An Investigation of the JAZ Family and the CwMYC2-like Protein to Reveal Their Regulation Roles in the MeJA-Induced Biosynthesis of β-Elemene in Curcuma wenyujin
Abstract
:1. Introduction
2. Results
2.1. Identification of the JAZ Family Genes in C. wenyujin
2.2. Phylogenetic Relationships and Motif Composition of JAZs in C. wenyujin
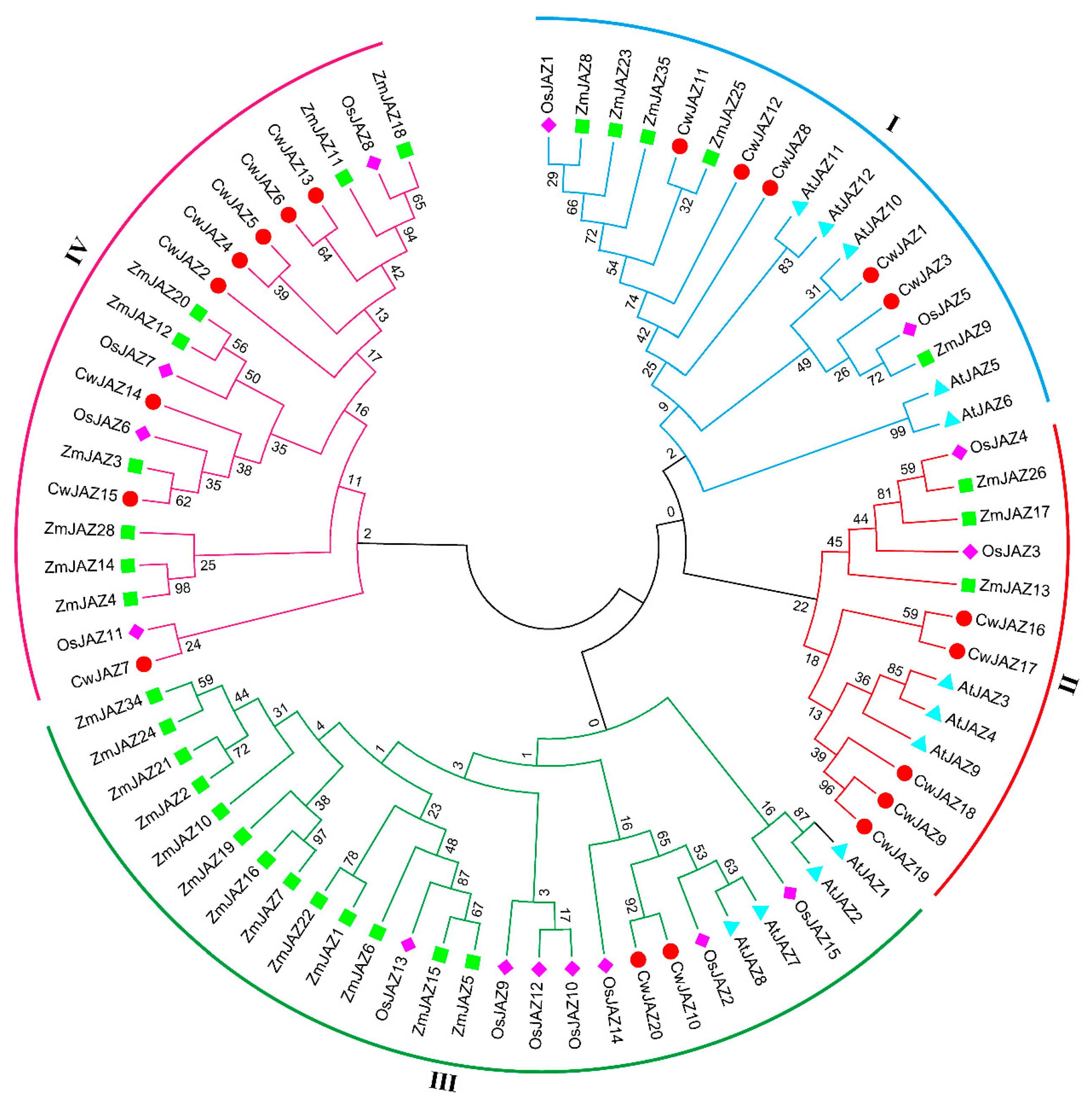
2.3. Phylogenetic Analysis of JAZ Proteins from Different Plant Species
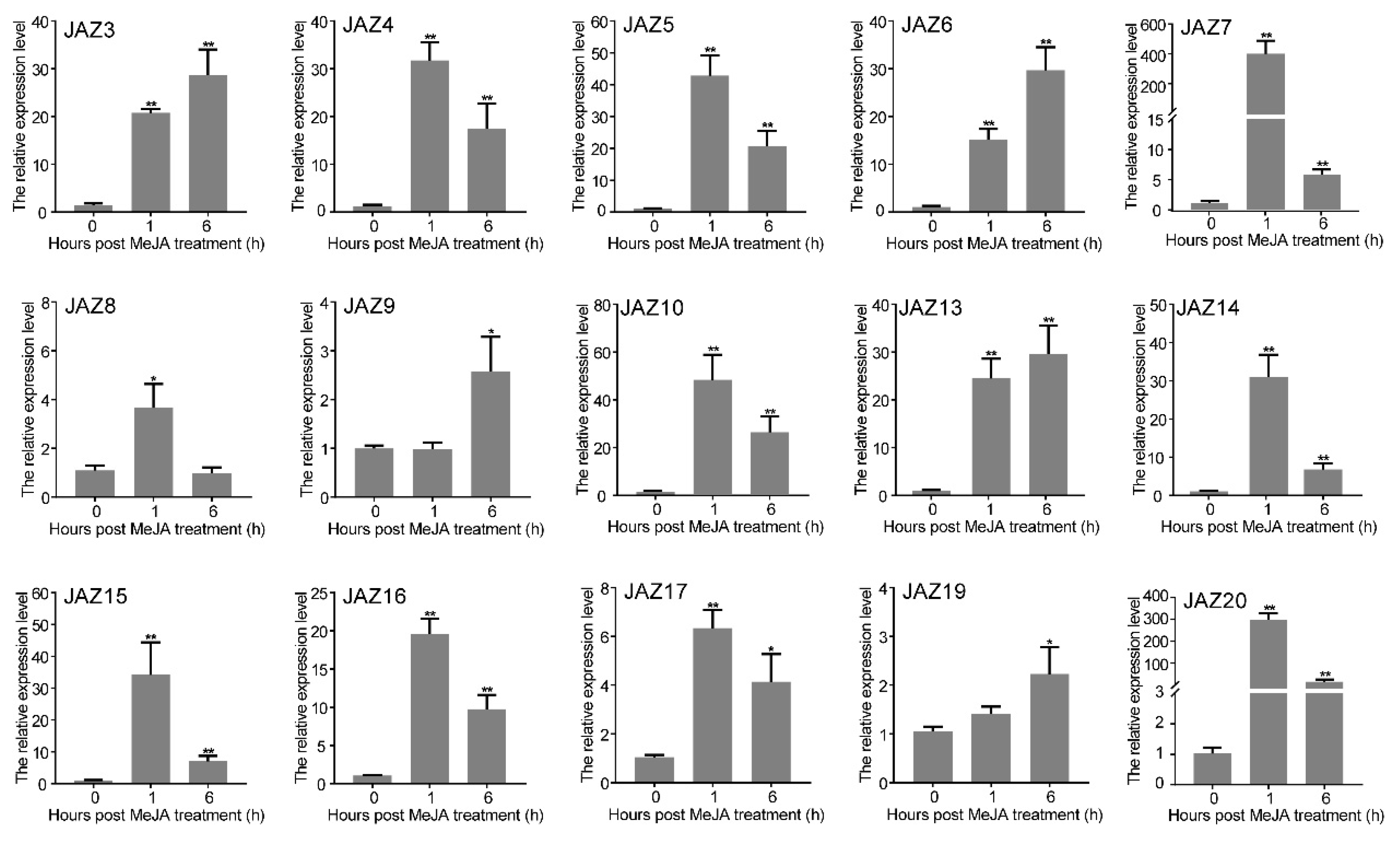
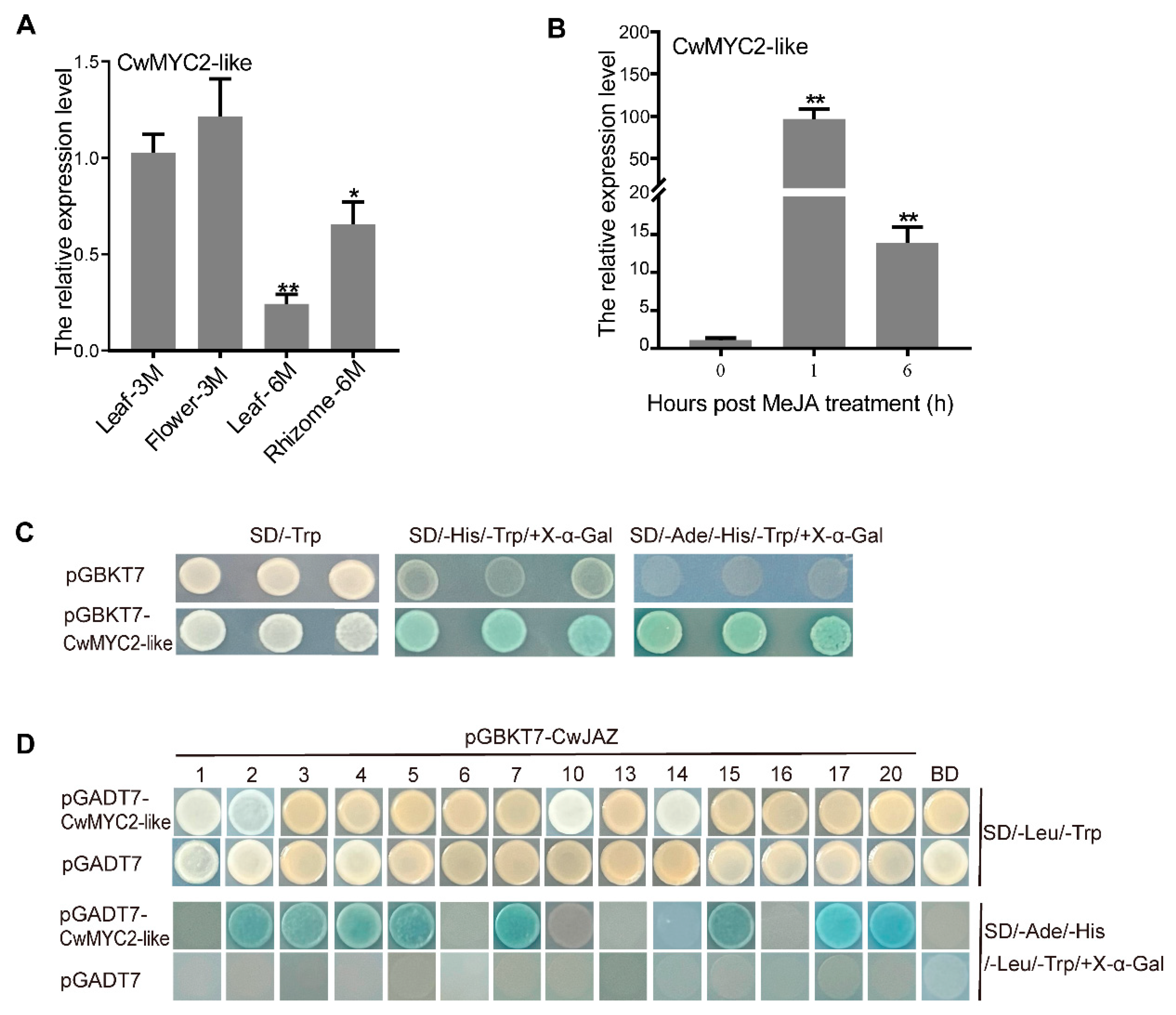
2.4. Expression Profiles of CwJAZ Genes
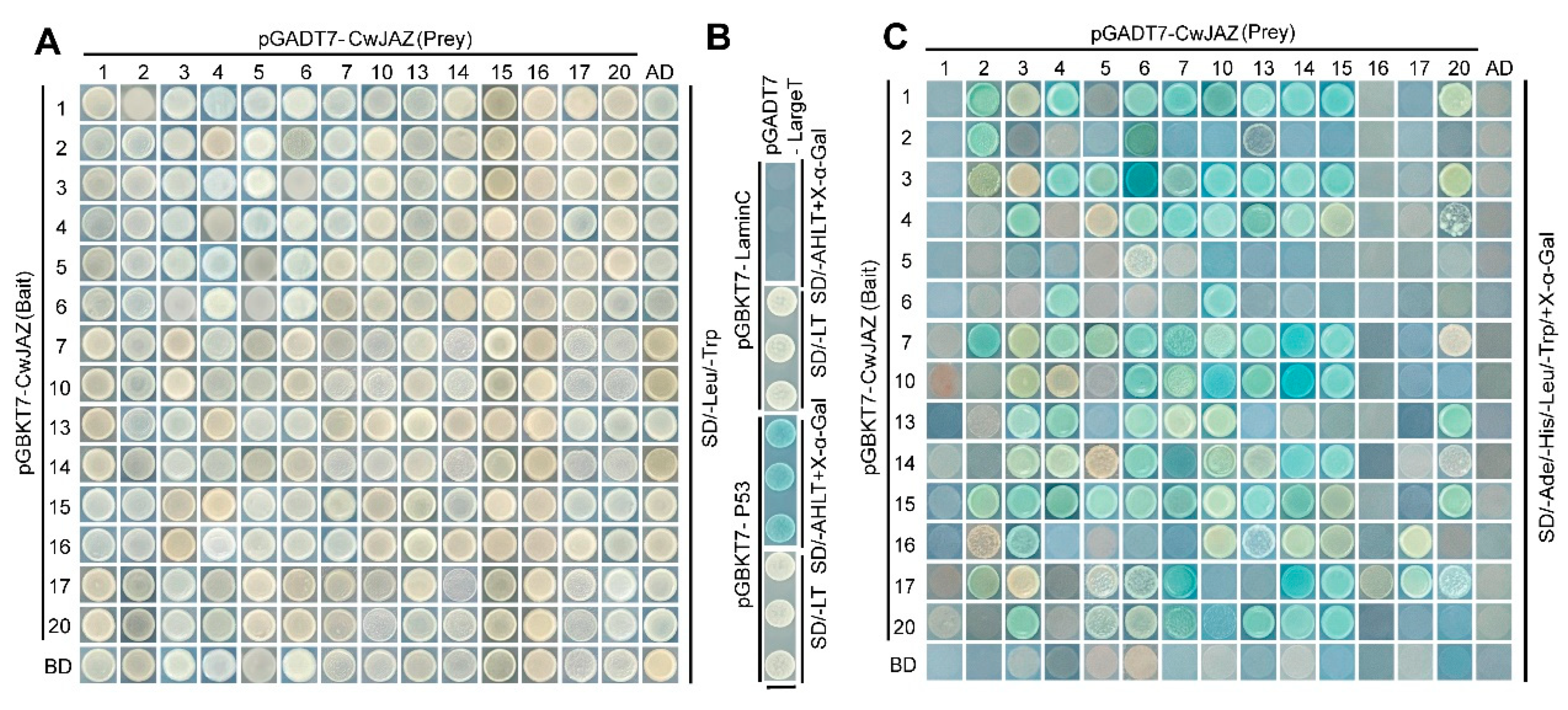
2.5. Interaction between CwJAZ Proteins
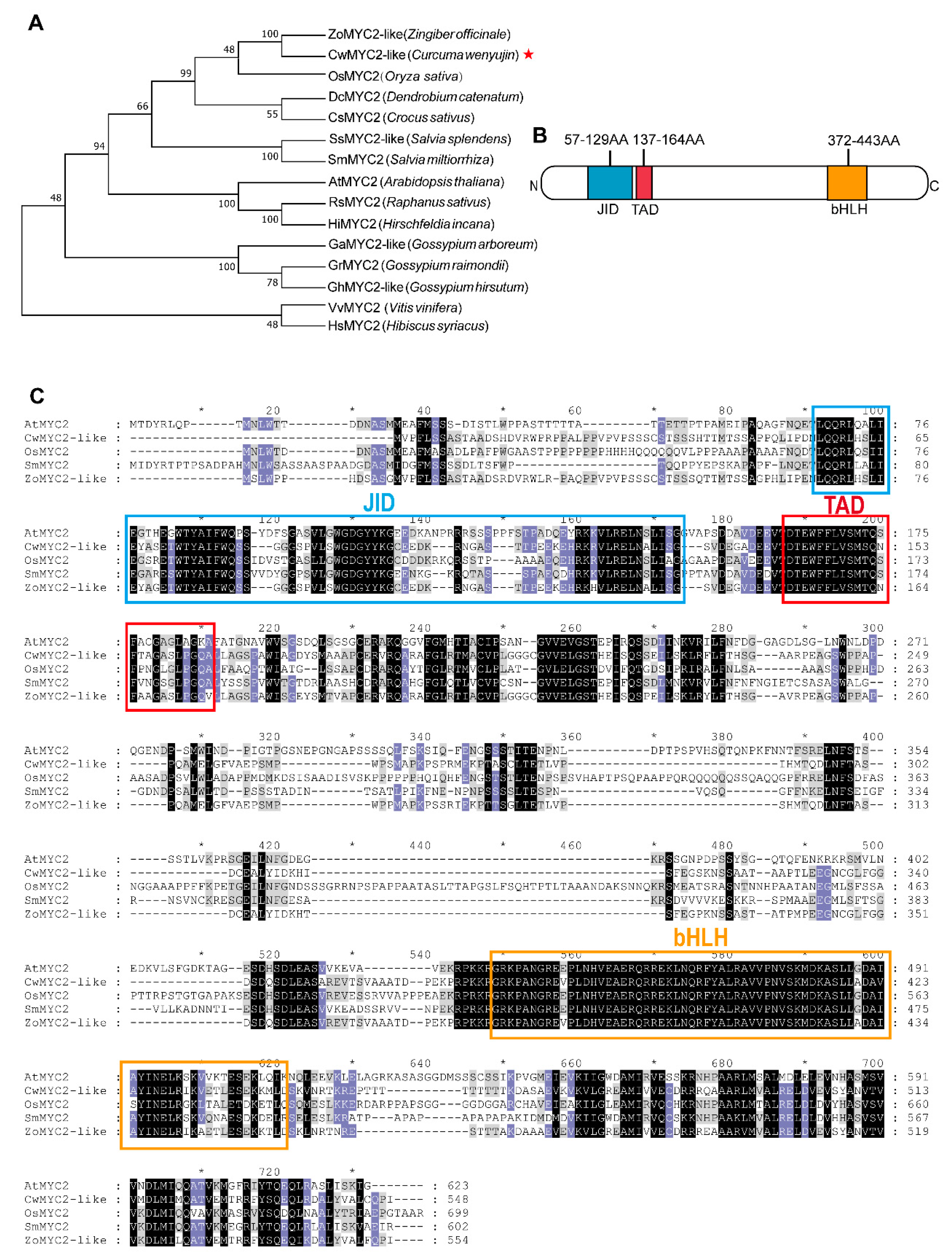
2.6. Characterization of CwMYC2-like Protein
2.7. CwMYC2-like Protein Interacted with CwJAZs
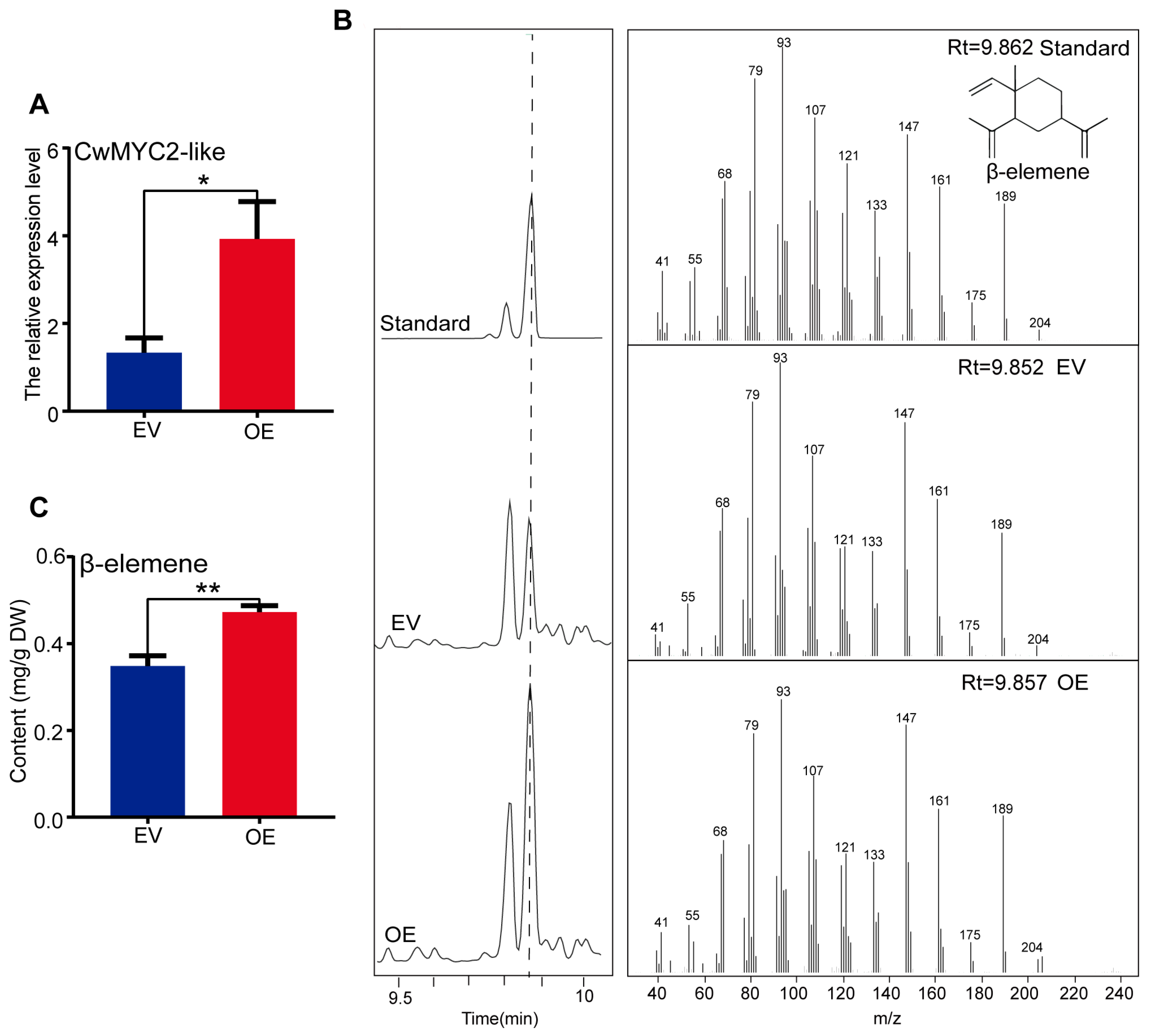
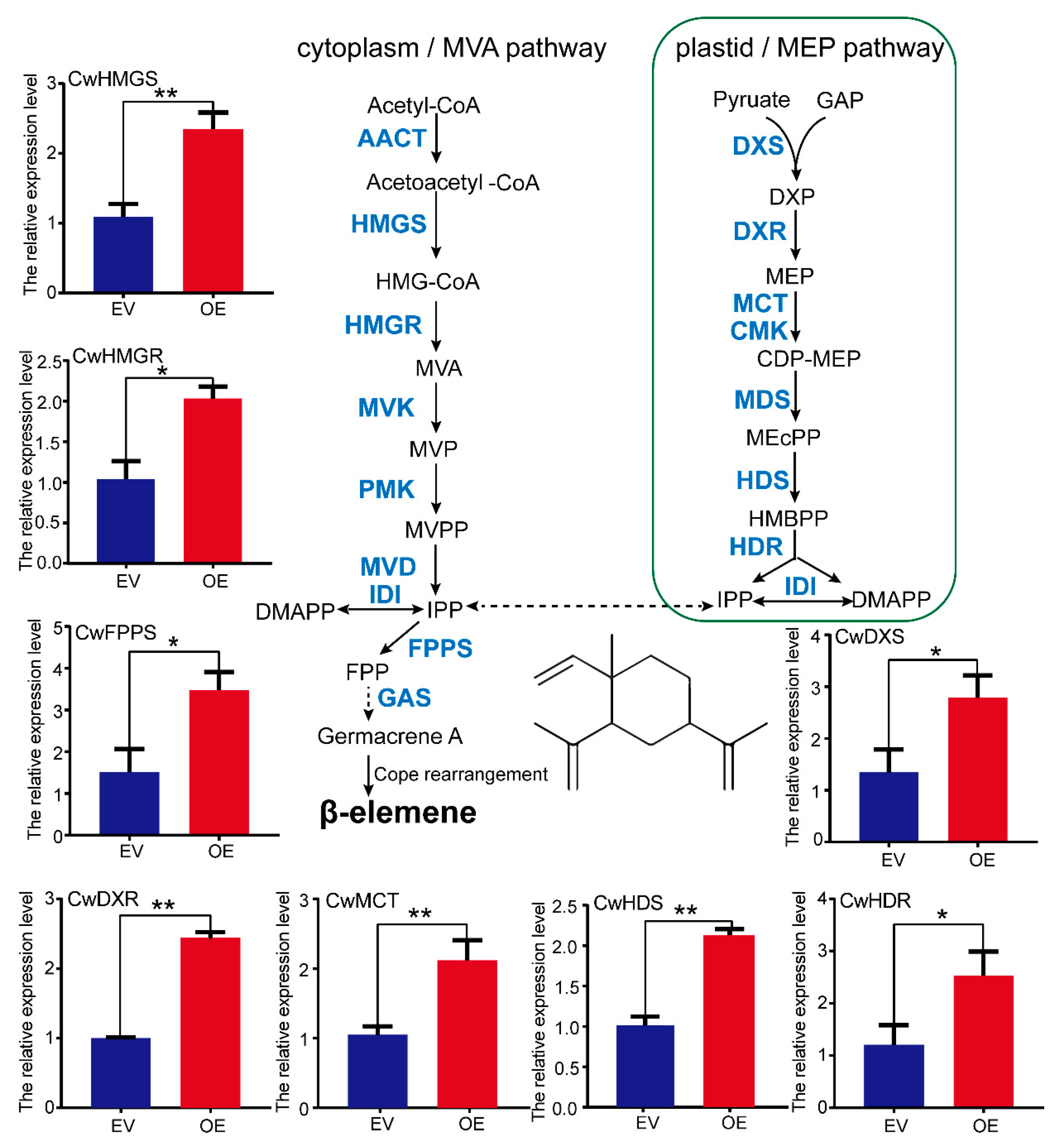
2.8. Transient Over-Expression of the CwMYC2-like Gene Enhanced the Accumulation of β-Elemene in C. wenyujin
3. Discussion
4. Materials and Methods
4.1. Plant Material and JA Treatment
4.2. Identification of the JAZ Genes in C. wenyujin
4.3. Phylogenetic Analysis and Conserved Motif Identification
4.4. RNA Extraction and qRT-PCR Analysis
4.5. Yeast Two-Hybrid (Y2H) Assay and Transactivation Activity Analysis
4.6. Identification and Transient Overexpression of CwMYC2-like Gene in C. wenyujin Leaves
4.7. Extraction and Analysis of β-Elemene
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, H.X.; Yang, T.Y.; Yang, T.L.; Ge, F.H.; Pan, W.S.; Yang, X.G.; Chen, J.M. Study on ingredients of essential oils of Curcuma wenyujin extracted by supercritical-CO2 fluid extraction and steam distillation. China J. Chin. Mater. Med. 2006, 31, 1445–1446. [Google Scholar]
- Chen, X.; Huang, C.; Li, K.; Liu, J.; Zheng, Y.; Feng, Y.; Kai, G.Y. Recent advances in biosynthesis and pharmacology of β-elemene. Phytochem. Rev. 2023, 22, 169–186. [Google Scholar] [CrossRef]
- Zhai, B.T.; Zeng, Y.Y.; Zeng, Z.W.; Zhang, N.N.; Li, C.X.; Zeng, Y.J.; You, Y.; Wang, S.L.; Chen, X.B.; Sui, X.B.; et al. Drug delivery systems for elemene, its main active ingredient beta-elemene, and its derivatives in cancer therapy. Int. J. Nanomed. 2018, 13, 6279–6296. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.T.; Zhou, Y.J.J.; Bao, J.C.; Huang, L.Q.; Nielsen, J.; Krivoruchko, A. Metabolic engineering of Saccharomyces cerevisiae for production of germacrene A, a precursor of beta-elemene. J. Ind. Microbiol. Biotechnol. 2017, 44, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, J.; Wang, Z.; Li, Y.; Meng, X.; Shen, Y.; Liu, W. Improved production of germacrene A, a direct precursor of β-elemene, in engineered Saccharomyces cerevisiae by expressing a cyanobacterial germacrene A synthase. Microb. Cell Fact. 2021, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.C. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 2007, 3, 387–395. [Google Scholar] [CrossRef]
- Broker, J.K.; Muller, B.; van Deenen, N.; Prufer, D.; Gronover, C.S. Upregulating the mevalonate pathway and repressing sterol synthesis in Saccharomyces cerevisiae enhances the production of triterpenes. Appl. Microbiol. Biotechnol. 2018, 102, 6923–6934. [Google Scholar] [CrossRef]
- Chen, R.; Liu, Y.H.; Chen, S.; Wang, M.; Zhu, Y.; Hu, T.Y.; Wei, Q.H.; Yin, X.P.; Xie, T. Protein engineering of a germacrene A synthase from Lactuca sativa and its application in high productivity of germacrene A in Escherichia coli. Front. Plant Sci. 2022, 13, 932966. [Google Scholar] [CrossRef]
- De Kraker, J.W.; Franssen, M.C.; de Groot, A.; Konig, W.A.; and Bouwmeester, H.J. (+)-Germacrene A biosynthesis. The committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol. 1998, 117, 1381–1392. [Google Scholar] [CrossRef]
- De Geyter, N.; Gholami, A.; Goormachtig, S.; Goossens, A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012, 17, 349–359. [Google Scholar] [CrossRef]
- Wang, H.H.; Ma, C.F.; Li, Z.Q.; Ma, L.Q.; Wang, H.; Ye, H.C.; Xu, G.W.; Liu, B.Y. Effects of exogenous methyl jasmonate on artemisinin biosynthesis and secondary metabolites in Artemisia annua L. Ind. Crops Prod. 2010, 31, 214–218. [Google Scholar] [CrossRef]
- Hao, X.L.; Shi, M.; Cui, L.J.; Xu, C.; Zhang, Y.J.; Kai, G.Y. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 2015, 62, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Vanden Bossche, R.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef]
- Suza, W.P.; Rowe, M.L.; Hamberg, M.; Staswick, P.E. A tomato enzyme synthesizes (+)-7-iso-jasmonoyl-L-isoleucine in wounded leaves. Planta 2010, 231, 717–728. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.H.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCO11 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Vanholme, B.; Grunewald, W.; Bateman, A.; Kohchi, T.; Gheysen, G. The tify family previously known as ZIM. Trends Plant Sci. 2007, 12, 239–244. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.; Chico, J.M.; Fernandez-Calvo, P.; Solano, R. The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 2009, 59, 77–87. [Google Scholar] [CrossRef]
- Shyu, C.; Figueroa, P.; DePew, C.L.; Cooke, T.F.; Sheard, L.B.; Moreno, J.E.; Katsir, L.; Zheng, N.; Browse, J.; Howe, G.A. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 2012, 24, 536–550. [Google Scholar] [CrossRef]
- Moreno, J.E.; Shyu, C.; Campos, M.L.; Patel, L.C.; Chung, H.S.; Yao, J.; He, S.Y.; Howe, G.A. Negative feedback control of Jasmonate signaling by an alternative splice variant of JAZ10. Plant Physiol. 2013, 162, 1006–1017. [Google Scholar] [CrossRef]
- Zhang, F.; Ke, J.Y.; Zhang, L.; Chen, R.Z.; Sugimoto, K.; Howe, G.A.; Xu, H.E.; Zhou, M.G.; He, S.Y.; Melcher, K. Structural insights into alternative splicing-mediated desensitization of jasmonate signaling. Proc. Natl. Acad. Sci. USA 2017, 114, 1720–1725. [Google Scholar] [CrossRef]
- Ma, Y.N.; Xu, D.B.; Li, L.; Zhang, F.; Fu, X.Q.; Shen, Q.; Lyu, X.Y.; Wu, Z.K.; Pan, Q.F.; Shi, P.; et al. Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 2018, 4, eaas9357. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.Q.; Peng, B.W.; Hassani, D.; Xie, L.H.; Liu, H.; Li, Y.P.; Chen, T.T.; Liu, P.; Tang, Y.L.; Li, L.; et al. AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in Artemisia annua. Plant Physiol. 2021, 231, 1858–1874. [Google Scholar] [CrossRef]
- Breeze, E. Master MYCs: MYC2, the jasmonate signaling “Master Switch”. Plant Cell 2019, 31, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martinez-Garcia, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Shen, Q.; Lu, X.; Yan, T.X.; Fu, X.Q.; Lv, Z.Y.; Zhang, F.Y.; Pan, Q.F.; Wang, G.F.; Sun, X.F.; Tang, K.X. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 2016, 210, 1269–1281. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Sun, W.; Chen, J.F.; Tan, H.X.; Xiao, Y.; Li, Q.; Ji, Q.; Gao, S.H.; Chen, L.; Chen, S.L.; et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 2020, 10, 22852. [Google Scholar] [CrossRef]
- Yang, N.; Zhou, W.; Su, J.; Wang, X.; Li, L.; Wang, L.; Cao, X.; Wang, Z. Overexpression of SmMYC2 increases the production of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 2017, 8, 1804. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.H.; Lan, K.R.; Liu, Y.Y.; Chen, R.; Hu, T.Y.; Zhao, S.J.; Yin, X.P.; Xie, T. Transcriptome analysis reveals regulation mechanism of methyl jasmonate-induced terpenes biosynthesis in Curcuma wenyujin. PLoS ONE 2022, 17, e0270309. [Google Scholar] [CrossRef]
- Sheard, L.B.; Tan, X.; Mao, H.B.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468, 400–405. [Google Scholar] [CrossRef]
- Takaoka, Y.; Suzuki, K.; Nozawa, A.; Takahashi, H.; Sawasaki, T.; Ueda, M. Protein-protein interactions between jasmonate-related master regulator MYC and transcriptional mediator MED25 depend on a short binding domain. J Biol. Chem. 2022, 298, 101504. [Google Scholar] [CrossRef]
- Cai, H.; Tian, S.; Dong, H. Large scale in silico identification of MYB family genes from wheat expressed sequence tags. Mol. Biotechnol. 2012, 52, 184–192. [Google Scholar] [CrossRef]
- Zabala, M.D.; Zhai, B.; Jayaraman, S.; Eleftheriadou, G.; Winsbury, R.; Yang, R.; Truman, W.; Tang, S.J.; Smirnoff, N.; Grant, M. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytol. 2016, 209, 1120–1134. [Google Scholar] [CrossRef]
- Boter, M.; Golz, J.F.; Gimenez-Ibanez, S.; Fernandez-Barbero, G.; Franco-Zorrilla, J.M.; Solano, R. FILAMENTOUS FLOWER Is a direct target of JAZ3 and modulates responses to Jasmonate. Plant Cell 2015, 27, 3160–3174. [Google Scholar] [CrossRef]
- Oblessuc, P.R.; Obulareddy, N.; DeMott, L.; Matiolli, C.C.; Thompson, B.K.; Melotto, M. JAZ4 is involved in plant defense, growth, and development in Arabidopsis. Plant J. 2020, 101, 371–383. [Google Scholar] [CrossRef]
- Qi, T.C.; Song, S.S.; Ren, Q.C.; Wu, D.W.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.M.; Xie, D.X. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef]
- Singh, A.P.; Pandey, B.K.; Mehra, P.; Heitz, T.; Giri, J. OsJAZ9 overexpression modulates jasmonic acid biosynthesis and potassium deficiency responses in rice. Plant Mol. Bio. 2020, 104, 397–410. [Google Scholar] [CrossRef]
- Lv, Y.; Yang, M.; Hu, D.; Yang, Z.Y.; Ma, S.Q.; Li, X.H.; Xiong, L.Z. The OsMYB30 transcription factor suppresses cold tolerance by interacting with a JAZ protein and suppressing β-amylase expression. Plant Physiol. 2017, 173, 1475–1491. [Google Scholar] [CrossRef]
- Wu, H.; Ye, H.Y.; Yao, R.F.; Zhang, T.; Xiong, L.Z. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Howe, G.A. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 2009, 21, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Geerinck, J.; Pauwels, L.; De Jaeger, G.; Goossens, A. Dissection of the one-MegaDalton JAZ1 protein complex. Plant Signal. Behav. 2010, 5, 1039–1041. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, Y.; Liu, B.; Chen, Y.; Song, H.; Yu, R.; Che, X.; Qu, X.; Liu, Y.; Hu, X.; et al. β-Elemene inhibits peritoneal metastasis of gastric cancer cells by modulating FAK/Claudin-1 signaling. Phytother. Res. 2019, 33, 2448–2456. [Google Scholar] [CrossRef]
- Han, B.; Wang, T.; Xue, Z.; Wen, T.; Lu, L.; Meng, J.; Liu, J.; Wu, S.; Yu, J.; Xu, H. Elemene nanoemulsion inhibits metastasis of breast cancer by ROS scavenging. Int. J. Nanomed. 2021, 16, 6035–6048. [Google Scholar] [CrossRef]
- Yu, Z.; Wang, R.; Xu, L.; Dong, J.; Jing, Y. N-(beta-Elemene-13-yl)tryptophan methyl ester induces apoptosis in human leukemia cells and synergizes with arsenic trioxide through a hydrogen peroxide dependent pathway. Cancer Lett. 2008, 269, 165–173. [Google Scholar] [CrossRef]
- Liu, J.P.; Hu, J.; Liu, Y.H.; Yang, C.P.; Zhuang, Y.F.; Guo, X.L.; Li, Y.J.; Zhang, L.S. Transcriptome analysis of Hevea brasiliensis in response to exogenous methyl jasmonate provides novel insights into regulation of jasmonate-elicited rubber biosynthesis. Physiol. Mol. Biol. Plants 2018, 24, 349–358. [Google Scholar] [CrossRef]
- Chen, X.Z.; Li, J.R.; Wang, X.B.; Zhong, L.T.; Tang, Y.; Zhou, X.X.; Liu, Y.T.; Zhan, R.T.; Zheng, H.; Chen, W.W.; et al. Full-length transcriptome sequencing and methyl jasmonate-induced expression profile analysis of genes related to patchoulol biosynthesis and regulation in Pogostemon cablin. BMC Plant Biol. 2019, 19, 266. [Google Scholar] [CrossRef]
- Ye, H.Y.; Du, H.; Tang, N.; Li, X.H.; Xiong, L.Z. Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 2009, 71, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.B.; Li, X.L.; Yu, R.; Han, M.; Wu, Z.Y. Isolation, structural analysis, and expression characteristics of the maize TIFY gene family. Mol. Genet. Genomics 2015, 290, 1849–1858. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Wu, L.Y.; Wang, Y.; Zhang, X.S.; Chen, L.N. Bridging protein local structures and protein functions. Amino Acids 2008, 35, 627–650. [Google Scholar] [CrossRef] [PubMed]
- Millard, P.S.; Kragelund, B.B.; Burow, M. R2R3 MYB transcription factors—Functions outside the DNA-binding domain. Trends Plant Sci. 2019, 24, 934–946. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, L.; Qi, T.; Zhang, B.; Peng, W.; Liu, Y.; Xie, D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant 2011, 4, 279–288. [Google Scholar] [CrossRef]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef]
- Pei, T.L.; Ma, P.D.; Ding, K.; Liu, S.J.; Jia, Y.Y.; Ru, M.; Dong, J.E.; Liang, Z.S. SmJAZ8 acts as a core repressor regulating JA-induced biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots. J. Exp. Bot. 2018, 69, 1663–1678. [Google Scholar] [CrossRef]
- Ma, P.; Pei, T.; Lv, B.; Wang, M.; Dong, J.; Liang, Z. Functional pleiotropism, diversity, and redundancy of Salvia miltiorrhiza Bunge JAZ family proteins in jasmonate-induced tanshinone and phenolic acid biosynthesis. Hortic. Res. 2022, 9, uhac166. [Google Scholar] [CrossRef]
- Sheng, Y.; Yu, H.; Pan, H.; Qiu, K.; Xie, Q.; Chen, H.; Fu, S.; Zhang, J.; Zhou, H. Genome-wide analysis of the gene structure, expression and protein interactions of the Peach (Prunus persica) TIFY gene family. Front. Plant Sci. 2022, 13, 792802. [Google Scholar] [CrossRef]
- Cui, Y.; Mao, R.; Chen, J.; Guo, Z. Regulation mechanism of MYC family transcription factors in jasmonic acid signalling pathway on taxol biosynthesis. Int. J. Mol. Sci. 2019, 20, 1843. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef]
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d’etre for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


| Gene Name | Protein (aa) | TIFY Motif TIF[F/Y]XG | Jas Motif SLX2FX2KRX2RX5PY | MW (kDa) | pI | Subcellular |
|---|---|---|---|---|---|---|
| CwJAZ1 | 176 | TIFYNG | SLX2FX2KRX2RX5GP | 19.40 | 9.76 | Chloroplast |
| CwJAZ2 | 253 | TIFYGG | canonical | 27.61 | 8.79 | Nucleus |
| CwJAZ3 | 200 | TIFYNG | canonical | 21.57 | 9.73 | Nucleus |
| CwJAZ4 | 230 | TIFYAG | canonical | 25.36 | 9.60 | Nucleus |
| CwJAZ5 | 224 | TIFYAG | canonical | 25.15 | 9.47 | Nucleus |
| CwJAZ6 | 235 | TIFYNG | canonical | 25.63 | 9.34 | Chloroplast |
| CwJAZ7 | 227 | TIVYGG | canonical | 24.99 | 8.51 | Chloroplast |
| CwJAZ8 | 133 | TIFYNG | canonical | 14.99 | 5.89 | Nucleus |
| CwJAZ9 | 390 | TIFYDG | canonical | 42.07 | 8.63 | Nucleus |
| CwJAZ10 | 164 | TILYKG | canonical | 18.73 | 9.82 | Nucleus |
| CwJAZ11 | 250 | IMFYGG | canonical | 26.40 | 9.49 | Plasma membrane |
| CwJAZ12 | 162 | TIFYGG | canonical | 17.28 | 9.66 | Nucleus |
| CwJAZ13 | 228 | TIFYDG | SLX2FX2KRX2RX5AY | 24.80 | 9.24 | Chloroplast |
| CwJAZ14 | 262 | TIFYGG | canonical | 28.10 | 8.70 | Chloroplast |
| CwJAZ15 | 238 | TIFYGG | canonical | 25.75 | 7.81 | Chloroplast |
| CwJAZ16 | 325 | TVFYGG | canonical | 35.70 | 9.63 | Nucleus |
| CwJAZ17 | 326 | TIFYGG | canonical | 35.41 | 10.28 | Nucleus |
| CwJAZ18 | 459 | TIFYAG | SIX2FX2NRX2RX5PY | 49.12 | 9.19 | Nucleus |
| CwJAZ19 | 397 | TIFYAG | canonical | 43.28 | 8.88 | Nucleus |
| CwJAZ20 | 114 | TIFYKG | SLX2FX2KRX2RX7PY | 12.97 | 9.55 | Mitochondria |
| pGADT7-CwJAZ (Prey) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pGBKT7-CwJAZ (Bait) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 10 | 13 | 14 | 15 | 16 | 17 | 20 | AD | |
| 1 | - | ++ | + | ++ | - | ++ | ++ | ++ | ++ | ++ | ++ | - | - | + | - | |
| 2 | - | ++ | - | - | - | - | - | - | + | - | - | - | - | - | - | |
| 3 | - | + | + | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | - | - | + | - | |
| 4 | - | - | ++ | - | + | ++ | ++ | ++ | ++ | ++ | + | - | - | + | - | |
| 5 | - | - | - | - | - | + | - | - | - | - | - | - | - | - | - | |
| 6 | - | - | - | ++ | - | - | - | ++ | - | - | - | - | - | - | - | |
| 7 | - | ++ | + | ++ | + | ++ | ++ | ++ | ++ | ++ | ++ | - | - | + | - | |
| 10 | - | - | + | + | - | ++ | ++ | ++ | ++ | ++ | ++ | - | - | - | - | |
| 13 | - | - | ++ | ++ | - | ++ | + | + | - | - | - | - | - | ++ | - | |
| 14 | - | - | + | + | + | ++ | + | + | + | ++ | ++ | - | - | + | - | |
| 15 | - | + | ++ | ++ | + | ++ | ++ | + | ++ | ++ | + | - | - | ++ | - | |
| 16 | - | + | ++ | - | - | - | - | + | + | + | + | - | + | - | - | |
| 17 | - | + | + | - | + | + | ++ | - | - | ++ | ++ | + | + | + | - | |
| 20 | - | - | ++ | - | + | + | ++ | + | ++ | ++ | ++ | - | - | - | - | |
| BD | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wu, S.; Lan, K.; Wang, Q.; Ye, T.; Jin, H.; Hu, T.; Xie, T.; Wei, Q.; Yin, X. An Investigation of the JAZ Family and the CwMYC2-like Protein to Reveal Their Regulation Roles in the MeJA-Induced Biosynthesis of β-Elemene in Curcuma wenyujin. Int. J. Mol. Sci. 2023, 24, 15004. https://doi.org/10.3390/ijms241915004
Liu Y, Wu S, Lan K, Wang Q, Ye T, Jin H, Hu T, Xie T, Wei Q, Yin X. An Investigation of the JAZ Family and the CwMYC2-like Protein to Reveal Their Regulation Roles in the MeJA-Induced Biosynthesis of β-Elemene in Curcuma wenyujin. International Journal of Molecular Sciences. 2023; 24(19):15004. https://doi.org/10.3390/ijms241915004
Chicago/Turabian StyleLiu, Yuyang, Shiyi Wu, Kaer Lan, Qian Wang, Tingyu Ye, Huanan Jin, Tianyuan Hu, Tian Xie, Qiuhui Wei, and Xiaopu Yin. 2023. "An Investigation of the JAZ Family and the CwMYC2-like Protein to Reveal Their Regulation Roles in the MeJA-Induced Biosynthesis of β-Elemene in Curcuma wenyujin" International Journal of Molecular Sciences 24, no. 19: 15004. https://doi.org/10.3390/ijms241915004






