 13, 6606–6624. [Google Scholar] [CrossRef]
- Qiao, C.; Huang, W.; Chen, J.; Feng, W.; Zhang, T.; Wang, Y.; Liu, D.; Ji, X.; Xie, M.; Sun, M.; et al. IGF1-mediated HOXA13 overexpression promotes colorectal cancer metastasis through upregulating ACLY and IGF1R. Cell Death Dis. 2021, 12, 564. [Google Scholar] [CrossRef]
- Tang, B.; Qi, G.; Sun, X.; Tang, F.; Yuan, S.; Wang, Z.; Liang, X.; Li, B.; Yu, S.; Liu, J.; et al. HOXA7 plays a critical role in metastasis of liver cancer associated with activation of Snail. Mol. Cancer 2016, 15, 57. [Google Scholar] [CrossRef]
- Ji, F.; Du, R.; Chen, T.; Zhang, M.; Zhu, Y.; Luo, X.; Ding, Y. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol. Ther. Nucleic Acids 2020, 19, 413–420. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Zhou, X.; Wright, S.; Hyle, J.; Zhao, L.; An, J.; Zhao, X.; Shao, Y.; Xu, B.; et al. Functional interrogation of HOXA9 regulome in MLLr leukemia via reporter-based CRISPR/Cas9 screen. eLife 2020, 9, e57858. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.M.; Mouw, J.K.; Unger, M.A.; Lakins, J.N.; Gbegnon, M.K.; Clemmer, V.B.; Benezra, M.; Licht, J.D.; Boudreau, N.J.; Tsai, K.K.; et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J. Clin. Investig. 2010, 120, 1535–1550. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Z.; Tang, L.; Chen, L. LINC00355 promotes tumor progression in HNSCC by hindering MicroRNA-195-Mediated suppression of HOXA10 expression. Mol. Ther. Nucleic Acids 2020, 19, 61–71, Retraction published in Mol. Ther. Nucleic Acids 2022, 29, 218. [Google Scholar] [CrossRef]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Qian, F.; Huang, Q.; Wei, L.; Fu, Y.; Du, Y. HOXA4, down-regulated in lung cancer, inhibits the growth, motility and invasion of lung cancer cells. Cell Death Dis. 2018, 9, 465. [Google Scholar] [CrossRef]
- Klausen, C.; Leung, P.C.; Auersperg, N. Cell motility and spreading are suppressed by HOXA4 in ovarian cancer cells: Possible involvement of beta1 integrin. Mol. Cancer Res. 2009, 7, 1425–1437. [Google Scholar] [CrossRef]
- Rayner, E.; van Gool, I.C.; Palles, C.; Kearsey, S.E.; Bosse, T.; Tomlinson, I.; Church, D.N. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer 2016, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Li, K.; Luo, H.; Huang, L.; Luo, H.; Zhu, X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020, 20, 16. [Google Scholar] [CrossRef]
- Meyer, L.A.; Broaddus, R.R.; Lu, K.H. Endometrial cancer and Lynch syndrome: Clinical and pathologic considerations. Cancer Control 2009, 16, 14–22. [Google Scholar] [CrossRef]
- Esteller, M.; Levine, R.; Baylin, S.B.; Ellenson, L.H.; Herman, J.G. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene 1998, 17, 2413–2417. [Google Scholar] [CrossRef]
- Day, R.S.; McDade, K.K.; Chandran, U.R.; Lisovich, A.; Conrads, T.P.; Hood, B.L.; Kolli, V.S.; Kirchner, D.; Litzi, T.; Maxwell, G.L. Identifier mapping performance for integrating transcriptomics and proteomics experimental results. BMC Bioinform. 2011, 12, 213. [Google Scholar] [CrossRef] [PubMed]
- Day, R.S.; McDade, K.K. A decision theory paradigm for evaluating identifier mapping and filtering methods using data integration. BMC Bioinform. 2013, 14, 223. [Google Scholar] [CrossRef] [PubMed]
- Nagy, A.; Munkacsy, G.; Gyorffy, B. Pancancer survival analysis of cancer hallmark genes. Sci. Rep. 2021, 11, 6047. [Google Scholar] [CrossRef] [PubMed]

| Variables | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|
| HR † | 95% CI ‡ | p-Value | HR † | 95% CI ‡ | p-Value | ||
| HOXA5 | Lower | 1 | - | - | - | - | - |
| expression | Higher | 2.368 | 1.376–4.077 | 0.002 | 2.286 | 1.129–4.630 | 0.022 |
| Age | <60 | 1 | - | - | - | - | - |
| ≥60 | 1.678 | 0.923–3.049 | 0.09 | 0.872 | 0.436–1.746 | 0.699 | |
| Stage | 1, 2 | 1 | - | - | - | - | - |
| 3, 4 | 4.763 | 2.862–7.926 | <0.0001 | 3.275 | 1.640–6.541 | 0.001 | |
| Diabetes | No | 1 | - | - | - | - | - |
| Yes | 0.959 | 0.472–1.947 | 0.908 | - | - | - | |
| Hypertension | No | 1 | - | - | - | - | - |
| Yes | 0.924 | 0.508–1.680 | 0.796 | - | - | - | |
| HRT § | No | 1 | - | - | - | - | - |
| Yes | 0.891 | 0.339–2.344 | 0.815 | - | - | - | |
| Menopause | No | 1 | - | - | - | - | - |
| Yes | 1.202 | 0.481–3.004 | 0.693 | - | - | - | |
| Grade | 1, 2 | 1 | - | - | - | - | - |
| 3 | 3.405 | 1.809–6.410 | <0.0001 | 1.898 | 0.887–4.062 | 0.099 | |
| Cytology | Negative | 1 | - | - | - | - | - |
| Positive | 6.615 | 3.739–11.703 | <0.0001 | 2.734 | 1.367–5.470 | 0.004 | |
| Parameters | Number | Mean Gene Expression (FPKM †) | p-Value | |
|---|---|---|---|---|
| Age | <60 | 160 | 1.812 | 0.273 |
| ≥60 | 239 | 1.980 | ||
| Clinical stage | Stage I and II | 286 | 1.909 | 0.792 |
| Stage III and IV | 116 | 1.953 | ||
| Histological grade | Grade 1 | 97 | 1.655 | 0.014 |
| Grade 2 | 116 | 1.773 | ||
| Grade 3 | 189 | 2.150 | ||
| Hypertension | No | 120 | 1.818 | 0.995 |
| Yes | 174 | 1.819 | ||
| Diabetes | No | 189 | 1.877 | 0.335 |
| Yes | 79 | 1.700 | ||
| HRT ‡ | No | 184 | 1.679 | 0.154 |
| Yes | 28 | 2.047 | ||
| Menopausal status | Pre | 36 | 2.100 | 0.430 |
| Peri | 32 | 1.631 | ||
| Post | 313 | 1.919 | ||
| Cytology | negative | 271 | 1.937 | 0.807 |
| positive | 28 | 2.011 | ||
| Tumor status | Negative tumor | 310 | 1.797 | 0.055 |
| With tumor | 42 | 2.250 | ||
| Adjuvant treatment | No | 289 | 1.850 | 0.721 |
| Yes | 94 | 1.912 |
| Subgroup | Number | Mean Gene Expression (FPKM †) | p-Value | |
|---|---|---|---|---|
| Group 1 vs. 2 | Group 1 (grade 1) | 97 | 1.655 | 0.028 |
| Group 2 (grades 2 and 3) | 305 | 2.007 | ||
| Group 3 vs. 4 | Group 3 (grades 1 and 2) | 213 | 1.369 | 0.004 |
| Group 4 (grade 3) | 289 | 1.613 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Kim, K.B.; Lee, G.S.; Shin, S.; Kim, B. Is HOXA5 a Novel Prognostic Biomarker for Uterine Corpus Endometrioid Adenocarcinoma? Int. J. Mol. Sci. 2023, 24, 14758. https://doi.org/10.3390/ijms241914758
Song C, Kim KB, Lee GS, Shin S, Kim B. Is HOXA5 a Novel Prognostic Biomarker for Uterine Corpus Endometrioid Adenocarcinoma? International Journal of Molecular Sciences. 2023; 24(19):14758. https://doi.org/10.3390/ijms241914758
Chicago/Turabian StyleSong, Changho, Kyoung Bo Kim, Gi Su Lee, Soyoung Shin, and Byoungje Kim. 2023. "Is HOXA5 a Novel Prognostic Biomarker for Uterine Corpus Endometrioid Adenocarcinoma?" International Journal of Molecular Sciences 24, no. 19: 14758. https://doi.org/10.3390/ijms241914758
APA StyleSong, C., Kim, K. B., Lee, G. S., Shin, S., & Kim, B. (2023). Is HOXA5 a Novel Prognostic Biomarker for Uterine Corpus Endometrioid Adenocarcinoma? International Journal of Molecular Sciences, 24(19), 14758. https://doi.org/10.3390/ijms241914758






