β-Nicotinamide Mononucleotide Alleviates Hydrogen Peroxide-Induced Cell Cycle Arrest and Death in Ovarian Granulosa Cells
Abstract
:1. Introduction
2. Results
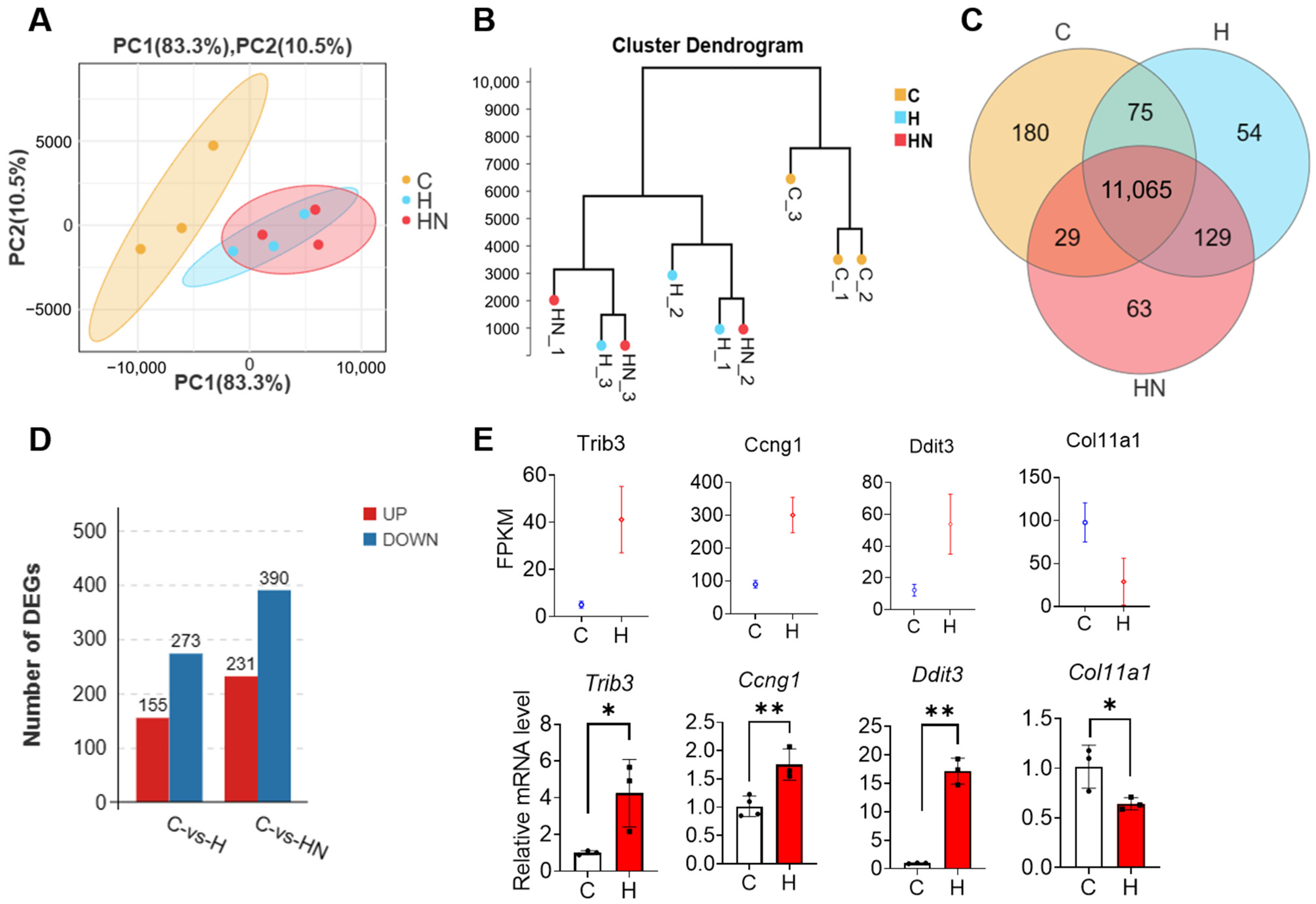
2.1. NMN Supplementation Shapes the Gene Expression Pattern in mGCs That Are Exposed to H2O2
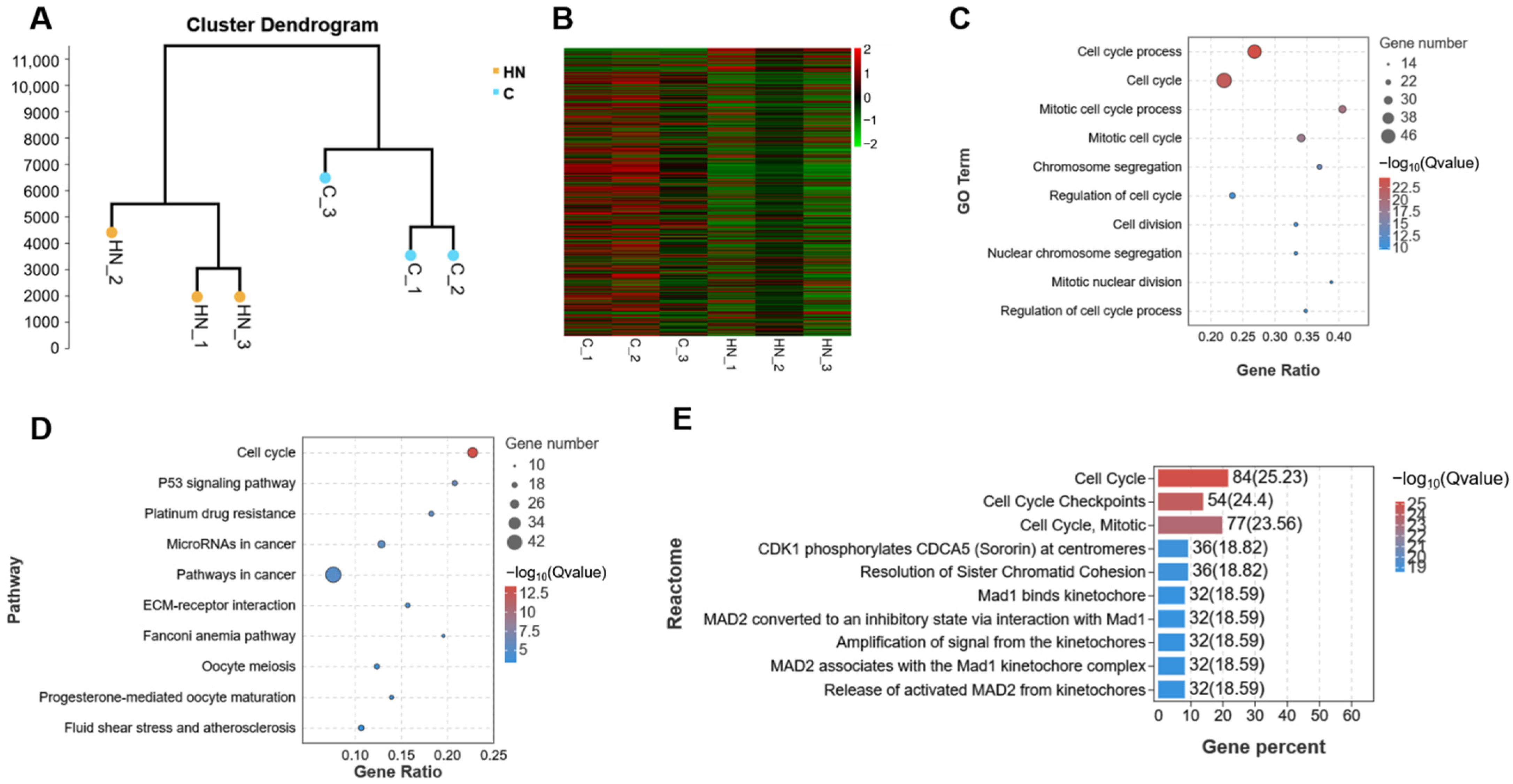
2.2. The Transcriptome Analysis of mGCs That Are Exposed to H2O2
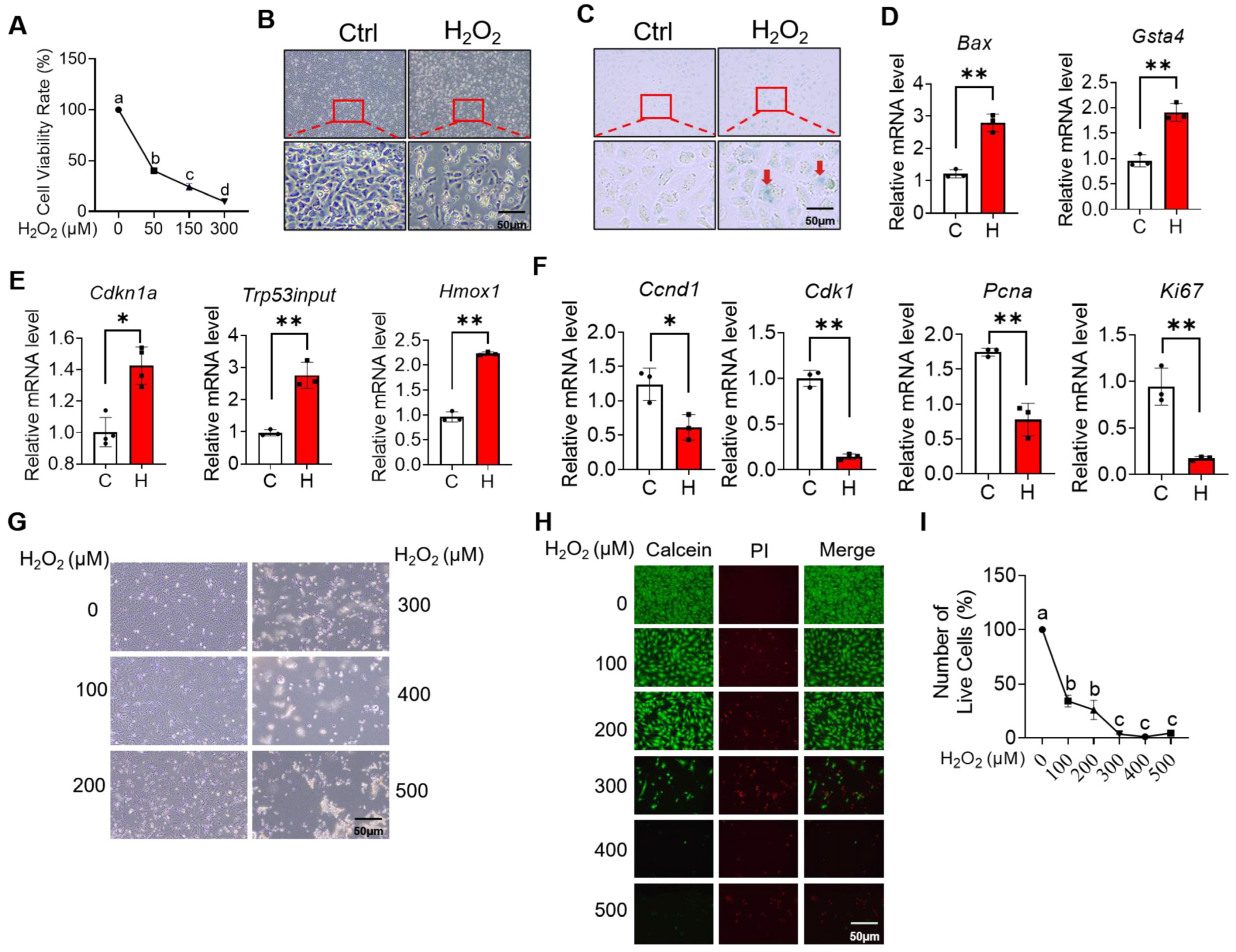
2.3. H2O2 Induces Ovarian Granulosa Cell Cycle Arrest and Apoptosis
2.4. Transcriptome Analysis of mGCs Induced by the Co-Addition of H2O2 and NMN
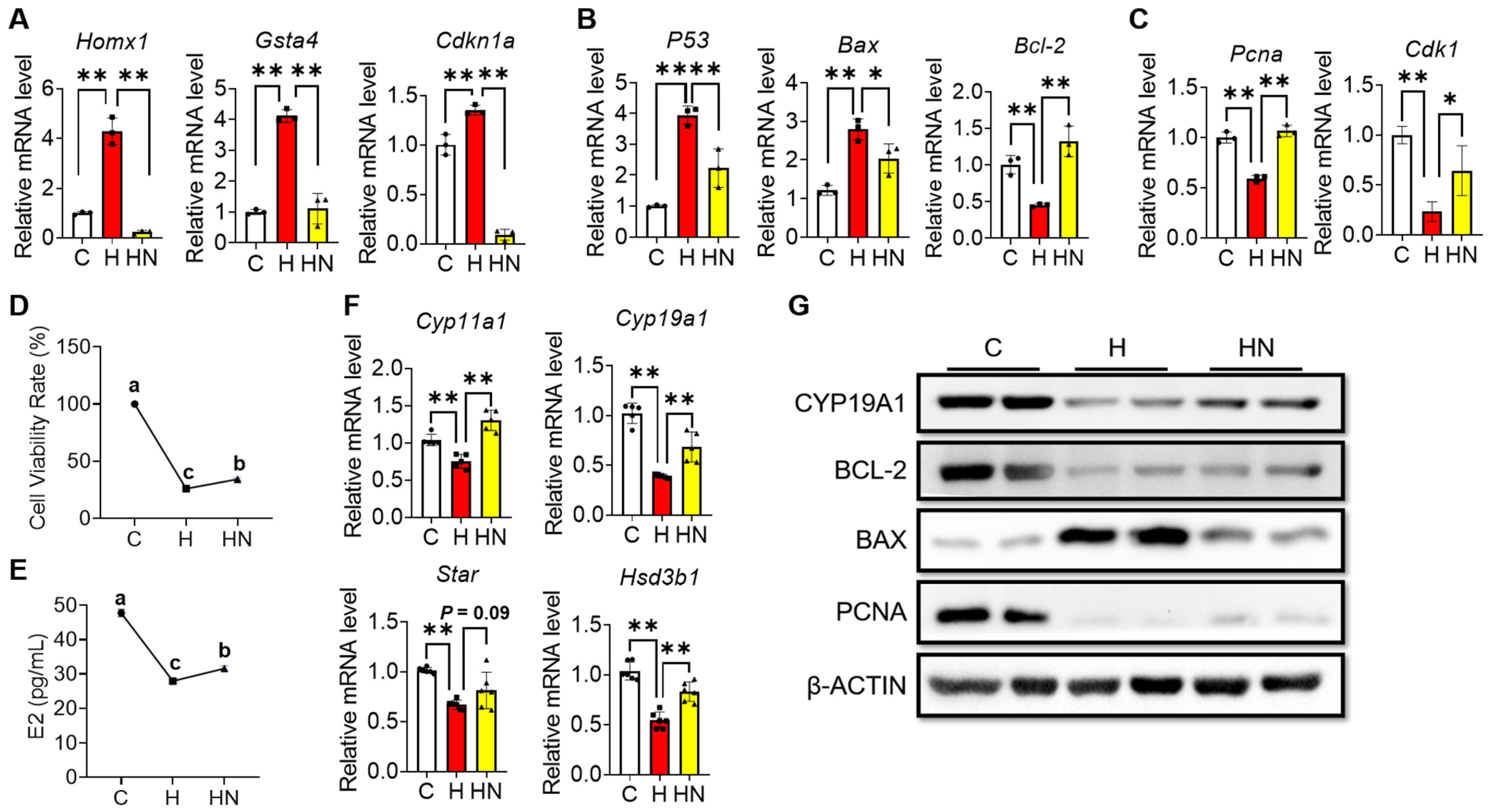
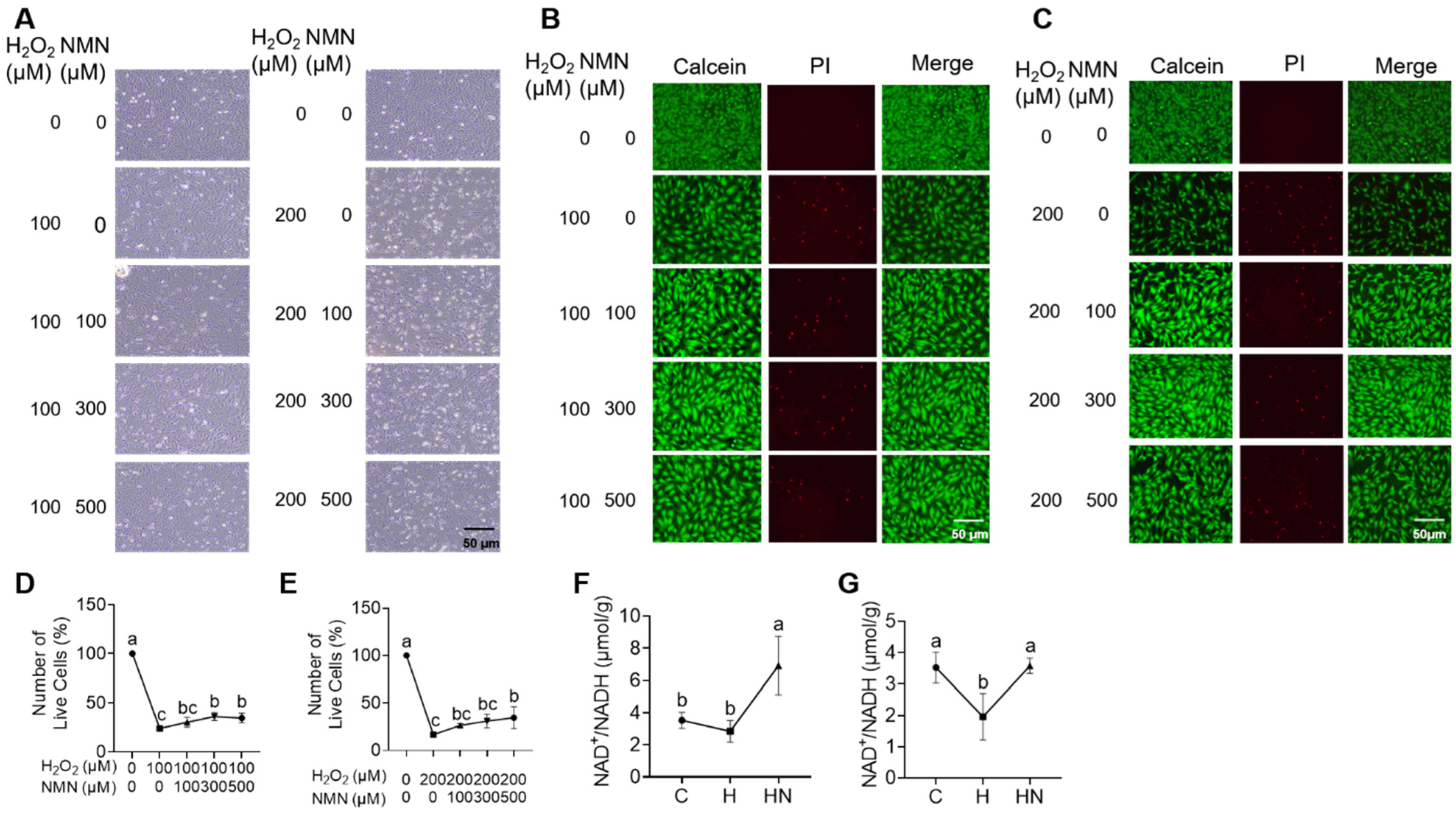
2.5. NMN Partly Rescues the Cell Cycle Arrest, Apoptosis and Impaired Estrogen Synthesis of Ovarian Granulosa Induced by H2O2
3. Discussion
4. Materials and Methods
4.1. Mouse Granulosa Cell Collection
4.2. GCs Cell Culture
4.3. High-Throughput RNA Sequencing and Bioinformatic Analysis
4.4. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.5. Cell SA-β-Gal Staining
4.6. Western Blots
4.7. The PI Staining
4.8. Total NAD+/NADH Detection
4.9. Cell Viability Determination
4.10. Detection of E2 Level
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hsueh, A.J.; Kawamura, K.; Cheng, Y.; Fauser, B.C. Intraovarian control of early folliculogenesis. Endocr. Rev. 2015, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.G. Oocyte maturation and ovum quality in pigs. Rev. Reprod. 2000, 5, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular growth and atresia in mammalian ovaries: Regulation by survival and death of granulosa cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [PubMed]
- McGee, E.A.; Hsueh, A.J. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [PubMed]
- Simpson, E.R.; Clyne, C.; Rubin, G.; Boon, W.C.; Robertson, K.; Britt, K.; Speed, C.; Jones, M. Aromatase—A brief overview. Annu. Rev. Physiol. 2002, 64, 93–127. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Prasad, S.; Tripathi, A.; Pandey, A.N.; Ali, I.; Singh, A.K.; Shrivastav, T.G.; Chaube, S.K. Apoptosis in mammalian oocytes: A review. Apoptosis 2015, 20, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Tajima, K.; Orisaka, M.; Yata, H.; Goto, K.; Hosokawa, K.; Kotsuji, F. Role of granulosa and theca cell interactions in ovarian follicular maturation. Microsc. Res. Tech. 2006, 69, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, C.; Chen, S.; Sun, L.; Li, H.; Chen, L.; Zhou, X. Effect of inhibin A on proliferation of porcine granulosa cells in vitro. Theriogenology 2018, 114, 136–142. [Google Scholar] [CrossRef]
- Jiang, Y.; Shen, M.; Chen, Y.; Wei, Y.; Tao, J.; Liu, H. Melatonin Represses Mitophagy to Protect Mouse Granulosa Cells from Oxidative Damage. Biomolecules 2021, 11, 968. [Google Scholar] [CrossRef]
- Nie, Z.; Hua, R.; Zhang, Y.; Zhang, N.; Zhang, Y.; Li, Q.; Wu, H. Resveratrol protects human luteinised granulosa cells against hydrogen peroxide-induced oxidative injury through the Sirt1. Reprod. Fertil. Dev. 2021, 33, 831–840. [Google Scholar] [CrossRef]
- Lai, Q.; Xiang, W.; Li, Q.; Zhang, H.; Li, Y.; Zhu, G.; Xiong, C.; Jin, L. Oxidative stress in granulosa cells contributes to poor oocyte quality and IVF-ET outcomes in women with polycystic ovary syndrome. Front. Med. 2018, 12, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Pathak, D.; Kriplani, A.; Ammini, A.C.; Talwar, P.; Dada, R. Nucleotide variations in mitochondrial DNA and supra-physiological ROS levels in cytogenetically normal cases of premature ovarian insufficiency. Arch. Gynecol. Obstet. 2010, 282, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.; Aponte-Mellado, A.; Premkumar, B.J.; Shaman, A.; Gupta, S. The effects of oxidative stress on female reproduction: A review. Reprod. Biol. Endocrinol. 2012, 10, 49. [Google Scholar] [CrossRef]
- Yang, H.; Xie, Y.; Yang, D.; Ren, D. Oxidative stress-induced apoptosis in granulosa cells involves JNK, p53 and Puma. Oncotarget 2017, 8, 25310–25322. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Q.; Hu, Y.; Xu, L.; Jiang, Y.; Zhang, C.; Ding, L.; Jiang, R.; Sun, J.; Sun, H.; et al. miR-181a increases FoxO1 acetylation and promotes granulosa cell apoptosis via SIRT1 downregulation. Cell Death Dis. 2017, 8, e3088. [Google Scholar] [CrossRef]
- Deng, D.; Yan, J.; Wu, Y.; Wu, K.; Li, W. Morroniside suppresses hydrogen peroxide-stimulated autophagy and apoptosis in rat ovarian granulosa cells through the PI3K/AKT/mTOR pathway. Hum. Exp. Toxicol. 2021, 40, 577–586. [Google Scholar] [CrossRef]
- Huang, B.; Jin, L.; Zhang, L.; Cui, X.; Zhang, Z.; Lu, Y.; Yu, L.; Ma, T.; Zhang, H. Aquaporin-8 transports hydrogen peroxide to regulate granulosa cell autophagy. Front. Cell Dev. Biol. 2022, 10, 897666. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.L.; Lin, T.M.; Wang, S.W.; Wang, P.S. Antisteroidogenic effects of hydrogen peroxide on rat granulosa cells. Free Radic. Res. 2012, 46, 718–725. [Google Scholar] [CrossRef]
- Wang, M.; Li, Y.; Molenaar, A.; Li, Q.; Cao, Y.; Shen, Y.; Chen, P.; Yan, J.; Gao, Y.; Li, J. Vitamin E and selenium supplementation synergistically alleviate the injury induced by hydrogen peroxide in bovine granulosa cells. Theriogenology 2021, 170, 91–106. [Google Scholar] [CrossRef]
- Ma, Y.; Hao, G.; Lin, X.; Zhao, Z.; Yang, A.; Cao, Y.; Zhang, S.; Fan, L.; Geng, J.; Zhang, Y.; et al. Morroniside Protects Human Granulosa Cells against H2O2-Induced Oxidative Damage by Regulating the Nrf2 and MAPK Signaling Pathways. Evid.-Based Complement. Altern. Med. 2022, 2022, 8099724. [Google Scholar] [CrossRef]
- Li, B.S.; Zhu, R.Z.; Lim, S.H.; Seo, J.H.; Choi, B.M. Apigenin Alleviates Oxidative Stress-Induced Cellular Senescence via Modulation of the SIRT1-NAD+-CD38 Axis. Am. J. Chin. Med. 2021, 49, 1235–1250. [Google Scholar] [CrossRef] [PubMed]
- Bertoldo, M.J.; Listijono, D.R.; Ho, W.J.; Riepsamen, A.H.; Goss, D.M.; Richani, D.; Jin, X.L.; Mahbub, S.; Campbell, J.M.; Habibalahi, A.; et al. NAD+ Repletion Rescues Female Fertility during Reproductive Aging. Cell Rep. 2020, 30, 1670–1681.e7. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Liang, X.; Yang, H.; Huang, Z.; Huang, X.; Liang, C.; Kuang, Y.; Qin, Y.; Lin, F.; Luo, Z. Nicotinamide mononucleotide (NMN) protects bEnd.3 cells against H2O2-induced damage via NAMPT and the NF-κB p65 signalling pathway. FEBS Open Bio 2021, 11, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Björvang, R.D.; Hassan, J.; Stefopoulou, M.; Gemzell-Danielsson, K.; Pedrelli, M.; Kiviranta, H.; Rantakokko, P.; Ruokojärvi, P.; Lindh, C.H.; Acharya, G.; et al. Persistent organic pollutants and the size of ovarian reserve in reproductive-aged women. Environ. Int. 2021, 155, 106589. [Google Scholar] [CrossRef] [PubMed]
- De Felici, M.; Scaldaferri, M.L.; Lobascio, M.; Iona, S.; Nazzicone, V.; Klinger, F.G.; Farini, D. Experimental approaches to the study of primordial germ cell lineage and proliferation. Hum. Reprod. Update 2004, 10, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Edson, M.A.; Nagaraja, A.K.; Matzuk, M.M. The mammalian ovary from genesis to revelation. Endocr. Rev. 2009, 30, 624–712. [Google Scholar] [CrossRef]
- May-Panloup, P.; Boucret, L.; Chao de la Barca, J.M.; Desquiret-Dumas, V.; Ferré-L’Hotellier, V.; Morinière, C.; Descamps, P.; Procaccio, V.; Reynier, P. Ovarian ageing: The role of mitochondria in oocytes and follicles. Hum. Reprod. Update 2016, 22, 725–743. [Google Scholar] [CrossRef]
- Baerwald, A.R.; Adams, G.P.; Pierson, R.A. Ovarian antral folliculogenesis during the human menstrual cycle: A review. Hum. Reprod. Update 2012, 18, 73–91. [Google Scholar] [CrossRef]
- Leopardo, N.P.; Velazquez, M.E.; Cortasa, S.; González, C.R.; Vitullo, A.D. A dual death/survival role of autophagy in the adult ovary of Lagostomus maximus (Mammalia-Rodentia). PLoS ONE 2020, 15, e0232819. [Google Scholar] [CrossRef]
- Agarwal, A.; Saleh, R.A.; Bedaiwy, M.A. Role of reactive oxygen species in the pathophysiology of human reproduction. Fertil. Steril. 2003, 79, 829–843. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fu, R.; Cheng, H.; Li, Y.; Sui, S. Analysis of the resistance of small peptides from Periplaneta americana to hydrogen peroxide-induced apoptosis in human ovarian granular cells based on RNA-seq. Gene 2022, 813, 146120. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Li, Q.; Cao, Q.; Wang, S.; Liu, H.; Li, Q. Integrated Analysis of miRNA-mRNA Interaction Network in Porcine Granulosa Cells Undergoing Oxidative Stress. Oxid. Med. Cell. Longev. 2019, 2019, 1041583. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.M.H.; Akyuz, B.; Konca, Y.; Arslan, K.; Sariozkan, S.; Cinar, M.U. Oxidative stress modulates the expression of apoptosis-associated microRNAs in bovine granulosa cells in vitro. Cell Tissue Res. 2019, 376, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Huis In ’t Veld, P.J.; Wohlgemuth, S.; Koerner, C.; Müller, F.; Janning, P.; Musacchio, A. Reconstitution and use of highly active human CDK1:Cyclin-B:CKS1 complexes. Protein Sci. 2022, 31, 528–537. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Cui, M.; Lin, H.; Zhao, L.; Wang, J.; Chen, S.; Shao, Z. Melatonin resists oxidative stress-induced apoptosis in nucleus pulposus cells. Life Sci. 2018, 199, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Pei, R.; Zhang, Z.; Liao, J.; Yu, W.; Qiao, N.; Han, Q.; Li, Y.; Hu, L.; Guo, J.; et al. Copper induces oxidative stress and apoptosis through mitochondria-mediated pathway in chicken hepatocytes. Toxicol. In Vitro 2019, 54, 310–316. [Google Scholar] [CrossRef]
- Luo, Z.; Xu, X.; Sho, T.; Zhang, J.; Xu, W.; Yao, J.; Xu, J. ROS-induced autophagy regulates porcine trophectoderm cell apoptosis, proliferation, and differentiation. Am. J. Physiol. Cell Physiol. 2019, 316, C198–C209. [Google Scholar] [CrossRef]
- Kang, R.; Li, R.; Dai, P.; Li, Z.; Li, Y.; Li, C. Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ. Pollut. 2019, 251, 689–698. [Google Scholar] [CrossRef]
- Jahagirdar, D.; Gore, C.R.; Patel, H.; Maria, K.; Tandon, I.; Sharma, N.K. Induction of Apoptotic Death and Cell Cycle Arrest in HeLa Cells by Extracellular Factors of Breast Cancer Cells. Asian Pac. J. Cancer Prev. 2018, 19, 3307–3316. [Google Scholar] [CrossRef]
- Frade, J.M.; Ovejero-Benito, M.C. Neuronal cell cycle: The neuron itself and its circumstances. Cell Cycle 2015, 14, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Lüschen, S.; Ussat, S.; Scherer, G.; Kabelitz, D.; Adam-Klages, S. Sensitization to death receptor cytotoxicity by inhibition of fas-associated death domain protein (FADD)/caspase signaling. Requirement of cell cycle progression. J. Biol. Chem. 2000, 275, 24670–24678. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.S.; Lee, B.Y.; Hwang, E.S. Dinstinct ROS and biochemical profiles in cells undergoing DNA damage-induced senescence and apoptosis. Mech. Ageing Dev. 2005, 126, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, J.A.; Sanoff, H.K.; Sharpless, N.E. Defining the toxicology of aging. Trends Mol. Med. 2014, 20, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Gómez-Lahoz, E.; DePinho, R.A.; Beach, D.; Bar-Sagi, D. Inhibition of RAS-induced proliferation and cellular transformation by p16INK4. Science 1995, 267, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Prives, C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018, 25, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Onder, T.T.; Young, J.J.; McFaline, J.L.; Pang, B.; Dedon, P.C.; Weinberg, R.A. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl. Acad. Sci. USA 2009, 106, 169–174. [Google Scholar] [CrossRef]
- Zager, R.A.; Johnson, A.C.; Becker, K. Plasma and urinary heme oxygenase-1 in AKI. J. Am. Soc. Nephrol. 2012, 23, 1048–1057. [Google Scholar] [CrossRef]
- Park, J.H.; Long, A.; Owens, K.; Kristian, T. Nicotinamide mononucleotide inhibits post-ischemic NAD+ degradation and dramatically ameliorates brain damage following global cerebral ischemia. Neurobiol. Dis. 2016, 95, 102–110. [Google Scholar] [CrossRef]
- Yoshino, J.; Mills, K.F.; Yoon, M.J.; Imai, S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011, 14, 528–536. [Google Scholar] [CrossRef]
- Hara, N.; Yamada, K.; Shibata, T.; Osago, H.; Hashimoto, T.; Tsuchiya, M. Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem. 2007, 282, 24574–24582. [Google Scholar] [CrossRef] [PubMed]
- Tarantini, S.; Valcarcel-Ares, M.N.; Toth, P.; Yabluchanskiy, A.; Tucsek, Z.; Kiss, T.; Hertelendy, P.; Kinter, M.; Ballabh, P.; Süle, Z.; et al. Nicotinamide mononucleotide (NMN) supplementation rescues cerebromicrovascular endothelial function and neurovascular coupling responses and improves cognitive function in aged mice. Redox Biol. 2019, 24, 101192. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yu, C.; Zhou, J.; Xiao, Q.; Shen, Q.; Xiong, Z.; Li, Z.; Fu, Z. Nicotinamide mononucleotide ameliorates the depression-like behaviors and is associated with attenuating the disruption of mitochondrial bioenergetics in depressed mice. J. Affect. Disord. 2020, 263, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Li, H.R.; Liu, Q.; Zhu, C.L.; Sun, X.Y.; Sun, C.Y.; Yu, C.M.; Li, P.; Deng, X.M.; Wang, J.F. β-Nicotinamide mononucleotide activates NAD+/SIRT1 pathway and attenuates inflammatory and oxidative responses in the hippocampus regions of septic mice. Redox Biol. 2023, 63, 102745. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.F.; Xu, J.; Wang, X.L.; Li, S.J.; Han, Z.Y. Nicotinamide mononucleotide alleviates heat stress-induced oxidative stress and apoptosis in BMECs through reducing mitochondrial damage and endoplasmic reticulum stress. Ecotoxicol. Environ. Saf. 2022, 235, 113441. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fu, W.; Zhao, X.; Chang, X.; Liu, H.; Zhou, L.; Li, J.; Cheng, R.; Wu, X.; Li, X.; et al. Zearalenone disturbs the reproductive-immune axis in pigs: The role of gut microbial metabolites. Microbiome 2022, 10, 234. [Google Scholar] [CrossRef]



| Gene | Primers | Sequences (5′-3′) |
|---|---|---|
| Cdkn1a | Forward | TCCAGACATTCAGAGCCACAG |
| Reversed | AAAGTTCCACCGTTCTCGGG | |
| Hmox1 | Forward | GCCCCACCAAGTTCAAACAG |
| Reversed | GCTCCTCAAACAGCTCAATGT | |
| Gsta4 | Forward | ACTTTAATGGCAGGGGACGG |
| Reversed | GCAGGTGTCCATCCTTTTGC | |
| Ccng1 | Forward | AAGACGTGGCTGTCAAGATGA |
| Reversed | TGTCTCCGTGTCAAAGCCAA | |
| Trp53inp1 | Forward | TAAGACTCACGGGCACAGAAATG |
| Reversed | AGTGTGGCAATCCCTGGTAAG | |
| Gapdh | Forward | TGCCCCCATGTTTGTGATG |
| Reversed | TGTGGTCATGAGCCCTTCC | |
| Ddit3 | Forward | CCTGAGGAGAGAGTGTTCCAG |
| Reversed | GACACCGTCTCCAAGGTGAA | |
| Col11a1 | Forward | ACAGTAGCACAAACAGAGGCAA |
| Reversed | AATCCCTGCCGTCTACTCCT | |
| P53 | Forward | TGATGGAGAGTATTTCACCC |
| Reversed | GGGCATCCTTTAACTCTAAGA | |
| Bax | Forward | CGGCGAATTGGAGATGAACTG |
| Reversed | GCAAAGTAGAAGAGGGCAACC | |
| Cdk1 | Forward | CGGTACTTACGGTGTGGTGTAT |
| Reversed | CTCGCTTTCAAGTCTGATCTTCT | |
| Ccnd1 | Forward | AGGGATGATGATGCTGGTATG |
| Reversed | AACACCACACCTGGGCTTAT | |
| Pcna | Forward | TAAAGAAGAGGAGGCGGTAA |
| Reversed | TAAGTGTCCCATGTCAGCAA | |
| Ki67 | Forward | AATCCAACTCAAGTAAACGGGG |
| Reversed | TTGGCTTGCTTCCATCCTCA | |
| β-action | Forward | GCAGAAGGAGATCACTGCCCT |
| Reversed | GCTGATCCACATCTGCTGGAA | |
| Cyp11a1 | Forward | ACCAAGAACTTTTTGCCCCT |
| Reversed | ATGTCCCCCGAGTAATTTCC | |
| Cyp19a1 | Forward | GACTTTGCCACTGAGTTGATTT |
| Reversed | CGATCAGCATTTCCAATATGCA | |
| Star | Forward | ACGTGGATTAACCAGGTTCG |
| Reversed | CAGCCCTCTTGGTTGCTAAG | |
| Hsd3b1 | Forward | CTCTTCTGTCCAGCTTTTAAC |
| Forward | ACCAAGAACTTTTTGCCCCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, Q.; Ma, Z.; Xu, H.; Peng, F.; Chen, B.; Ma, B.; Qin, L.; Lan, J.; Li, Y.; et al. β-Nicotinamide Mononucleotide Alleviates Hydrogen Peroxide-Induced Cell Cycle Arrest and Death in Ovarian Granulosa Cells. Int. J. Mol. Sci. 2023, 24, 15666. https://doi.org/10.3390/ijms242115666
Wang Y, Li Q, Ma Z, Xu H, Peng F, Chen B, Ma B, Qin L, Lan J, Li Y, et al. β-Nicotinamide Mononucleotide Alleviates Hydrogen Peroxide-Induced Cell Cycle Arrest and Death in Ovarian Granulosa Cells. International Journal of Molecular Sciences. 2023; 24(21):15666. https://doi.org/10.3390/ijms242115666
Chicago/Turabian StyleWang, Yunduan, Qiao Li, Zifeng Ma, Hongmei Xu, Feiyu Peng, Bin Chen, Bo Ma, Linmei Qin, Jiachen Lan, Yueyue Li, and et al. 2023. "β-Nicotinamide Mononucleotide Alleviates Hydrogen Peroxide-Induced Cell Cycle Arrest and Death in Ovarian Granulosa Cells" International Journal of Molecular Sciences 24, no. 21: 15666. https://doi.org/10.3390/ijms242115666
APA StyleWang, Y., Li, Q., Ma, Z., Xu, H., Peng, F., Chen, B., Ma, B., Qin, L., Lan, J., Li, Y., Lan, D., Li, J., Wang, S., & Fu, W. (2023). β-Nicotinamide Mononucleotide Alleviates Hydrogen Peroxide-Induced Cell Cycle Arrest and Death in Ovarian Granulosa Cells. International Journal of Molecular Sciences, 24(21), 15666. https://doi.org/10.3390/ijms242115666




