Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis
Abstract
:1. Introduction
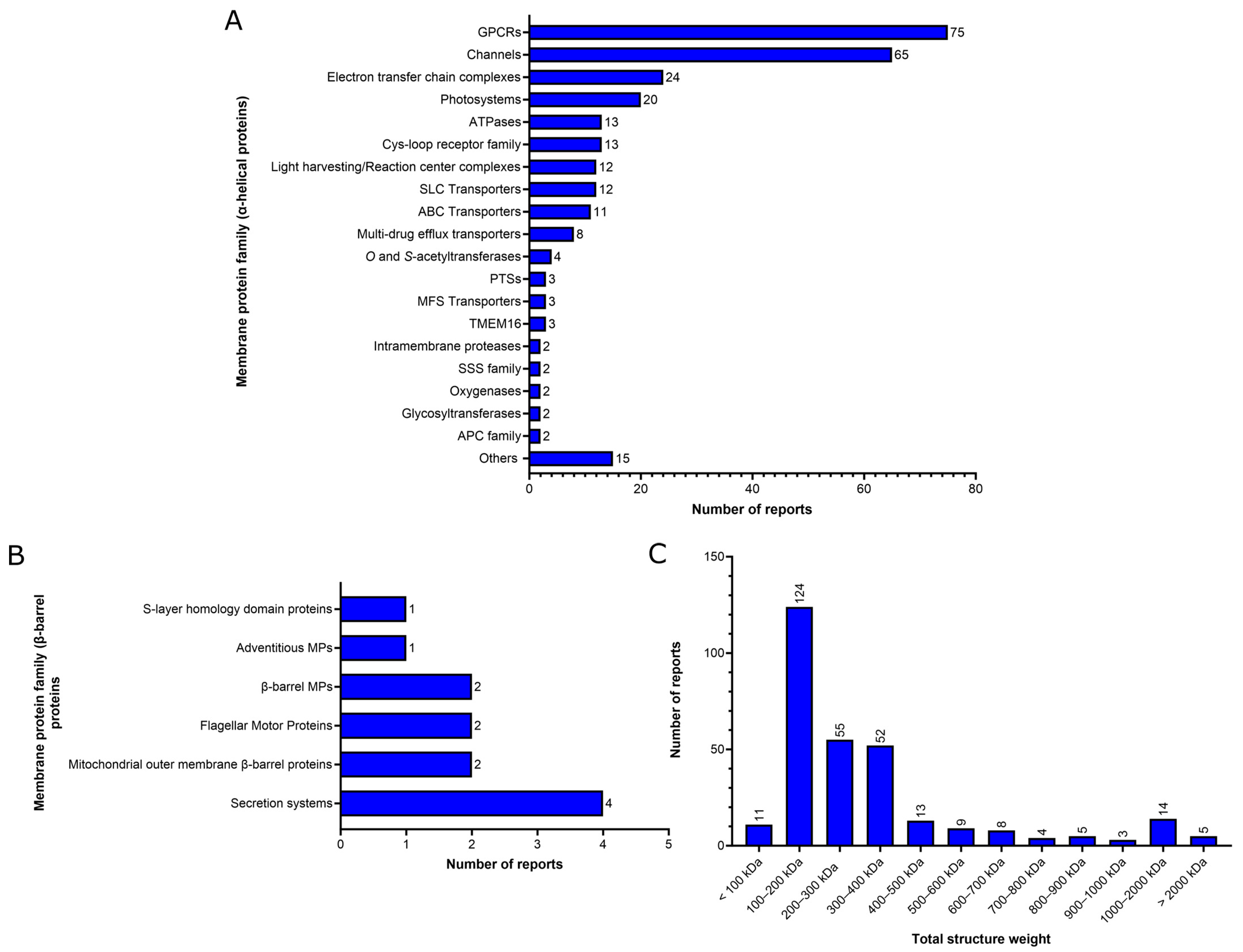
2. Functional Analysis of the MP Structural Reports
3. Sample Preparation
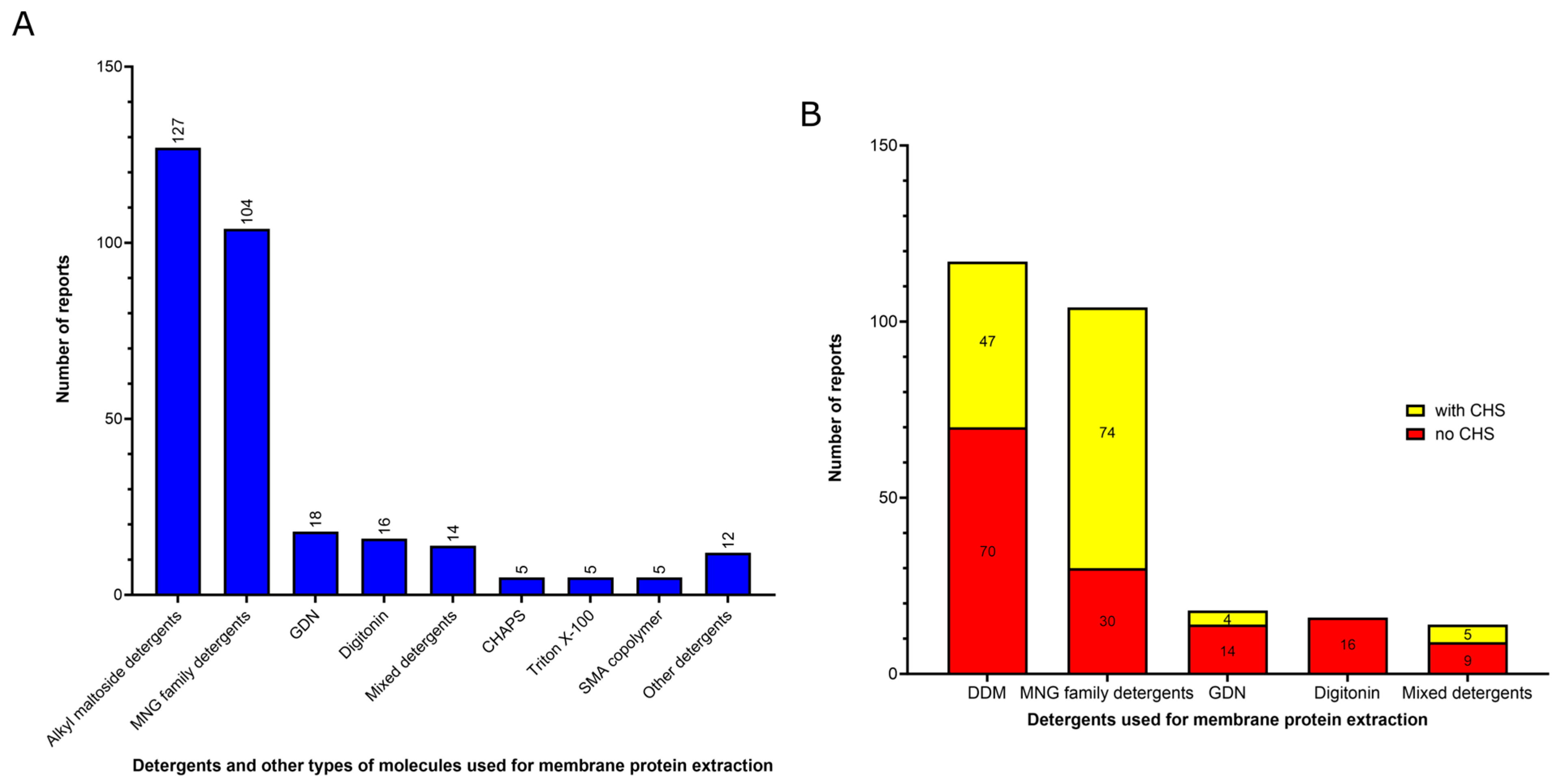
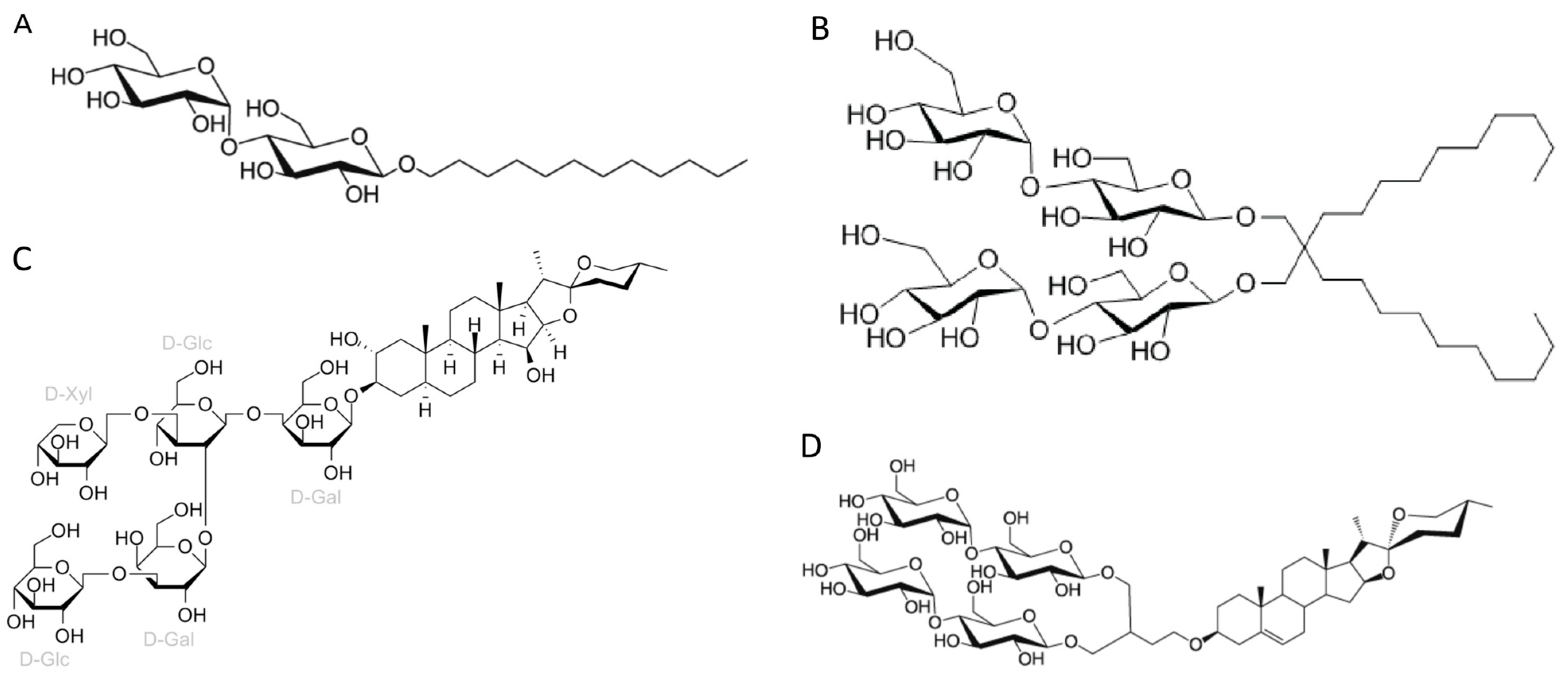
3.1. Amphiphiles Used for Membrane Protein Extraction
- (1)
- Detergents with a single maltose-based polar moiety and single alkyl chain (DDM, UDM, DM): 127 of 306 reports (41%). In this group, the most widely used detergent is DDM, which is present in 117 of 127 reports of this detergent class. DM is present in 9 of 131 reports and UDM in 1 of 131 reports;
- (2)
- Detergents belonging to the maltoside–neopentyl glycol (MNG) family (MNG, LMNG and DMNG): 104 of 306 reports (34%). In this group, the most widely used detergent is LMNG, which is present in 101 of 104 reports of this detergent class. MNG is present in 2 of 104 reports and DMNG is present in 1 of 107 reports;
- (3)
- Glyco-diosgenin (GDN): 18 of 306 reports (6%);
- (4)
- Digitonin: 16 of 306 reports (5%).
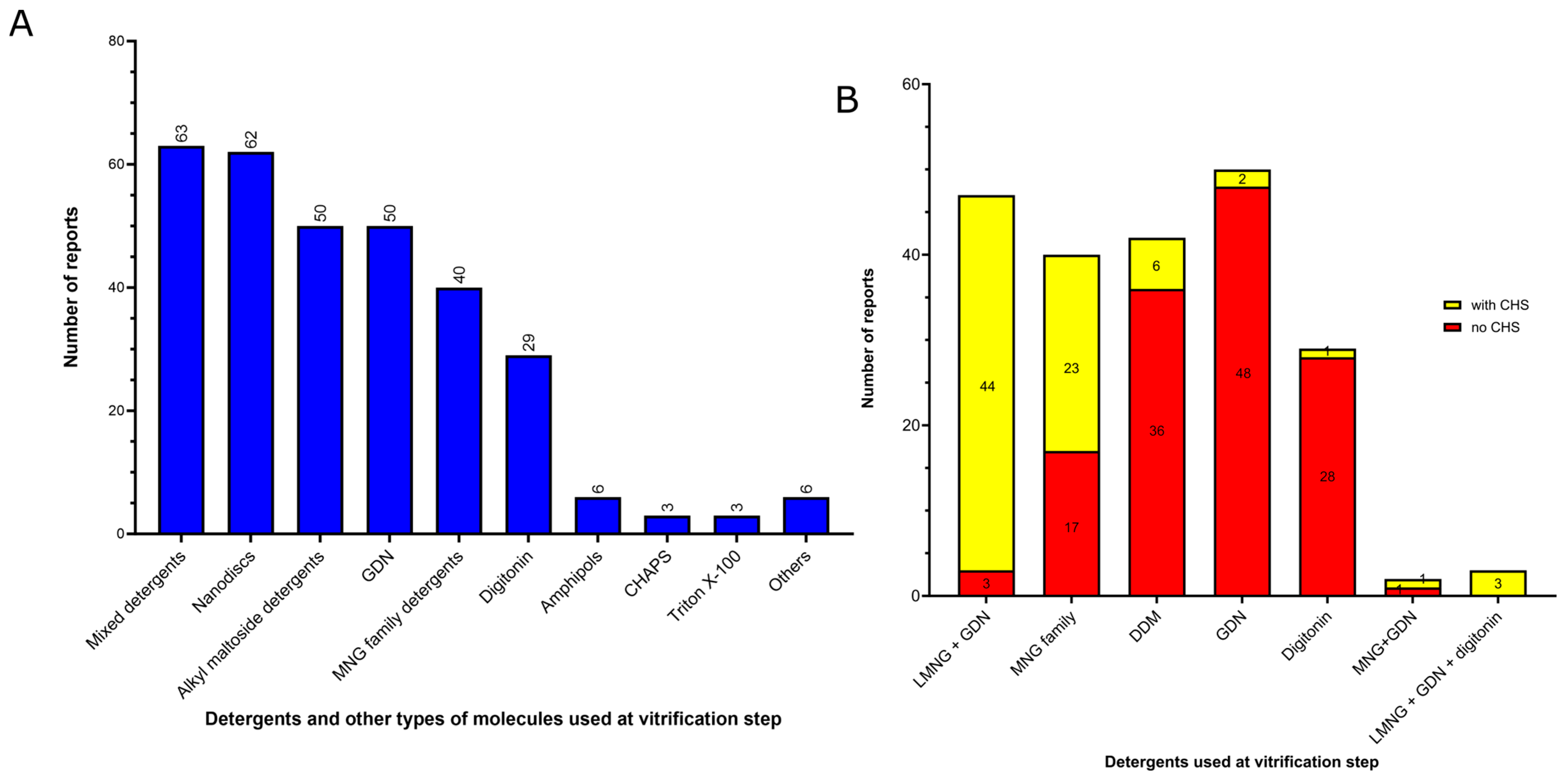
3.2. Amphiphiles and Other Molecules Used in the Vitrification Step
- (1)
- Mixed detergents: 63 of 312 reports (20%). In this group, the most widely used association of detergents was the combination of LMNG and GDN, which is present in 47 of 63 reports.
- (2)
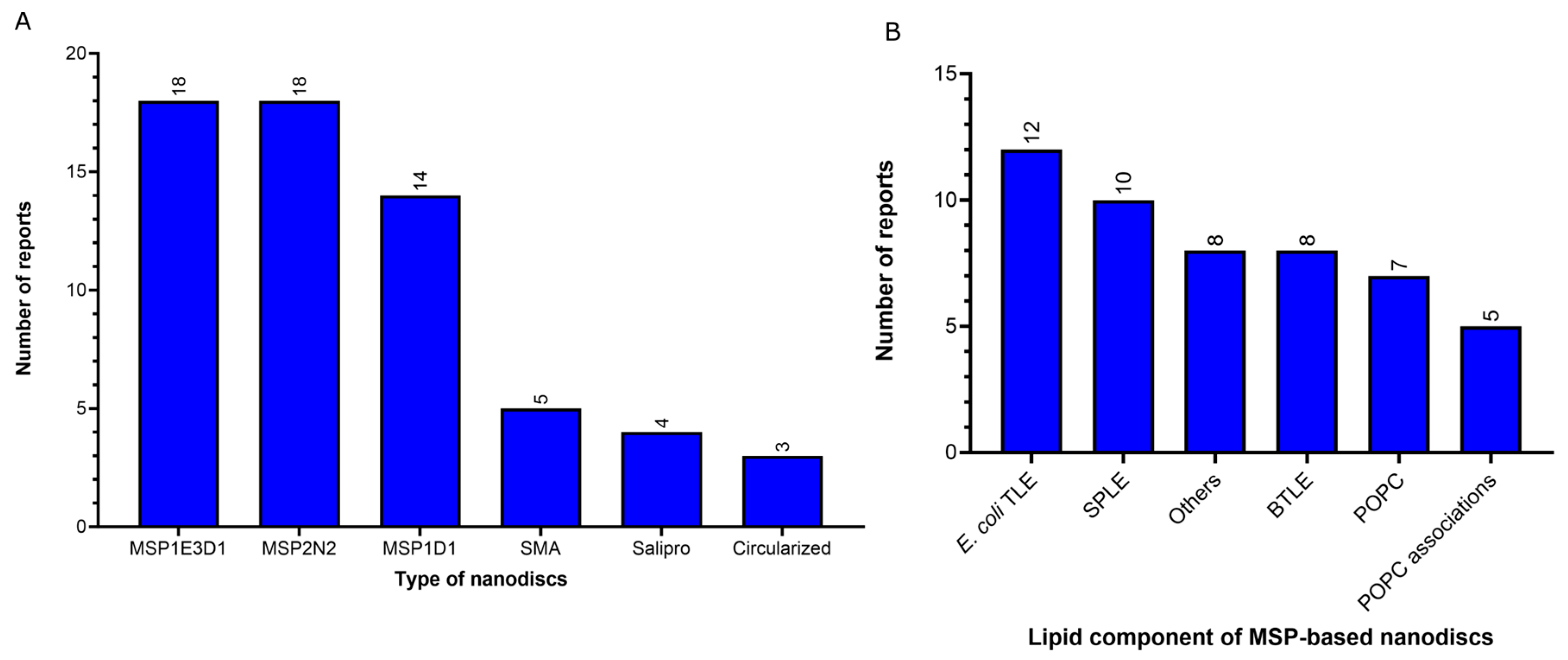
- Nanodiscs: 62 of 312 reports (20%). In this group, the most widely used type is the membrane scaffold proteins (MSP)-based nanodiscs, which is present in 50 of 62 reports.
- (3)
- Detergents with a single maltose-based polar moiety and single alkyl chain (DDM, UDM, DM): 50 of 312 reports (16%). In this group, the most widely used detergent is DDM, which is present in 42 of the 50 reports of this detergent class. DM is responsible for the eight remaining reports;
- (4)
- Glyco-diosgenin (GDN): 50 of 312 reports (16%).
- (5)
- Detergents belonging to the maltoside–neopentyl glycol (MNG) family (MNG and LMNG): 40 of 312 reports (13%). In this group, the most widely used detergent is LMNG, which is present in 38 of the 40 reports of this detergent class. MNG is responsible for the two remaining reports.
- (6)
- Digitonin: 29 of 312 reports (9%).
4. Sample Vitrification
5. Type of the Grids
- (1)
- Regular holey-carbon coated copper mesh grids (Quantifoil®): 112 of 310 reports (36%);
- (2)
- Regular holey-carbon coated gold mesh grids (Quantifoil®): 82 of 310 reports (26.5%);
- (3)
- Regular holey-gold coated gold mesh grids (UltrAuFoil®): 75 of 310 reports (24%).
6. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| APC | Amino Acid/Polyamine/Organocation |
| ApoA1 | Apolipoprotein A1 |
| C12E9 | Nonaethylene glycol monododecyl ether |
| C12E8 | Octaethylene glycol monododecyl ether |
| CCD | Charged-coupled device |
| CHAPS | 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate |
| CHS | Cholesteryl hemisuccinate |
| CMC | Critical micelle concentration |
| CMOS | Complementary metal–oxide–semiconductor |
| Cryo-EM | Cryo-electron microscopy |
| Cymal-4 | 4-cyclohexyl-1-butyl-β-D-maltoside |
| Cymal-6 | 6-cyclohexyl-1-hexyl-β-D-maltoside |
| DDD | Direct detector device |
| DDM | n-Dodecyl-β-D-maltopyranoside |
| DM | Decyl-b-D-Maltopyranoside |
| DQE | Detective quantum efficiency |
| FOM | Fluorinated octyl maltoside |
| FAB | Fragment antigen binding |
| GDN | Glyco-diosgenin |
| GPCR | G-protein coupled receptor |
| LDAO | N,N-Dimethyl-n-dodecylamine N-oxide |
| LMNG | Lauryl Maltose Neopentyl Glycol |
| MFS | Major facilitator superfamily |
| MNG | Maltose-Neopentyl Glycol |
| MP | Membrane Protein |
| MSP | Membrane Scaffold Protein |
| OGNG | Octyl glucose neopentyl glycol |
| t-PCCαM | 4-trans-(4-trans-Propylcyclohexyl)-cyclohexyl α-maltoside |
| PDB | Protein Data Bank |
| POPC | 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (16:0–18:1PC) |
| POPE | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (16:0–18:1PE) |
| POPG | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (16:0–18:1PG) |
| PTS | Phosphoenolpyruvate-dependent phosphotransferase |
| Salipro | Saposin–lipid–protein complexes |
| SLC | Solute carrier |
| SMA | Styrene maleic acid copolymer |
| SPA | Single-particle analysis |
| SPME | Soybean polar lipid extract |
| SNR | Signal-to-noise ratio |
| SSS | Solute sodium symporter |
| TEM: | Transmission electron microscopy |
| TLE: | Total lipid extract |
| UDM | n-Undecyl-β-D-maltopyranoside |
References
- Pandey, A.; Shin, K.; Patterson, R.E.; Liu, X.-Q.; Rainey, J.K. Current Strategies for Protein Production and Purification Enabling Membrane Protein Structural Biology. Biochem. Cell Biol. 2016, 94, 507–527. [Google Scholar] [CrossRef]
- Gong, J.; Chen, Y.; Pu, F.; Sun, P.; He, F.; Zhang, L.; Li, Y.; Ma, Z.; Wang, H. Understanding Membrane Protein Drug Targets in Computational Perspective. Curr. Drug Targets 2019, 20, 551–564. [Google Scholar] [CrossRef]
- Arinaminpathy, Y.; Khurana, E.; Engelman, D.M.; Gerstein, M.B. Computational Analysis of Membrane Proteins: The Largest Class of Drug Targets. Drug Discov. Today 2009, 14, 1130–1135. [Google Scholar] [CrossRef]
- Vénien-Bryan, C.; Li, Z.; Vuillard, L.; Boutin, J.A. Cryo-Electron Microscopy and X-Ray Crystallography: Complementary Approaches to Structural Biology and Drug Discovery. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 174–183. [Google Scholar] [CrossRef]
- Carpenter, E.P.; Beis, K.; Cameron, A.D.; Iwata, S. Overcoming the Challenges of Membrane Protein Crystallography. Curr. Opin. Struct. Biol. 2008, 18, 581–586. [Google Scholar] [CrossRef]
- Sgro, G.G.; Costa, T.R.D. Cryo-EM Grid Preparation of Membrane Protein Samples for Single Particle Analysis. Front. Mol. Biosci. 2018, 5, 74. [Google Scholar] [CrossRef]
- Xu, Y.; Dang, S. Recent Technical Advances in Sample Preparation for Single-Particle Cryo-EM. Front. Mol. Biosci. 2022, 9, 892459. [Google Scholar] [CrossRef]
- Liao, M.; Cao, E.; Julius, D.; Cheng, Y. Structure of the TRPV1 Ion Channel Determined by Electron Cryo-Microscopy. Nature 2013, 504, 107–112. [Google Scholar] [CrossRef]
- Cao, E.; Liao, M.; Cheng, Y.; Julius, D. TRPV1 Structures in Distinct Conformations Reveal Activation Mechanisms. Nature 2013, 504, 113–118. [Google Scholar] [CrossRef]
- Wu, S.; Armache, J.-P.; Cheng, Y. Single-Particle Cryo-EM Data Acquisition by Using Direct Electron Detection Camera. Microscopy 2016, 65, 35–41. [Google Scholar] [CrossRef]
- Weissenberger, G.; Henderikx, R.J.M.; Peters, P.J. Understanding the Invisible Hands of Sample Preparation for Cryo-EM. Nat. Methods 2021, 18, 463–471. [Google Scholar] [CrossRef]
- Cheng, Y.; Grigorieff, N.; Penczek, P.A.; Walz, T. A Primer to Single-Particle Cryo-Electron Microscopy. Cell 2015, 161, 438–449. [Google Scholar] [CrossRef]
- Kühlbrandt, W. The Resolution Revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- McMullan, G.; Faruqi, A.R.; Henderson, R. Direct Electron Detectors. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 579, pp. 1–17. ISBN 978-0-12-805382-9. [Google Scholar]
- Brilot, A.F.; Chen, J.Z.; Cheng, A.; Pan, J.; Harrison, S.C.; Potter, C.S.; Carragher, B.; Henderson, R.; Grigorieff, N. Beam-Induced Motion of Vitrified Specimen on Holey Carbon Film. J. Struct. Biol. 2012, 177, 630–637. [Google Scholar] [CrossRef]
- Li, X.; Mooney, P.; Zheng, S.; Booth, C.R.; Braunfeld, M.B.; Gubbens, S.; Agard, D.A.; Cheng, Y. Electron Counting and Beam-Induced Motion Correction Enable near-Atomic-Resolution Single-Particle Cryo-EM. Nat. Methods 2013, 10, 584–590. [Google Scholar] [CrossRef]
- Scheres, S.H.W. RELION: Implementation of a Bayesian Approach to Cryo-EM Structure Determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for Rapid Unsupervised Cryo-EM Structure Determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]
- Gómez-Blanco, J.; De La Rosa-Trevín, J.M.; Marabini, R.; Del Cano, L.; Jiménez, A.; Martínez, M.; Melero, R.; Majtner, T.; Maluenda, D.; Mota, J.; et al. Using Scipion for Stream Image Processing at Cryo-EM Facilities. J. Struct. Biol. 2018, 204, 457–463. [Google Scholar] [CrossRef]
- Nakane, T.; Kotecha, A.; Sente, A.; McMullan, G.; Masiulis, S.; Brown, P.M.G.E.; Grigoras, I.T.; Malinauskaite, L.; Malinauskas, T.; Miehling, J.; et al. Single-Particle Cryo-EM at Atomic Resolution. Nature 2020, 587, 152–156. [Google Scholar] [CrossRef]
- Guo, H.; Franken, E.; Deng, Y.; Benlekbir, S.; Singla Lezcano, G.; Janssen, B.; Yu, L.; Ripstein, Z.A.; Tan, Y.Z.; Rubinstein, J.L. Electron-Event Representation Data Enable Efficient cryoEM File Storage with Full Preservation of Spatial and Temporal Resolution. Int. Union Crystallogr. J. 2020, 7, 860–869. [Google Scholar] [CrossRef]
- Le Bon, C.; Michon, B.; Popot, J.-L.; Zoonens, M. Amphipathic Environments for Determining the Structure of Membrane Proteins by Single-Particle Electron Cryo-Microscopy. Quart. Rev. Biophys. 2021, 54, e6. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-C.; Lyu, M.; Morgan, C.E.; Bolla, J.R.; Robinson, C.V.; Yu, E.W. A ‘Build and Retrieve’ Methodology to Simultaneously Solve Cryo-EM Structures of Membrane Proteins. Nat. Methods 2021, 18, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Hagino, T.; Kato, T.; Kasuya, G.; Kobayashi, K.; Kusakizako, T.; Hamamoto, S.; Sobajima, T.; Fujiwara, Y.; Yamashita, K.; Kawasaki, H.; et al. Cryo-EM Structures of Thylakoid-Located Voltage-Dependent Chloride Channel VCCN1. Nat. Commun. 2022, 13, 2505. [Google Scholar] [CrossRef]
- He, Y.; Wang, K.; Yan, N. The Recombinant Expression Systems for Structure Determination of Eukaryotic Membrane Proteins. Protein Cell 2014, 5, 658–672. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, H.S.; Youn, T.; Byrne, B.; Chae, P.S. Impact of Novel Detergents on Membrane Protein Studies. Chem 2022, 8, 980–1013. [Google Scholar] [CrossRef]
- Stetsenko, A.; Guskov, A. An Overview of the Top Ten Detergents Used for Membrane Protein Crystallization. Crystals 2017, 7, 197. [Google Scholar] [CrossRef]
- Dörr, J.M.; Scheidelaar, S.; Koorengevel, M.C.; Dominguez, J.J.; Schäfer, M.; Van Walree, C.A.; Killian, J.A. The Styrene–Maleic Acid Copolymer: A Versatile Tool in Membrane Research. Eur. Biophys. J. 2016, 45, 3–21. [Google Scholar] [CrossRef]
- Lindhoud, S.; Carvalho, V.; Pronk, J.W.; Aubin-Tam, M.-E. SMA-SH: Modified Styrene–Maleic Acid Copolymer for Functionalization of Lipid Nanodiscs. Biomacromolecules 2016, 17, 1516–1522. [Google Scholar] [CrossRef]
- Lee, S.; Ghosh, S.; Jana, S.; Robertson, N.; Tate, C.G.; Vaidehi, N. How Do Branched Detergents Stabilize GPCRs in Micelles? Biochemistry 2020, 59, 2125–2134. [Google Scholar] [CrossRef]
- Breyton, C.; Javed, W.; Vermot, A.; Arnaud, C.-A.; Hajjar, C.; Dupuy, J.; Petit-Hartlein, I.; Le Roy, A.; Martel, A.; Thépaut, M.; et al. Assemblies of Lauryl Maltose Neopentyl Glycol (LMNG) and LMNG-Solubilized Membrane Proteins. Biochim. Biophys. Acta (BBA)—Biomembr. 2019, 1861, 939–957. [Google Scholar] [CrossRef]
- Magnani, F.; Shibata, Y.; Serrano-Vega, M.J.; Tate, C.G. Co-Evolving Stability and Conformational Homogeneity of the Human Adenosine A 2a Receptor. Proc. Natl. Acad. Sci. USA 2008, 105, 10744–10749. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.A.; Liu, J.J.; Chun, E.; Wacker, D.; Wu, H.; Cherezov, V.; Stevens, R.C. GPCR Stabilization Using the Bicelle-like Architecture of Mixed Sterol-Detergent Micelles. Methods 2011, 55, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, K.; Szabo, L.; Fales, H.M.; Pitha, J. 2-Hydroxypropyldigitonin: Synthesis and Properties of Preparations Differing in Degree of Substitution. J. Pharm. Sci. 1988, 77, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Otzen, D.E. Protein Unfolding in Detergents: Effect of Micelle Structure, Ionic Strength, pH, and Temperature. Biophys. J. 2002, 83, 2219–2230. [Google Scholar] [CrossRef] [PubMed]
- Khao, J.; Arce-Lopera, J.; Sturgis, J.N.; Duneau, J.-P. Structure of a Protein–Detergent Complex: The Balance between Detergent Cohesion and Binding. Eur. Biophys. J. 2011, 40, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.O.C.; Reis, R.; Siligardi, G.; Hussain, R.; Cheruvara, H.; Moraes, I. Selection of Biophysical Methods for Characterisation of Membrane Proteins. Int. J. Mol. Sci. 2019, 20, 2605. [Google Scholar] [CrossRef]
- Gimpl, K.; Klement, J.; Keller, S. Characterising Protein/Detergent Complexes by Triple-Detection Size-Exclusion Chromatography. Biol. Proced. Online 2016, 18, 4. [Google Scholar] [CrossRef]
- Sligar, S.G.; Denisov, I.G. Nanodiscs: A Toolkit for Membrane Protein Science. Protein Sci. 2021, 30, 297–315. [Google Scholar] [CrossRef]
- Bayburt, T.H.; Grinkova, Y.V.; Sligar, S.G. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Lett. 2002, 2, 853–856. [Google Scholar] [CrossRef]
- Denisov, I.G.; Grinkova, Y.V.; Lazarides, A.A.; Sligar, S.G. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004, 126, 3477–3487. [Google Scholar] [CrossRef]
- Li, M.J.; Atkins, W.M.; McClary, W.D. Preparation of Lipid Nanodiscs with Lipid Mixtures. CP Protein Sci. 2019, 98, e100. [Google Scholar] [CrossRef] [PubMed]
- Luthra, A.; Gregory, M.; Grinkova, Y.V.; Denisov, I.G.; Sligar, S.G. Nanodiscs in the Studies of Membrane-Bound Cytochrome P450 Enzymes. In Cytochrome P450 Protocols; Phillips, I.R., Shephard, E.A., Ortiz De Montellano, P.R., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 987, pp. 115–127. ISBN 978-1-62703-320-6. [Google Scholar]
- Nasr, M.L.; Baptista, D.; Strauss, M.; Sun, Z.-Y.J.; Grigoriu, S.; Huser, S.; Plückthun, A.; Hagn, F.; Walz, T.; Hogle, J.M.; et al. Covalently Circularized Nanodiscs for Studying Membrane Proteins and Viral Entry. Nat. Methods 2017, 14, 49–52. [Google Scholar] [CrossRef]
- Nasr, M.L.; Wagner, G. Covalently Circularized Nanodiscs; Challenges and Applications. Curr. Opin. Struct. Biol. 2018, 51, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Fan, X.; Yan, N. Cryo-EM Analysis of a Membrane Protein Embedded in the Liposome. Proc. Natl. Acad. Sci. USA 2020, 117, 18497–18503. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Zhao, C.; MacKinnon, R. Membrane Protein Isolation and Structure Determination in Cell-Derived Membrane Vesicles. Proc. Natl. Acad. Sci. USA 2023, 120, e2302325120. [Google Scholar] [CrossRef]
- Zoonens, M.; Popot, J.-L. Amphipols for Each Season. J. Membr. Biol 2014, 247, 759–796. [Google Scholar] [CrossRef] [PubMed]
- Popot, J.-L.; Althoff, T.; Bagnard, D.; Banères, J.-L.; Bazzacco, P.; Billon-Denis, E.; Catoire, L.J.; Champeil, P.; Charvolin, D.; Cocco, M.J.; et al. Amphipols From A to Z. Annu. Rev. Biophys. 2011, 40, 379–408. [Google Scholar] [CrossRef]
- Le Maire, M.; Champeil, P.; Møller, J.V. Interaction of Membrane Proteins and Lipids with Solubilizing Detergents. Biochim. Biophys. Acta (BBA)—Biomembr. 2000, 1508, 86–111. [Google Scholar] [CrossRef]
- Le Bon, C.; Marconnet, A.; Masscheleyn, S.; Popot, J.-L.; Zoonens, M. Folding and Stabilizing Membrane Proteins in Amphipol A8-35. Methods 2018, 147, 95–105. [Google Scholar] [CrossRef]
- Carlson, M.L.; Young, J.W.; Zhao, Z.; Fabre, L.; Jun, D.; Li, J.; Li, J.; Dhupar, H.S.; Wason, I.; Mills, A.T.; et al. The Peptidisc, a Simple Method for Stabilizing Membrane Proteins in Detergent-Free Solution. eLife 2018, 7, e34085. [Google Scholar] [CrossRef]
- Kampjut, D.; Steiner, J.; Sazanov, L.A. Cryo-EM Grid Optimization for Membrane Proteins. iScience 2021, 24, 102139. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Avila-Sakar, A.; Kim, J.; Booth, D.S.; Greenberg, C.H.; Rossi, A.; Liao, M.; Li, X.; Alian, A.; Griner, S.L.; et al. Fabs Enable Single Particle cryoEM Studies of Small Proteins. Structure 2012, 20, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Wentinck, K.; Gogou, C.; Meijer, D.H. Putting on Molecular Weight: Enabling Cryo-EM Structure Determination of Sub-100-kDa Proteins. Curr. Res. Struct. Biol. 2022, 4, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Uchański, T.; Masiulis, S.; Fischer, B.; Kalichuk, V.; López-Sánchez, U.; Zarkadas, E.; Weckener, M.; Sente, A.; Ward, P.; Wohlkönig, A.; et al. Megabodies Expand the Nanobody Toolkit for Protein Structure Determination by Single-Particle Cryo-EM. Nat. Methods 2021, 18, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, R.M. Preparing Better Samples for Cryo–Electron Microscopy: Biochemical Challenges Do Not End with Isolation and Purification. Annu. Rev. Biochem. 2021, 90, 451–474. [Google Scholar] [CrossRef]
- Naydenova, K.; Russo, C.J. Measuring the Effects of Particle Orientation to Improve the Efficiency of Electron Cryomicroscopy. Nat. Commun. 2017, 8, 629. [Google Scholar] [CrossRef]
- Sorzano, C.O.S.; Semchonok, D.; Lin, S.-C.; Lo, Y.-C.; Vilas, J.L.; Jiménez-Moreno, A.; Gragera, M.; Vacca, S.; Maluenda, D.; Martínez, M.; et al. Algorithmic Robustness to Preferred Orientations in Single Particle Analysis by CryoEM. J. Struct. Biol. 2021, 213, 107695. [Google Scholar] [CrossRef]
- Armstrong, M.; Han, B.-G.; Gomez, S.; Turner, J.; Fletcher, D.A.; Glaeser, R.M. Microscale Fluid Behavior during Cryo-EM Sample Blotting. Biophys. J. 2020, 118, 708–719. [Google Scholar] [CrossRef]
- Noble, A.J.; Wei, H.; Dandey, V.P.; Zhang, Z.; Tan, Y.Z.; Potter, C.S.; Carragher, B. Reducing Effects of Particle Adsorption to the Air–Water Interface in Cryo-EM. Nat. Methods 2018, 15, 793–795. [Google Scholar] [CrossRef]
- Razinkov, I.; Dandey, V.P.; Wei, H.; Zhang, Z.; Melnekoff, D.; Rice, W.J.; Wigge, C.; Potter, C.S.; Carragher, B. A New Method for Vitrifying Samples for cryoEM. J. Struct. Biol. 2016, 195, 190–198. [Google Scholar] [CrossRef]
- Kaledhonkar, S.; Fu, Z.; White, H.; Frank, J. Time-Resolved Cryo-Electron Microscopy Using a Microfluidic Chip. In Protein Complex Assembly; Marsh, J.A., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1764, pp. 59–71. ISBN 978-1-4939-7758-1. [Google Scholar]
- Kontziampasis, D.; Klebl, D.P.; Iadanza, M.G.; Scarff, C.A.; Kopf, F.; Sobott, F.; Monteiro, D.C.F.; Trebbin, M.; Muench, S.P.; White, H.D. A Cryo-EM Grid Preparation Device for Time-Resolved Structural Studies. Int. Union Crystallogr. J. 2019, 6, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, J.L.; Guo, H.; Ripstein, Z.A.; Haydaroglu, A.; Au, A.; Yip, C.M.; Di Trani, J.M.; Benlekbir, S.; Kwok, T. Shake-It-off: A Simple Ultrasonic Cryo-EM Specimen-Preparation Device. Acta Crystallogr. D Struct. Biol. 2019, 75, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.Z.; Rubinstein, J.L. Through-Grid Wicking Enables High-Speed cryoEM Specimen Preparation. Acta Crystallogr. D Struct. Biol. 2020, 76, 1092–1103. [Google Scholar] [CrossRef]
- Ashtiani, D.; De Marco, A.; Neild, A. Tailoring Surface Acoustic Wave Atomisation for Cryo-Electron Microscopy Sample Preparation. Lab Chip 2019, 19, 1378–1385. [Google Scholar] [CrossRef] [PubMed]
- White, H.D.; Thirumurugan, K.; Walker, M.L.; Trinick, J. A Second Generation Apparatus for Time-Resolved Electron Cryo-Microscopy Using Stepper Motors and Electrospray. J. Struct. Biol. 2003, 144, 246–252. [Google Scholar] [CrossRef]
- Ravelli, R.B.G.; Nijpels, F.J.T.; Henderikx, R.J.M.; Weissenberger, G.; Thewessem, S.; Gijsbers, A.; Beulen, B.W.A.M.M.; López-Iglesias, C.; Peters, P.J. Cryo-EM Structures from Sub-Nl Volumes Using Pin-Printing and Jet Vitrification. Nat. Commun. 2020, 11, 2563. [Google Scholar] [CrossRef]
- Rima, L.; Zimmermann, M.; Fränkl, A.; Clairfeuille, T.; Lauer, M.; Engel, A.; Engel, H.-A.; Braun, T. cryoWriter: A Blotting Free Cryo-EM Preparation System with a Climate Jet and Cover-Slip Injector. Faraday Discuss. 2022, 240, 55–66. [Google Scholar] [CrossRef]
- Darrow, M.C.; Moore, J.P.; Walker, R.J.; Doering, K.; King, R.S. Chameleon: Next Generation Sample Preparation for CryoEM Based on Spotiton. Microsc. Microanal. 2019, 25, 994–995. [Google Scholar] [CrossRef]
- Dandey, V.P.; Wei, H.; Zhang, Z.; Tan, Y.Z.; Acharya, P.; Eng, E.T.; Rice, W.J.; Kahn, P.A.; Potter, C.S.; Carragher, B. Spotiton: New Features and Applications. J. Struct. Biol. 2018, 202, 161–169. [Google Scholar] [CrossRef]
- Wei, H.; Dandey, V.P.; Zhang, Z.; Raczkowski, A.; Rice, W.J.; Carragher, B.; Potter, C.S. Optimizing “Self-Wicking” Nanowire Grids. J. Struct. Biol. 2018, 202, 170–174. [Google Scholar] [CrossRef]
- Levitz, T.S.; Weckener, M.; Fong, I.; Naismith, J.H.; Drennan, C.L.; Brignole, E.J.; Clare, D.K.; Darrow, M.C. Approaches to Using the Chameleon: Robust, Automated, Fast-Plunge cryoEM Specimen Preparation. Front. Mol. Biosci. 2022, 9, 903148. [Google Scholar] [CrossRef] [PubMed]
- Vilella, P. Étude des Mécanismes Moléculaires Régulant L’activité des Complexes Formés par le Récepteur Nucléaire ERRa. Ph.D. Thesis, University of Strasbourg, Strasbourg, France, 2023. [Google Scholar]
- Russo, C.J.; Passmore, L.A. Progress towards an Optimal Specimen Support for Electron Cryomicroscopy. Curr. Opin. Struct. Biol. 2016, 37, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.J.; Passmore, L.A. Ultrastable Gold Substrates for Electron Cryomicroscopy. Science 2014, 346, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.J.; Passmore, L.A. Ultrastable Gold Substrates: Properties of a Support for High-Resolution Electron Cryomicroscopy of Biological Specimens. J. Struct. Biol. 2016, 193, 33–44. [Google Scholar] [CrossRef]
- Quispe, J.; Damiano, J.; Mick, S.E.; Nackashi, D.P.; Fellmann, D.; Ajero, T.G.; Carragher, B.; Potter, C.S. An Improved Holey Carbon Film for Cryo-Electron Microscopy. Microsc. Microanal. 2007, 13, 365–371. [Google Scholar] [CrossRef]
- Cho, H.-J.; Hyun, J.-K.; Kim, J.-G.; Jeong, H.S.; Park, H.N.; You, D.-J.; Jung, H.S. Measurement of Ice Thickness on Vitreous Ice Embedded Cryo-EM Grids: Investigation of Optimizing Condition for Visualizing Macromolecules. J. Anal. Sci. Technol. 2013, 4, 7. [Google Scholar] [CrossRef]
- Park, J.; Park, J.; Lee, S.; Roh, S.-H. Grid Selection Strategy for High-Resolution Cryo-EM. Korean Soc. Struct. Biol. 2020, 8, 41–48. [Google Scholar] [CrossRef]
- Naydenova, K.; Jia, P.; Russo, C.J. Cryo-EM with Sub–1 Å Specimen Movement. Science 2020, 370, 223–226. [Google Scholar] [CrossRef]
- Naydenova, K.; Peet, M.J.; Russo, C.J. Multifunctional Graphene Supports for Electron Cryomicroscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 11718–11724. [Google Scholar] [CrossRef]
- Guo, F.; Jiang, W. Single Particle Cryo-Electron Microscopy and 3-D Reconstruction of Viruses. In Electron Microscopy; Kuo, J., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1117, pp. 401–443. ISBN 978-1-62703-775-4. [Google Scholar]
- Mahler, F.; Meister, A.; Vargas, C.; Durand, G.; Keller, S. Self-Assembly of Protein-Containing Lipid-Bilayer Nanodiscs from Small-Molecule Amphiphiles. Small 2021, 17, 2103603. [Google Scholar] [CrossRef]
- Flötenmeyer, M.; Weiss, H.; Tribet, C.; Popot, J.-L.; Leonard, K. The Use of Amphipathic Polymers for Cryo Electron Microscopy of NADH:Ubiquinone Oxidoreductase (Complex I). J. Microsc. 2007, 227, 229–235. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, T.; Huang, S.-Y. Improvement of Cryo-EM Maps by Simultaneous Local and Non-Local Deep Learning. Nat. Commun. 2023, 14, 3217. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.F.; Abeyrathne, P.D.; Dukovski, D.; Walz, T. The Affinity Grid: A Pre-Fabricated EM Grid for Monolayer Purification. J. Mol. Biol. 2008, 382, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Llaguno, M.C.; Xu, H.; Shi, L.; Huang, N.; Zhang, H.; Liu, Q.; Jiang, Q.-X. Chemically Functionalized Carbon Films for Single Molecule Imaging. J. Struct. Biol. 2014, 185, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Han, B.-G.; Watson, Z.; Kang, H.; Pulk, A.; Downing, K.H.; Cate, J.; Glaeser, R.M. Long Shelf-Life Streptavidin Support-Films Suitable for Electron Microscopy of Biological Macromolecules. J. Struct. Biol. 2016, 195, 238–244. [Google Scholar] [CrossRef]
- Meyerson, J.R.; Rao, P.; Kumar, J.; Chittori, S.; Banerjee, S.; Pierson, J.; Mayer, M.L.; Subramaniam, S. Self-Assembled Monolayers Improve Protein Distribution on Holey Carbon Cryo-EM Supports. Sci. Rep. 2014, 4, 7084. [Google Scholar] [CrossRef]
- Zhang, Z.; Shigematsu, H.; Shimizu, T.; Ohto, U. Improving Particle Quality in Cryo-EM Analysis Using a PEGylation Method. Structure 2021, 29, 1192–1199.e4. [Google Scholar] [CrossRef]
- Murata, K.; Wolf, M. Cryo-Electron Microscopy for Structural Analysis of Dynamic Biological Macromolecules. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2018, 1862, 324–334. [Google Scholar] [CrossRef]
- Punjani, A.; Fleet, D.J. 3D Variability Analysis: Resolving Continuous Flexibility and Discrete Heterogeneity from Single Particle Cryo-EM. J. Struct. Biol. 2021, 213, 107702. [Google Scholar] [CrossRef]
- Vuillemot, R.; Mirzaei, A.; Harastani, M.; Hamitouche, I.; Fréchin, L.; Klaholz, B.P.; Miyashita, O.; Tama, F.; Rouiller, I.; Jonic, S. MDSPACE: Extracting Continuous Conformational Landscapes from Cryo-EM Single Particle Datasets Using 3D-to-2D Flexible Fitting Based on Molecular Dynamics Simulation. J. Mol. Biol. 2023, 435, 167951. [Google Scholar] [CrossRef]
- Chen, M.; Ludtke, S.J. Deep Learning-Based Mixed-Dimensional Gaussian Mixture Model for Characterizing Variability in Cryo-EM. Nat. Methods 2021, 18, 930–936. [Google Scholar] [CrossRef] [PubMed]
- Krieger, J.M.; Sorzano, C.O.S.; Carazo, J.M. Scipion-EM-ProDy: A Graphical Interface for the ProDy Python Package within the Scipion Workflow Engine Enabling Integration of Databases, Simulations and Cryo-Electron Microscopy Image Processing. IJMS 2023, 24, 14245. [Google Scholar] [CrossRef] [PubMed]
- Kidmose, R.T.; Juhl, J.; Nissen, P.; Boesen, T.; Karlsen, J.L.; Pedersen, B.P. Namdinator—Automatic Molecular Dynamics Flexible Fitting of Structural Models into Cryo-EM and Crystallography Experimental Maps. Int. Union Crystallogr. J. 2019, 6, 526–531. [Google Scholar] [CrossRef]
- Vuillemot, R.; Miyashita, O.; Tama, F.; Rouiller, I.; Jonic, S. NMMD: Efficient Cryo-EM Flexible Fitting Based on Simultaneous Normal Mode and Molecular Dynamics Atomic Displacements. J. Mol. Biol. 2022, 434, 167483. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.G.S.; Fagnen, C.; Vénien-Bryan, C.; Perahia, D. A New Strategy for Atomic Flexible Fitting in Cryo-EM Maps by Molecular Dynamics with Excited Normal Modes (MDeNM-EMfit). J. Chem. Inf. Model. 2020, 60, 2419–2423. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vénien-Bryan, C.; Fernandes, C.A.H. Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis. Int. J. Mol. Sci. 2023, 24, 14785. https://doi.org/10.3390/ijms241914785
Vénien-Bryan C, Fernandes CAH. Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis. International Journal of Molecular Sciences. 2023; 24(19):14785. https://doi.org/10.3390/ijms241914785
Chicago/Turabian StyleVénien-Bryan, Catherine, and Carlos A. H. Fernandes. 2023. "Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis" International Journal of Molecular Sciences 24, no. 19: 14785. https://doi.org/10.3390/ijms241914785
APA StyleVénien-Bryan, C., & Fernandes, C. A. H. (2023). Overview of Membrane Protein Sample Preparation for Single-Particle Cryo-Electron Microscopy Analysis. International Journal of Molecular Sciences, 24(19), 14785. https://doi.org/10.3390/ijms241914785




