A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize
Abstract
1. Introduction
2. Results
2.1. Bibliometric Analysis of Kernel Size-Related Traits in Maize
2.2. Characterization of Cloned Genes Controlling Maize Kernel Size-Related Traits
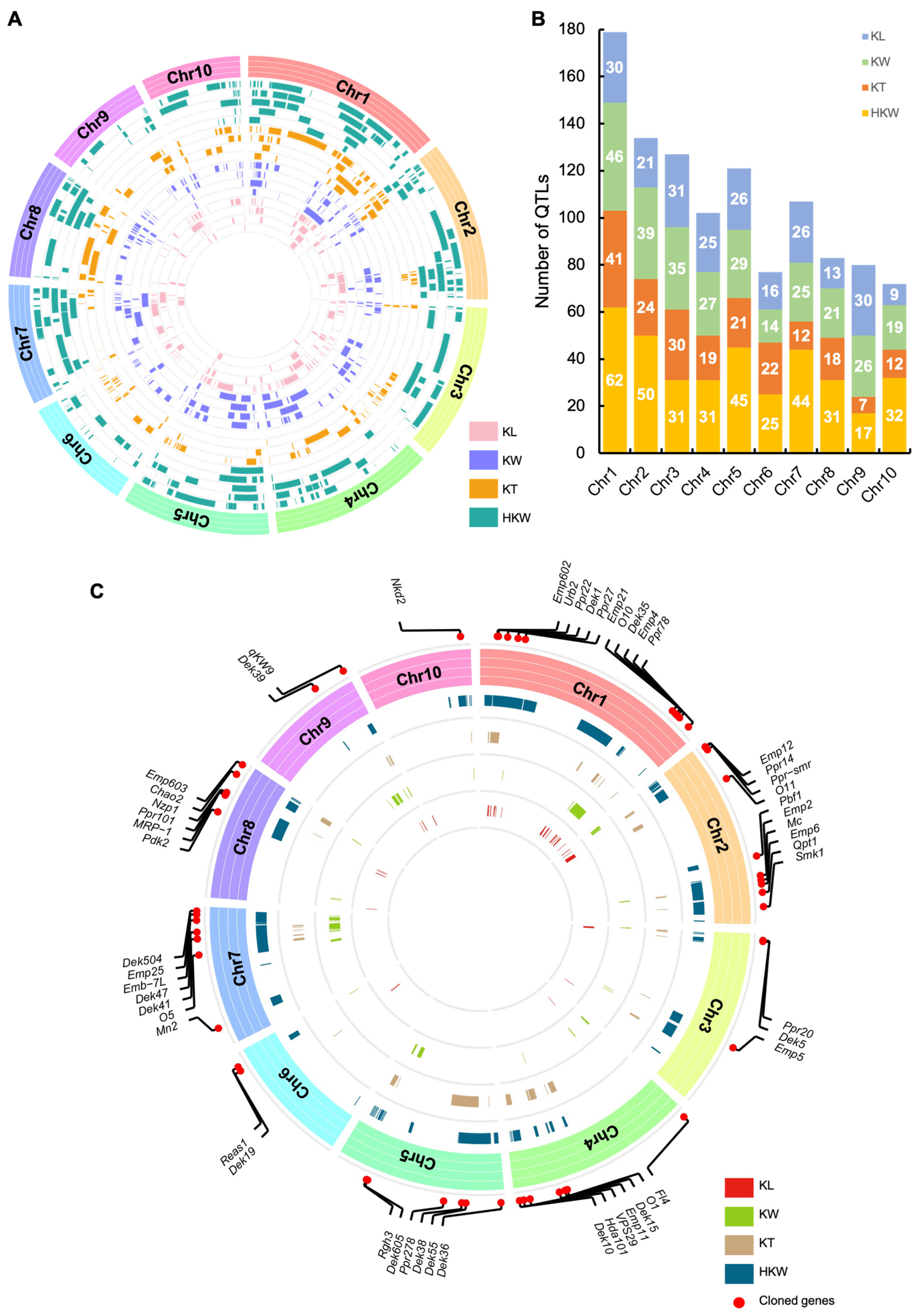
2.3. Characterization of QTL Clusters for Kernel Size-Related Traits in Maize
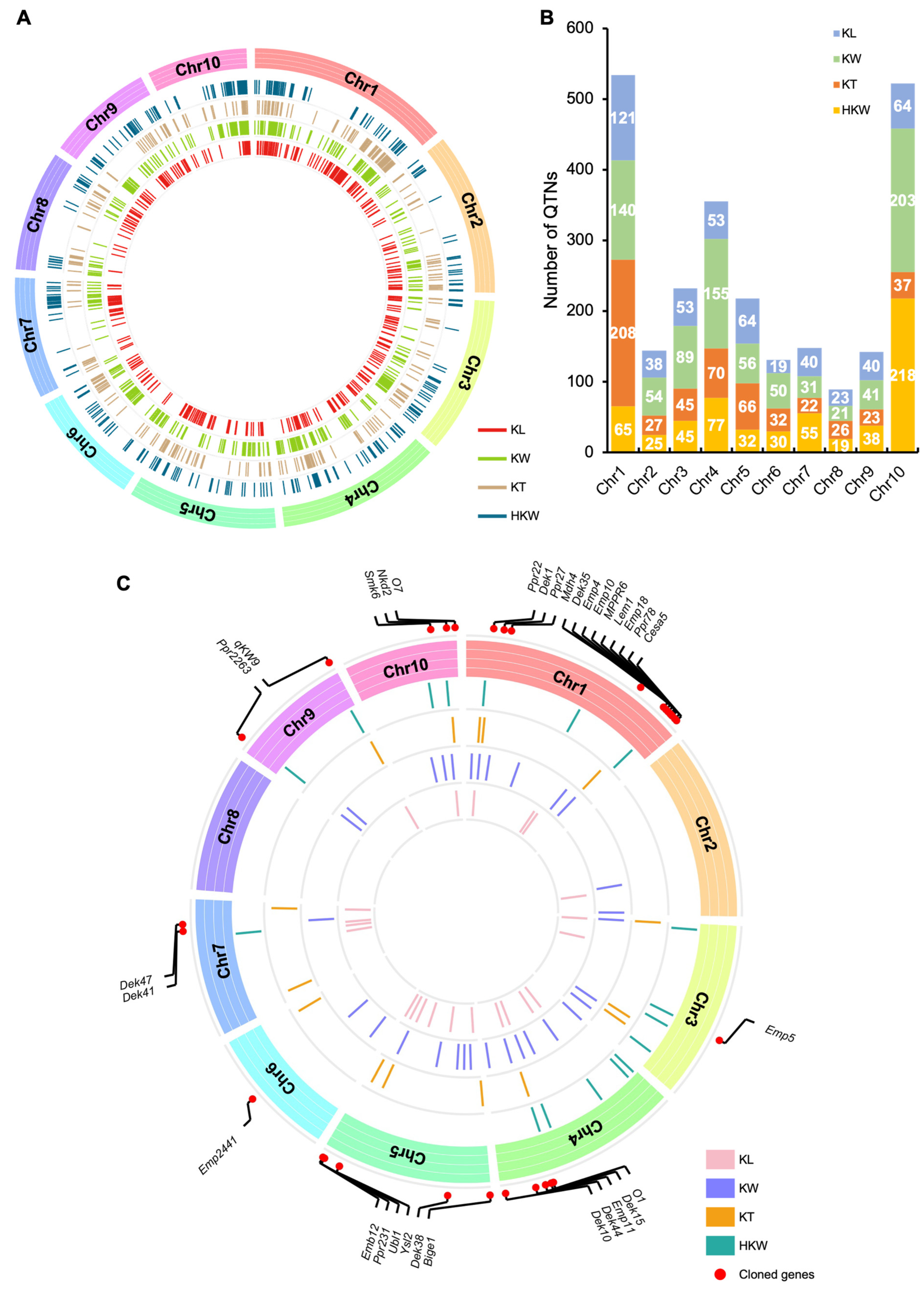
2.4. Characterization of QTN Clusters for Kernel Size-Related Traits in Maize
2.5. Integrating QTL and QTN Clusters Related to Kernel Size-Related Traits in Maize
2.6. Identification of Candidate Genes Controlling Kernel Development in Maize
3. Discussion
4. Materials and Methods
4.1. Bibliometric Analysis
4.2. Gene, QTL and QTN Data Collection
4.3. Projection of QTL, QTNs, and Genes on Reference Genome
4.4. Identification of QTL and QTN Clusters
4.5. Integration of QTL and QTN Hotspots
4.6. GO Enrichment Analysis
4.7. In Silico Gene Expression Analysis
5. Conclusions and Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Russell, S.D. Double Fertilization. In International Review of Cytology; Russell, S.D., Dumas, C., Eds.; Academic Press: San Diego, CA, USA, 1992; Volume 140, pp. 357–388. [Google Scholar]
- Elhiti, M.; Stasolla, C. Plant Embryogenesis, Genetics of. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Maloy, S., Hughes, K., Eds.; Academic Press: San Diego, CA, USA, 2013; pp. 343–345. [Google Scholar]
- Goldberg, R.B.; de Paiva, G.; Yadegari, R. Plant embryogenesis: Zygote to seed. Science 1994, 266, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Van Lammeren, A.A.M. Developmental Morphology and Cytology of the Young Maize Embryo (Zea Mays L.). Acta Bot. Neerl. 1986, 35, 169–188. [Google Scholar] [CrossRef]
- Lopes, M.A.; Larkins, B.A. Endosperm origin, development, and function. Plant Cell 1993, 5, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Ma, Z.; Song, R. Maize endosperm development. J. Integr. Plant Biol. 2021, 63, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Leroux, B.M.; Goodyke, A.J.; Schumacher, K.I.; Abbott, C.P.; Clore, A.M.; Yadegari, R.; Larkins, B.A.; Dannenhoffer, J.M. Maize early endosperm growth and development: From fertilization through cell type differentiation. Am. J. Bot. 2014, 101, 1259–1274. [Google Scholar] [CrossRef]
- Ma, C.; Li, B.; Wang, L.; Xu, M.L.; Lizhu, E.; Jin, H.; Wang, Z.; Ye, J.R. Characterization of phytohormone and transcriptome reprogramming profiles during maize early kernel development. BMC Plant Biol. 2019, 19, 197. [Google Scholar] [CrossRef]
- Neuffer, M.G.; Sheridan, W.F. Defective Kernel Mutants of Maize. I. Genetic and lethality Studies. Genetics 1980, 95, 929–944. [Google Scholar] [CrossRef]
- Sheridan, W.F.; Neuffer, M.G. Defective Kernel Mutants of Maize II. Morphological and Embryo Culture Studies. Genetics 1980, 95, 945–960. [Google Scholar] [CrossRef]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. The Miniature1 Seed Locus of Maize Encodes a Cell Wall Invertase Required for Normal Development of Endosperm and Maternal Cells in the Pedicel. Plant Cell 1996, 8, 971–983. [Google Scholar] [CrossRef]
- Heckel, T.; Werner, K.; Sheridan, W.F.; Dumas, C.; Rogowsky, P.M. Novel phenotypes and developmental arrest in early embryo specific mutants of maize. Planta 1999, 210, 1–8. [Google Scholar] [CrossRef]
- Yi, F.; Gu, W.; Li, J.; Chen, J.; Hu, L.; Cui, Y.; Zhao, H.; Guo, Y.; Lai, J.; Song, W. Miniature Seed6, encoding an endoplasmic reticulum signal peptidase, is critical in seed development. Plant Physiol. 2021, 185, 985–1001. [Google Scholar] [CrossRef]
- Dolfini, S.; Consonni, G.; Viotti, C.; Dal Prà, M.; Saltini, G.; Giulini, A.; Pilu, R.; Malgioglio, A.; Gavazzi, G. A mutational approach to the study of seed development in maize. J. Exp. Bot. 2007, 58, 1197–1205. [Google Scholar] [CrossRef]
- Schmidt, R.J.; Burr, F.A.; Burr, B. Transposon tagging and molecular analysis of the maize regulatory locus opaque-2. Science 1987, 238, 960–963. [Google Scholar] [CrossRef]
- Chourey, P.S.; Nelson, O.E. The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem. Genet. 1976, 14, 1041–1055. [Google Scholar] [CrossRef]
- Lin, T.-S.; Song, Y.; Lawrence, P.; Kheshgi, H.S.; Jain, A.K. Worldwide Maize and Soybean Yield Response to Environmental and Management Factors Over the 20th and 21st Centuries. J. Geophys. Res. Biogeosci. 2021, 126, e2021JG006304. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, L.E.I.; Jia, A.; Rong, T. Identification of QTL for maize grain yield and kernel-related traits. J. Genet. 2016, 95, 239–247. [Google Scholar] [CrossRef]
- Wan, X.Y.; Wan, J.M.; Weng, J.F.; Jiang, L.; Bi, J.C.; Wang, C.M.; Zhai, H.Q. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 2005, 110, 1334–1346. [Google Scholar] [CrossRef]
- Liu, C.; Ma, T.; Yuan, D.; Zhou, Y.; Long, Y.; Li, Z.; Dong, Z.; Duan, M.; Yu, D.; Jing, Y.; et al. The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice. Plant Biotechnol. J. 2022, 20, 1470–1486. [Google Scholar] [CrossRef]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Wan, X.; Weng, J.; Zhai, H.; Wang, J.; Lei, C.; Liu, X.; Guo, T.; Jiang, L.; Su, N.; Wan, J. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics 2008, 179, 2239–2252. [Google Scholar] [CrossRef]
- Wan, X.Y.; Wan, J.M.; Jiang, L.; Wang, J.K.; Zhai, H.Q.; Weng, J.F.; Wang, H.L.; Lei, C.L.; Wang, J.L.; Zhang, X.; et al. QTL analysis for rice grain length and fine mapping of an identified QTL with stable and major effects. Theor. Appl. Genet. 2006, 112, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Beral, A.; Rincent, R.; Le Gouis, J.; Girousse, C.; Allard, V. Wheat individual grain-size variance originates from crop development and from specific genetic determinism. PLoS ONE 2020, 15, e0230689. [Google Scholar] [CrossRef] [PubMed]
- Mangini, G.; Blanco, A.; Nigro, D.; Signorile, M.A.; Simeone, R. Candidate Genes and Quantitative Trait Loci for Grain Yield and Seed Size in Durum Wheat. Plants 2021, 10, 312. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Nguyen, A.T.; Yoshioka, M.; Iehisa, J.C.; Takumi, S. Identification of quantitative trait loci controlling grain size and shape in the D genome of synthetic hexaploid wheat lines. Breed Sci. 2013, 63, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Z.; Hu, Y.; Li, W.; Fu, Z.; Ding, D.; Li, H.; Qiao, M.; Tang, J. QTL analysis of Kernel-related traits in maize using an immortalized F2 population. PLoS ONE 2014, 9, e89645. [Google Scholar] [CrossRef]
- Pang, J.; Fu, J.; Zong, N.; Wang, J.; Song, D.; Zhang, X.; He, C.; Fang, T.; Zhang, H.; Fan, Y.; et al. Kernel size-related genes revealed by an integrated eQTL analysis during early maize kernel development. Plant J. 2019, 98, 19–32. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, L.; Liu, S.; Li, Z.; Huang, R.; Li, Y.; Cheng, H.; Li, X.; Zhou, B.; Wu, S.; et al. The Genetic Basis of Natural Variation in Kernel Size and Related Traits Using a Four-Way Cross Population in Maize. PLoS ONE 2016, 11, e0153428. [Google Scholar] [CrossRef]
- Gupta, P.K.; Rustgi, S.; Kumar, N. Genetic and molecular basis of grain size and grain number and its relevance to grain productivity in higher plants. Genome 2006, 49, 565–571. [Google Scholar] [CrossRef]
- Revilla, P.; Butrón, A.; Malvar, R.A.; Ordás, R.A. Relationship among Kernel Weight, Early Vigor, and Growth in Maize. Crop Sci. 1999, 39, 654–658. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, G.; Tan, S.; Li, D.; Weiß, T.M.; Wang, X.; Chen, S.; Würschum, T.; Liu, W. A QTL atlas for grain yield and its component traits in maize (Zea mays). Plant Breed. 2020, 139, 562–574. [Google Scholar] [CrossRef]
- Dai, D.; Ma, Z.; Song, R. Maize kernel development. Mol. Breed. 2021, 41, 2. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Wang, X.; Deng, D. Integrated Meta-QTL and Genome-Wide Association Study Analyses Reveal Candidate Genes for Maize Yield. J. Plant Growth Regul. 2020, 39, 229–238. [Google Scholar] [CrossRef]
- Kong, M.; Qiao, Q.; Ma, X.; Tao, Y.; Maharjan Raj, P.; Zhen, W. Isolation and functional analysis of the zmARM4 locus in a novel maize (Zea mays) grain-filling mutant. Plant Breed. 2020, 139, 217–226. [Google Scholar] [CrossRef]
- Takacs, E.M.; Suzuki, M.; Scanlon, M.J. Discolored1 (DSC1) is an ADP-Ribosylation Factor-GTPase Activating Protein Required to Maintain Differentiation of Maize Kernel Structures. Front. Plant Sci. 2012, 3, 115. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, W.; Wang, H.; Du, Q.; Fu, Z.; Li, W.X.; Tang, J. ZmEHD1 Is Required for Kernel Development and Vegetative Growth through Regulating Auxin Homeostasis. Plant Physiol. 2020, 182, 1467–1480. [Google Scholar] [CrossRef]
- Suzuki, M.; Wu, S.; Mimura, M.; Alseekh, S.; Fernie, A.R.; Hanson, A.D.; McCarty, D.R. Construction and applications of a B vitamin genetic resource for investigation of vitamin-dependent metabolism in maize. Plant J. 2020, 101, 442–454. [Google Scholar] [CrossRef]
- Bai, F.; Corll, J.; Shodja, D.N.; Davenport, R.; Feng, G.; Mudunkothge, J.; Brigolin, C.J.; Martin, F.; Spielbauer, G.; Tseung, C.W.; et al. RNA Binding Motif Protein 48 Is Required for U12 Splicing and Maize Endosperm Differentiation. Plant Cell 2019, 31, 715–733. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, F.; Liu, B.; Xu, C.; He, Q.; Cheng, W.; Zhao, X.; Ding, Z.; Zhang, W.; Zhang, K.; et al. ZmSKS13, a cupredoxin domain-containing protein, is required for maize kernel development via modulation of redox homeostasis. New Phytol. 2021, 229, 2163–2178. [Google Scholar] [CrossRef]
- Dai, D.; Jin, L.; Huo, Z.; Yan, S.; Ma, Z.; Qi, W.; Song, R. Maize pentatricopeptide repeat protein DEK53 is required for mitochondrial RNA editing at multiple sites and seed development. J. Exp. Bot. 2020, 71, 6246–6261. [Google Scholar] [CrossRef]
- Feng, Y.; Ma, Y.; Feng, F.; Chen, X.; Qi, W.; Ma, Z.; Song, R. Accumulation of 22 kDa α-zein-mediated nonzein protein in protein body of maize endosperm. New Phytol. 2022, 233, 265–281. [Google Scholar] [CrossRef]
- Qi, W.; Zhu, J.; Wu, Q.; Wang, Q.; Li, X.; Yao, D.; Jin, Y.; Wang, G.; Wang, G.; Song, R. Maize reas1 Mutant Stimulates Ribosome Use Efficiency and Triggers Distinct Transcriptional and Translational Responses. Plant Physiol. 2016, 170, 971–988. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.K.; Liu, R.; Sayyed, A.; Sun, F.; Song, R.; Wang, X.; Xiu, Z.; Li, X.; Tan, B.C. Regulator of Chromosome Condensation 1-Domain Protein DEK47 Functions on the Intron Splicing of Mitochondrial Nad2 and Seed Development in Maize. Front. Plant Sci. 2021, 12, 695249. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Cao, S.K.; Yang, H.; Liu, R.; Sun, F.; Wang, L.; Wang, M.; Tan, B.C. DEK48 Is Required for RNA Editing at Multiple Mitochondrial Sites and Seed Development in Maize. Int. J. Mol. Sci. 2022, 23, 3064. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, W.; Zhang, S.; Lu, J.; Chen, R.; Fu, J.; Gu, R.; Wang, G.; Wang, J.; Cui, Y. Dek504 Encodes a Mitochondrion-Targeted E+-Type Pentatricopeptide Repeat Protein Essential for RNA Editing and Seed Development in Maize. Int. J. Mol. Sci. 2022, 23, 2513. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.C.; Yan, X.W.; Zhao, Y.J.; Wei, Y.M.; Lu, X.; Zang, J.; Wu, J.W.; Zheng, G.M.; Ding, X.H.; Zhang, X.S.; et al. The novel E-subgroup pentatricopeptide repeat protein DEK55 is responsible for RNA editing at multiple sites and for the splicing of nad1 and nad4 in maize. BMC Plant Biol. 2020, 20, 553. [Google Scholar] [CrossRef] [PubMed]
- Lid, S.E.; Gruis, D.; Jung, R.; Lorentzen, J.A.; Ananiev, E.; Chamberlin, M.; Niu, X.; Meeley, R.; Nichols, S.; Olsen, O.A. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 2002, 99, 5460–5465. [Google Scholar] [CrossRef]
- Qi, W.; Tian, Z.; Lu, L.; Chen, X.; Chen, X.; Zhang, W.; Song, R. Editing of Mitochondrial Transcripts nad3 and cox2 by Dek10 Is Essential for Mitochondrial Function and Maize Plant Development. Genetics 2017, 205, 1489–1501. [Google Scholar] [CrossRef]
- He, Y.; Wang, J.; Qi, W.; Song, R. Maize Dek15 Encodes the Cohesin-Loading Complex Subunit SCC4 and Is Essential for Chromosome Segregation and Kernel Development. Plant Cell 2019, 31, 465–485. [Google Scholar] [CrossRef]
- Dong, J.; Tu, M.; Feng, Y.; Zdepski, A.; Ge, F.; Kumar, D.; Slovin, J.P.; Messing, J. Candidate gene identification of existing or induced mutations with pipelines applicable to large genomes. Plant J. 2019, 97, 673–682. [Google Scholar] [CrossRef]
- Qi, W.; Yang, Y.; Feng, X.; Zhang, M.; Song, R. Mitochondrial Function and Maize Kernel Development Requires Dek2, a Pentatricopeptide Repeat Protein Involved in nad1 mRNA Splicing. Genetics 2017, 205, 239–249. [Google Scholar] [CrossRef]
- Dai, D.; Tong, H.; Cheng, L.; Peng, F.; Zhang, T.; Qi, W.; Song, R. Maize Dek33 encodes a pyrimidine reductase in riboflavin biosynthesis that is essential for oil-body formation and ABA biosynthesis during seed development. J. Exp. Bot. 2019, 70, 5173–5187. [Google Scholar] [CrossRef]
- Chen, X.; Feng, F.; Qi, W.; Xu, L.; Yao, D.; Wang, Q.; Song, R. Dek35 Encodes a PPR Protein that Affects cis-Splicing of Mitochondrial nad4 Intron 1 and Seed Development in Maize. Mol. Plant 2017, 10, 427–441. [Google Scholar] [CrossRef]
- Wang, G.; Zhong, M.; Shuai, B.; Song, J.; Zhang, J.; Han, L.; Ling, H.; Tang, Y.; Wang, G.; Song, R. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 2017, 214, 1563–1578. [Google Scholar] [CrossRef]
- Dai, D.; Luan, S.; Chen, X.; Wang, Q.; Feng, Y.; Zhu, C.; Qi, W.; Song, R. Maize Dek37 Encodes a P-type PPR Protein That Affects cis-Splicing of Mitochondrial nad2 Intron 1 and Seed Development. Genetics 2018, 208, 1069–1082. [Google Scholar] [CrossRef]
- Garcia, N.; Li, Y.; Dooner, H.K.; Messing, J. Maize defective kernel mutant generated by insertion of a Ds element in a gene encoding a highly conserved TTI2 cochaperone. Proc. Natl. Acad. Sci. USA 2017, 114, 5165–5170. [Google Scholar] [CrossRef]
- Li, X.; Gu, W.; Sun, S.; Chen, Z.; Chen, J.; Song, W.; Zhao, H.; Lai, J. Defective Kernel 39 encodes a PPR protein required for seed development in maize. J. Integr. Plant Biol. 2018, 60, 45–64. [Google Scholar] [CrossRef]
- Wang, G.; Fan, W.; Ou, M.; Wang, X.; Qin, H.; Feng, F.; Du, Y.; Ni, J.; Tang, J.; Song, R.; et al. Dek40 Encodes a PBAC4 Protein Required for 20S Proteasome Biogenesis and Seed Development. Plant Physiol. 2019, 180, 2120–2132. [Google Scholar] [CrossRef]
- Zhu, C.; Jin, G.; Fang, P.; Zhang, Y.; Feng, X.; Tang, Y.; Qi, W.; Song, R. Maize pentatricopeptide repeat protein DEK41 affects cis-splicing of mitochondrial nad4 intron 3 and is required for normal seed development. J. Exp. Bot. 2019, 70, 3795–3808. [Google Scholar] [CrossRef]
- Zuo, Y.; Feng, F.; Qi, W.; Song, R. Dek42 encodes an RNA-binding protein that affects alternative pre-mRNA splicing and maize kernel development. J. Integr. Plant Biol. 2019, 61, 728–748. [Google Scholar] [CrossRef]
- Qi, W.; Lu, L.; Huang, S.; Song, R. Maize Dek44 Encodes Mitochondrial Ribosomal Protein L9 and Is Required for Seed Development. Plant Physiol. 2019, 180, 2106–2119. [Google Scholar] [CrossRef]
- Ren, R.C.; Lu, X.; Zhao, Y.J.; Wei, Y.M.; Wang, L.L.; Zhang, L.; Zhang, W.T.; Zhang, C.; Zhang, X.S.; Zhao, X.Y. Pentatricopeptide repeat protein DEK40 is required for mitochondrial function and kernel development in maize. J. Exp. Bot. 2019, 70, 6163–6179. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Song, S.; Yang, Y.Z.; Lu, F.; Zhang, M.D.; Sun, F.; Jia, R.; Song, R.; Tan, B.C. DEK46 performs C-to-U editing of a specific site in mitochondrial nad7 introns that is critical for intron splicing and seed development in maize. Plant J. 2020, 103, 1767–1782. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, S.; Boehlein, S.K.; McCarty, D.R.; Song, G.; Walley, J.W.; Myers, A.; Settles, A.M. Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. J. Cell Biol. 2019, 218, 2638–2658. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Peng, Y.; Ren, Z.; Li, D.; Zhen, S.; Hey, S.; Cui, Y.; Fu, J.; Gu, R.; Wang, J.; et al. Maize Defective Kernel605 Encodes a Canonical DYW-Type PPR Protein that Edits a Conserved Site of nad1 and Is Essential for Seed Nutritional Quality. Plant Cell Physiol. 2020, 61, 1954–1966. [Google Scholar] [CrossRef] [PubMed]
- Manavski, N.; Guyon, V.; Meurer, J.; Wienand, U.; Brettschneider, R. An essential pentatricopeptide repeat protein facilitates 5’ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell 2012, 24, 3087–3105. [Google Scholar] [CrossRef]
- Gontarek, B.C.; Neelakandan, A.K.; Wu, H.; Becraft, P.W. NKD Transcription Factors Are Central Regulators of Maize Endosperm Development. Plant Cell 2016, 28, 2916–2936. [Google Scholar] [CrossRef]
- Mimura, M.; Kudo, T.; Wu, S.; McCarty, D.R.; Suzuki, M. Autonomous and non-autonomous functions of the maize Shohai1 gene, encoding a RWP-RK putative transcription factor, in regulation of embryo and endosperm development. Plant J. 2018, 95, 892–908. [Google Scholar] [CrossRef]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20. [Google Scholar] [CrossRef]
- Suzuki, M.; Sato, Y.; Wu, S.; Kang, B.-H.; McCarty, D.R. Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. Plant Cell 2015, 27, 2288–2300. [Google Scholar] [CrossRef]
- Shen, Y.; Li, C.; McCarty, D.R.; Meeley, R.; Tan, B.C. Embryo defective12 encodes the plastid initiation factor 3 and is essential for embryogenesis in maize. Plant J. 2013, 74, 792–804. [Google Scholar] [CrossRef]
- Li, C.; Shen, Y.; Meeley, R.; McCarty, D.R.; Tan, B.C. Embryo defective 14 encodes a plastid-targeted cGTPase essential for embryogenesis in maize. Plant J. 2015, 84, 785–799. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Hou, M.M.; Tan, B.C. The requirement of WHIRLY1 for embryogenesis is dependent on genetic background in maize. PLoS ONE 2013, 8, e67369. [Google Scholar] [CrossRef]
- Yuan, N.; Wang, J.; Zhou, Y.; An, D.; Xiao, Q.; Wang, W.; Wu, Y. EMB-7L is required for embryogenesis and plant development in maize involved in RNA splicing of multiple chloroplast genes. Plant Sci. 2019, 287, 110203. [Google Scholar] [CrossRef]
- Ma, Z.; Dooner, H.K. A mutation in the nuclear-encoded plastid ribosomal protein S9 leads to early embryo lethality in maize. Plant J. 2004, 37, 92–103. [Google Scholar] [CrossRef]
- Sosso, D.; Canut, M.; Gendrot, G.; Dedieu, A.; Chambrier, P.; Barkan, A.; Consonni, G.; Rogowsky, P.M. PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. J. Exp. Bot. 2012, 63, 5843–5857. [Google Scholar] [CrossRef]
- Magnard, J.L.; Heckel, T.; Massonneau, A.; Wisniewski, J.P.; Cordelier, S.; Lassagne, H.; Perez, P.; Dumas, C.; Rogowsky, P.M. Morphogenesis of maize embryos requires ZmPRPL35-1 encoding a plastid ribosomal protein. Plant Physiol. 2004, 134, 649–663. [Google Scholar] [CrossRef]
- Chen, W.; Cui, Y.; Wang, Z.; Chen, R.; He, C.; Liu, Y.; Du, X.; Liu, Y.; Fu, J.; Wang, G.; et al. Nuclear-Encoded Maturase Protein 3 Is Required for the Splicing of Various Group II Introns in Mitochondria during Maize (Zea mays L.) Seed Development. Plant Cell Physiol. 2021, 62, 293–305. [Google Scholar] [CrossRef]
- Wang, H.C.; Chen, Z.; Yang, Y.Z.; Sun, F.; Ding, S.; Li, X.L.; Xu, C.; Tan, B.C. PPR14 Interacts With PPR-SMR1 and CRM Protein Zm-mCSF1 to Facilitate Mitochondrial Intron Splicing in Maize. Front. Plant Sci. 2020, 11, 814. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.Y.; Huang, Z.Q.; Li, Y.Y.; Yang, Y.Z.; Sayyed, A.; Sun, F.; Gu, Z.Q.; Wang, X.; Tan, B.C. PPR-DYW Protein EMP17 Is Required for Mitochondrial RNA Editing, Complex III Biogenesis, and Seed Development in Maize. Front. Plant Sci. 2021, 12, 693272. [Google Scholar] [CrossRef]
- Liu, R.; Cao, S.K.; Sayyed, A.; Yang, H.H.; Zhao, J.; Wang, X.; Jia, R.X.; Sun, F.; Tan, B.C. The DYW-subgroup pentatricopeptide repeat protein PPR27 interacts with ZmMORF1 to facilitate mitochondrial RNA editing and seed development in maize. J. Exp. Bot. 2020, 71, 5495–5505. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.C.; Shen, J.; Sun, F.; Wang, M.; Xu, C.; Tan, B.C. PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J. Exp. Bot. 2019, 70, 5245–5258. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Z.; Peng, L.; Wang, Y.; Yang, H.; Sun, F.; Wang, X.; Cao, S.K.; Jiang, R.; Wang, L.; Chen, B.Y.; et al. Empty Pericarp24 and Empty Pericarp25 Are Required for the Splicing of Mitochondrial Introns, Complex I Assembly, and Seed Development in Maize. Front. Plant Sci. 2020, 11, 608550. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Ren, Z.; Zhang, X.; Liu, Y.; Fu, J.; Qi, C.; Tatar, W.; Rasmusson, A.G.; Wang, G.; Liu, Y. The pentatricopeptide repeat protein EMP603 is required for the splicing of mitochondrial Nad1 intron 2 and seed development in maize. J. Exp. Bot. 2021, 72, 6933–6948. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cao, S.K.; Li, X.L.; Liu, R.; Sun, F.; Jiang, R.C.; Xu, C.; Tan, B.C. EMP80 mediates the C-to-U editing of nad7 and atp4 and interacts with ZmDYW2 in maize mitochondria. New Phytol. 2022, 234, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Pan, Z.; Zhao, H.; Zhao, J.; Cai, M.; Li, J.; Zhang, Z.; Qiu, F. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. J. Exp. Bot. 2017, 68, 4571–4581. [Google Scholar] [CrossRef]
- Sun, F.; Xiu, Z.; Jiang, R.; Liu, Y.; Zhang, X.; Yang, Y.Z.; Li, X.; Zhang, X.; Wang, Y.; Tan, B.C. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. J. Exp. Bot. 2019, 70, 963–972. [Google Scholar] [CrossRef]
- Xiu, Z.; Sun, F.; Shen, Y.; Zhang, X.; Jiang, R.; Bonnard, G.; Zhang, J.; Tan, B.C. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. Plant J. 2016, 85, 507–519. [Google Scholar] [CrossRef]
- Li, X.L.; Huang, W.L.; Yang, H.H.; Jiang, R.C.; Sun, F.; Wang, H.C.; Zhao, J.; Xu, C.H.; Tan, B.C. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytol. 2019, 221, 896–907. [Google Scholar] [CrossRef]
- Cai, M.; Li, S.; Sun, F.; Sun, Q.; Zhao, H.; Ren, X.; Zhao, Y.; Tan, B.C.; Zhang, Z.; Qiu, F. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. Plant J. 2017, 91, 132–144. [Google Scholar] [CrossRef]
- Fu, S.; Meeley, R.; Scanlon, M.J. Empty pericarp2 encodes a negative regulator of the heat shock response and is required for maize embryogenesis. Plant Cell 2002, 14, 3119–3132. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.Y.; Yang, Y.Z.; Huang, J.; Sun, F.; Lin, J.; Gu, Z.Q.; Sayyed, A.; Xu, C.; Tan, B.C. Empty Pericarp21 encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize. PLoS Genet. 2019, 15, e1008305. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Ding, S.; Liu, X.Y.; Tang, J.J.; Wang, Y.; Sun, F.; Xu, C.; Tan, B.C. EMP32 is required for the cis-splicing of nad7 intron 2 and seed development in maize. RNA Biol. 2021, 18, 499–509. [Google Scholar] [CrossRef]
- Gutiérrez-Marcos, J.F.; Dal Prà, M.; Giulini, A.; Costa, L.M.; Gavazzi, G.; Cordelier, S.; Sellam, O.; Tatout, C.; Paul, W.; Perez, P.; et al. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. Plant Cell 2007, 19, 196–210. [Google Scholar] [CrossRef]
- Chettoor, A.M.; Yi, G.; Gomez, E.; Hueros, G.; Meeley, R.B.; Becraft, P.W. A putative plant organelle RNA recognition protein gene is essential for maize kernel development. J. Integr. Plant Biol. 2015, 57, 236–246. [Google Scholar] [CrossRef]
- Ren, Z.; Fan, K.; Fang, T.; Zhang, J.; Yang, L.; Wang, J.; Wang, G.; Liu, Y. Maize Empty Pericarp602 Encodes a P-Type PPR Protein That Is Essential for Seed Development. Plant Cell Physiol. 2019, 60, 1734–1746. [Google Scholar] [CrossRef]
- Sun, F.; Wang, X.; Bonnard, G.; Shen, Y.; Xiu, Z.; Li, X.; Gao, D.; Zhang, Z.; Tan, B.C. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 2015, 84, 283–295. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, X.; Shen, Y.; Wang, H.; Liu, R.; Wang, X.; Gao, D.; Yang, Y.Z.; Liu, Y.; Tan, B.C. The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize. Plant J. 2018, 95, 919–932. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Ding, S.; Wang, H.C.; Sun, F.; Huang, W.L.; Song, S.; Xu, C.; Tan, B.C. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytol. 2017, 214, 782–795. [Google Scholar] [CrossRef]
- Yang, H.; Xiu, Z.; Wang, L.; Cao, S.K.; Li, X.; Sun, F.; Tan, B.C. Two Pentatricopeptide Repeat Proteins Are Required for the Splicing of nad5 Introns in Maize. Front. Plant Sci. 2020, 11, 732. [Google Scholar] [CrossRef]
- Liu, R.; Cao, S.K.; Sayyed, A.; Xu, C.; Sun, F.; Wang, X.; Tan, B.C. The Mitochondrial Pentatricopeptide Repeat Protein PPR18 Is Required for the cis-Splicing of nad4 Intron 1 and Essential to Seed Development in Maize. Int. J. Mol. Sci. 2020, 21, 4047. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Ding, S.; Wang, Y.; Wang, H.C.; Liu, X.Y.; Sun, F.; Xu, C.; Liu, B.; Tan, B.C. PPR20 Is Required for the cis-Splicing of Mitochondrial nad2 Intron 3 and Seed Development in Maize. Plant Cell Physiol. 2020, 61, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Xiu, Z.H.; Meeley, R.; Tan, B.C. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 2013, 25, 868–883. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Li, C.; Min, Z.; Meeley, R.B.; Tarczynski, M.C.; Olsen, O.A. sal1 determines the number of aleurone cell layers in maize endosperm and encodes a class E vacuolar sorting protein. Proc. Natl. Acad. Sci. USA 2003, 100, 6552–6557. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.; Li, Q.B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, M.; Chen, J.; Qi, W.; Lai, J.; Ma, Z.; Song, R. ENB1 encodes a cellulose synthase 5 that directs synthesis of cell wall ingrowths in maize basal endosperm transfer cells. Plant Cell 2022, 34, 1054–1074. [Google Scholar] [CrossRef]
- Becraft, P.W.; Stinard, P.S.; McCarty, D.R. CRINKLY4: A TNFR-like receptor kinase involved in maize epidermal differentiation. Science 1996, 273, 1406–1409. [Google Scholar] [CrossRef]
- Bernardi, J.; Lanubile, A.; Li, Q.B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYuc1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012, 160, 1318–1328. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, Z.; Zhang, H.; Tian, R.; Yang, H.; Sun, C.; Wang, L.; Zhang, W.; Guo, Z.; Zhang, X.; et al. Cytosolic malate dehydrogenase 4 modulates cellular energetics and storage reserve accumulation in maize endosperm. Plant Biotechnol. J. 2020, 18, 2420–2435. [Google Scholar] [CrossRef]
- Feng, F.; Qi, W.; Lv, Y.; Yan, S.; Xu, L.; Yang, W.; Yuan, Y.; Chen, Y.; Zhao, H.; Song, R. OPAQUE11 Is a Central Hub of the Regulatory Network for Maize Endosperm Development and Nutrient Metabolism. Plant Cell 2018, 30, 375–396. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Ji, C.; Huang, Y.; Wu, Y. Maize Oxalyl-CoA Decarboxylase1 Degrades Oxalate and Affects the Seed Metabolome and Nutritional Quality. Plant Cell 2018, 30, 2447–2462. [Google Scholar] [CrossRef]
- Holding, D.R.; Otegui, M.S.; Li, B.; Meeley, R.B.; Dam, T.; Hunter, B.G.; Jung, R.; Larkins, B.A. The maize floury1 gene encodes a novel endoplasmic reticulum protein involved in zein protein body formation. Plant Cell 2007, 19, 2569–2582. [Google Scholar] [CrossRef]
- Lopes, M.A.; Coleman, C.E.; Kodrzycki, R.; Lending, C.R.; Larkins, B.A. Synthesis of an unusual alpha-zein protein is correlated with the phenotypic effects of the floury2 mutation in maize. Mol. Gen. Genet. 1994, 245, 537–547. [Google Scholar] [CrossRef]
- Li, Q.; Wang, J.; Ye, J.; Zheng, X.; Xiang, X.; Li, C.; Fu, M.; Wang, Q.; Zhang, Z.; Wu, Y. The Maize Imprinted Gene Floury3 Encodes a PLATZ Protein Required for tRNA and 5S rRNA Transcription through Interaction with RNA Polymerase III. Plant Cell 2017, 29, 2661–2675. [Google Scholar] [CrossRef]
- Wang, G.; Qi, W.; Wu, Q.; Yao, D.; Zhang, J.; Zhu, J.; Wang, G.; Wang, G.; Tang, Y.; Song, R. Identification and Characterization of Maize floury4 as a Novel Semidominant Opaque Mutant That Disrupts Protein Body Assembly. Plant Physiol. 2014, 165, 582–594. [Google Scholar] [CrossRef]
- Kim, C.S.; Gibbon, B.C.; Gillikin, J.W.; Larkins, B.A.; Boston, R.S.; Jung, R. The maize Mucronate mutation is a deletion in the 16-kDa gamma-zein gene that induces the unfolded protein response. Plant J. 2006, 48, 440–451. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Wang, G.; Wang, F.; Zhang, X.; Zhong, M.; Zhang, J.; Lin, D.; Tang, Y.; Xu, Z.; et al. Opaque1 encodes a myosin XI motor protein that is required for endoplasmic reticulum motility and protein body formation in maize endosperm. Plant Cell 2012, 24, 3447–3462. [Google Scholar] [CrossRef]
- Yao, D.; Qi, W.; Li, X.; Yang, Q.; Yan, S.; Ling, H.; Wang, G.; Wang, G.; Song, R. Maize opaque10 Encodes a Cereal-Specific Protein That Is Essential for the Proper Distribution of Zeins in Endosperm Protein Bodies. PLoS Genet. 2016, 12, e1006270. [Google Scholar] [CrossRef]
- Myers, A.M.; James, M.G.; Lin, Q.; Yi, G.; Stinard, P.S.; Hennen-Bierwagen, T.A.; Becraft, P.W. Maize opaque5 encodes monogalactosyldiacylglycerol synthase and specifically affects galactolipids necessary for amyloplast and chloroplast function. Plant Cell 2011, 23, 2331–2347. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, J.; Wang, G.; Fan, X.; Sun, X.; Qin, H.; Xu, N.; Zhong, M.; Qiao, Z.; Tang, Y.; et al. Proline responding1 Plays a Critical Role in Regulating General Protein Synthesis and the Cell Cycle in Maize. Plant Cell 2014, 26, 2582–2600. [Google Scholar] [CrossRef]
- Wang, G.; Sun, X.; Wang, G.; Wang, F.; Gao, Q.; Sun, X.; Tang, Y.; Chang, C.; Lai, J.; Zhu, L.; et al. Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 2011, 189, 1281–1295. [Google Scholar] [CrossRef]
- Vicente-Carbajosa, J.; Moose, S.P.; Parsons, R.L.; Schmidt, R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 1997, 94, 7685–7690. [Google Scholar] [CrossRef] [PubMed]
- Lappe, R.R.; Baier, J.W.; Boehlein, S.K.; Huffman, R.; Lin, Q.; Wattebled, F.; Settles, A.M.; Hannah, L.C.; Borisjuk, L.; Rolletschek, H.; et al. Functions of maize genes encoding pyruvate phosphate dikinase in developing endosperm. Proc. Natl. Acad. Sci. USA 2018, 115, E24–E33. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Kim, C.S.; Nguyen, H.N.; Meeley, R.B.; Larkins, B.A. The maize zmsmu2 gene encodes a putative RNA-splicing factor that affects protein synthesis and RNA processing during endosperm development. Plant Physiol. 2007, 144, 821–835. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.N.; Fisher, D.K.; Gao, M.; Guiltinan, M.J. Molecular cloning and characterization of the Amylose-Extender gene encoding starch branching enzyme IIB in maize. Plant Mol. Biol. 1998, 38, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Preiss, J.; Danner, S.; Summers, P.S.; Morell, M.; Barton, C.R.; Yang, L.; Nieder, M. Molecular Characterization of the Brittle-2 Gene Effect on Maize Endosperm ADPglucose Pyrophosphorylase Subunits. Plant Physiol. 1990, 92, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mogel, K.; Lor, V.S.; Hirsch, C.N.; De Vries, B.; Kaeppler, H.F.; Tracy, W.F.; Kaeppler, S.M. Maize sugary enhancer1 (se1) is a gene affecting endosperm starch metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 20776–20785. [Google Scholar] [CrossRef]
- Bhave, M.R.; Lawrence, S.; Barton, C.; Hannah, L.C. Identification and molecular characterization of shrunken-2 cDNA clones of maize. Plant Cell 1990, 2, 581–588. [Google Scholar] [CrossRef]
- James, M.G.; Robertson, D.S.; Myers, A.M. Characterization of the maize gene sugary1, a determinant of starch composition in kernels. Plant Cell 1995, 7, 417–429. [Google Scholar] [CrossRef]
- Zhang, Z.; Dong, J.; Ji, C.; Wu, Y.; Messing, J. NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. USA 2019, 116, 11223–11228. [Google Scholar] [CrossRef]
- Li, Q.; Zhou, S.; Liu, W.; Zhai, Z.; Pan, Y.; Liu, C.; Chern, M.; Wang, H.; Huang, M.; Zhang, Z. A chlorophyll a oxygenase 1 gene ZmCAO1 contributes to grain yield and waterlogging tolerance in maize. J. Exp. Bot. 2021, 72, 3155–3167. [Google Scholar] [CrossRef]
- Sun, H.; Xu, H.; Li, B.; Shang, Y.; Wei, M.; Zhang, S.; Zhao, C.; Qin, R.; Cui, F.; Wu, Y. The brassinosteroid biosynthesis gene, ZmD11, increases seed size and quality in rice and maize. Plant Physiol. Biochem. 2021, 160, 281–293. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, X.; Zhang, H.; Gu, Y.; Huang, X.; Huang, H.; Liu, H.; Zhang, J.; Hu, Y.; Li, Y.; et al. The miR164-dependent regulatory pathway in developing maize seed. Mol. Genet. Genom. 2019, 294, 501–517. [Google Scholar] [CrossRef]
- Huang, J.; Lu, G.; Liu, L.; Raihan, M.S.; Xu, J.; Jian, L.; Zhao, L.; Tran, T.M.; Zhang, Q.; Liu, J.; et al. The Kernel Size-Related Quantitative Trait Locus qKW9 Encodes a Pentatricopeptide Repeat Protein That Aaffects Photosynthesis and Grain Filling. Plant Physiol. 2020, 183, 1696–1709. [Google Scholar] [CrossRef]
- Zhang, Y.F.; Suzuki, M.; Sun, F.; Tan, B.C. The Mitochondrion-Targeted PENTATRICOPEPTIDE REPEAT78 Protein Is Required for nad5 Mature mRNA Stability and Seed Development in Maize. Mol. Plant 2017, 10, 1321–1333. [Google Scholar] [CrossRef]
- Yang, J.; Cui, Y.; Zhang, X.; Yang, Z.; Lai, J.; Song, W.; Liang, J.; Li, X. Maize PPR278 Functions in Mitochondrial RNA Splicing and Editing. Int. J. Mol. Sci. 2022, 23, 3035. [Google Scholar] [CrossRef]
- Li, X.J.; Zhang, Y.F.; Hou, M.; Sun, F.; Shen, Y.; Xiu, Z.H.; Wang, X.; Chen, Z.L.; Sun, S.S.; Small, I.; et al. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 2014, 79, 797–809. [Google Scholar] [CrossRef]
- Hu, M.; Zhao, H.; Yang, B.; Yang, S.; Liu, H.; Tian, H.; Shui, G.; Chen, Z.; E, L.; Lai, J.; et al. ZmCTLP1 is required for the maintenance of lipid homeostasis and the basal endosperm transfer layer in maize kernels. New Phytol. 2021, 232, 2384–2399. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, J.; Wang, J.; Xie, Y.; Cui, Y.; Du, X.; Li, L.; Fu, J.; Liu, Y.; Wang, J.; et al. Small kernel 501 (smk501) encodes the RUBylation activating enzyme E1 subunit ECR1 (E1 C-TERMINAL RELATED 1) and is essential for multiple aspects of cellular events during kernel development in maize. New Phytol. 2021, 230, 2337–2354. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, H.; Huang, X.; Wang, Q.; Wang, J.; An, D.; Li, J.; Wang, W.; Wu, Y. Maize VKS1 Regulates Mitosis and Cytokinesis During Early Endosperm Development. Plant Cell 2019, 31, 1238–1256. [Google Scholar] [CrossRef]
- Wei, Y.M.; Ren, Z.J.; Wang, B.H.; Zhang, L.; Zhao, Y.J.; Wu, J.W.; Li, L.G.; Zhang, X.S.; Zhao, X.Y. A nitrate transporter encoded by ZmNPF7.9 is essential for maize seed development. Plant Sci. 2021, 308, 110901. [Google Scholar] [CrossRef]
- Chourey, P.S.; Jain, M.; Li, Q.B.; Carlson, S.J. Genetic control of cell wall invertases in developing endosperm of maize. Planta 2006, 223, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Locatelli, S.; Varotto, S.; Donn, G.; Pirona, R.; Henderson, D.A.; Hartings, H.; Motto, M. Maize histone deacetylase hda101 is involved in plant development, gene transcription, and sequence-specific modulation of histone modification of genes and repeats. Plant Cell 2007, 19, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Gómez, E.; Royo, J.; Guo, Y.; Thompson, R.; Hueros, G. Establishment of cereal endosperm expression domains: Identification and properties of a maize transfer cell-specific transcription factor, ZmMRP-1. Plant Cell 2002, 14, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Mbelo, S.; Vernoud, V.; Gendrot, G.; Dedieu, A.; Chambrier, P.; Dauzat, M.; Heurtevin, L.; Guyon, V.; Takenaka, M.; et al. PPR2263, a DYW-Subgroup Pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 2012, 24, 676–691. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, Y.X.; Li, C.; Shi, Y.; Song, Y.; Zhang, D.; Wang, H.; Li, Y.; Wang, T. The retromer protein ZmVPS29 regulates maize kernel morphology likely through an auxin-dependent process(es). Plant Biotechnol. J. 2020, 18, 1004–1014. [Google Scholar] [CrossRef]
- Yang, Y.Z.; Ding, S.; Wang, Y.; Li, C.L.; Shen, Y.; Meeley, R.; McCarty, D.R.; Tan, B.C. Small kernel2 Encodes a Glutaminase in Vitamin B6 Biosynthesis Essential for Maize Seed Development. Plant Physiol. 2017, 174, 1127–1138. [Google Scholar] [CrossRef]
- Pan, Z.; Ren, X.; Zhao, H.; Liu, L.; Tan, Z.; Qiu, F. A Mitochondrial Transcription Termination Factor, ZmSmk3, Is Required for nad1 Intron4 and nad4 Intron1 Splicing and Kernel Development in Maize. G3 Genes Genomes Genet. 2019, 9, 2677–2686. [Google Scholar] [CrossRef]
- Wang, H.C.; Sayyed, A.; Liu, X.Y.; Yang, Y.Z.; Sun, F.; Wang, Y.; Wang, M.; Tan, B.C. SMALL KERNEL4 is required for mitochondrial cox1 transcript editing and seed development in maize. J. Integr. Plant Biol. 2020, 62, 777–792. [Google Scholar] [CrossRef]
- Ding, S.; Liu, X.Y.; Wang, H.C.; Wang, Y.; Tang, J.J.; Yang, Y.Z.; Tan, B.C. SMK6 mediates the C-to-U editing at multiple sites in maize mitochondria. J. Plant Physiol. 2019, 240, 152992. [Google Scholar] [CrossRef]
- Zhao, H.; Qin, Y.; Xiao, Z.; Li, Q.; Yang, N.; Pan, Z.; Gong, D.; Sun, Q.; Yang, F.; Zhang, Z.; et al. Loss of Function of an RNA Polymerase III Subunit Leads to Impaired Maize Kernel Development. Plant Physiol. 2020, 184, 359–373. [Google Scholar] [CrossRef]
- Li, J.; Fu, J.; Chen, Y.; Fan, K.; He, C.; Zhang, Z.; Li, L.; Liu, Y.; Zheng, J.; Ren, D.; et al. The U6 Biogenesis-Like 1 Plays an Important Role in Maize Kernel and Seedling Development by Affecting the 3’ End Processing of U6 snRNA. Mol. Plant 2017, 10, 470–482. [Google Scholar] [CrossRef]
- Wang, H.; Wang, K.; Du, Q.; Wang, Y.; Fu, Z.; Guo, Z.; Kang, D.; Li, W.X.; Tang, J. Maize Urb2 protein is required for kernel development and vegetative growth by affecting pre-ribosomal RNA processing. New Phytol. 2018, 218, 1233–1246. [Google Scholar] [CrossRef]
- Zang, J.; Huo, Y.; Liu, J.; Zhang, H.; Liu, J.; Chen, H. Maize YSL2 is required for iron distribution and development in kernels. J. Exp. Bot. 2020, 71, 5896–5910. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Beló, A.; Zheng, P.; Luck, S.; Shen, B.; Meyer, D.J.; Li, B.; Tingey, S.; Rafalski, A. Whole genome scan detects an allelic variant of fad2 associated with increased oleic acid levels in maize. Mol. Genet. Genom. 2008, 279, 1–10. [Google Scholar] [CrossRef]
- Hu, G.; Li, Z.; Lu, Y.; Li, C.; Gong, S.; Yan, S.; Li, G.; Wang, M.; Ren, H.; Guan, H.; et al. Genome-wide association study Identified multiple Genetic Loci on Chilling Resistance During Germination in Maize. Sci. Rep. 2017, 7, 10840. [Google Scholar] [CrossRef]
- Deng, M.; Li, D.Q.; Luo, J.Y.; Xiao, Y.J.; Liu, H.J.; Pan, Q.C.; Zhang, X.H.; Jin, M.L.; Zhao, M.C.; Yan, J.B. The genetic architecture of amino acids dissection by association and linkage analysis in maize. Plant Biotechnol. J. 2017, 15, 1250–1263. [Google Scholar] [CrossRef]
- Sitonik, C.; Suresh, L.M.; Beyene, Y.; Olsen, M.S.; Makumbi, D.; Oliver, K.; Das, B.; Bright, J.M.; Mugo, S.; Crossa, J.; et al. Genetic architecture of maize chlorotic mottle virus and maize lethal necrosis through GWAS, linkage analysis and genomic prediction in tropical maize germplasm. Theor. Appl. Genet. 2019, 132, 2381–2399. [Google Scholar] [CrossRef]
- Xue, Y.; Warburton, M.L.; Sawkins, M.; Zhang, X.; Setter, T.; Xu, Y.; Grudloyma, P.; Gethi, J.; Ribaut, J.M.; Li, W.; et al. Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor. Appl. Genet. 2013, 126, 2587–2596. [Google Scholar] [CrossRef]
- Xiao, Y.J.; Liu, H.J.; Wu, L.J.; Warburton, M.; Yan, J.B. Genome-wide Association Studies in Maize: Praise and Stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, Y.; Meng, Y.; Xiao, Y.; Zhao, J.; Xiao, B.; An, C.; Gao, Y. PPR proteins in the tea plant (Camellia sinensis) and their potential roles in the leaf color changes. Sci. Hortic. 2022, 293, 110745. [Google Scholar] [CrossRef]
- Li, J.Z.; Zhang, Z.W.; Li, Y.L.; Wang, Q.L.; Zhou, Y.G. QTL consistency and meta-analysis for grain yield components in three generations in maize. Theor. Appl. Genet. 2011, 122, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Swamy, B.P.; Vikram, P.; Dixit, S.; Ahmed, H.U.; Kumar, A. Meta-analysis of grain yield QTL identified during agricultural drought in grasses showed consensus. BMC Genom. 2011, 12, 319. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chen, L.; Li, Y.; Shi, Y.; Song, Y.; Zhang, D.; Li, Y.; Wang, T.; Yang, D.; Li, C. Meta-QTL analysis and identification of candidate genes related to root traits in maize. Euphytica 2018, 214, 223. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.; Jiang, Y.; Yan, T.; Fang, C.; Hou, Q.; Wu, S.; Xie, K.; An, X.; Wan, X. Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize. Cells 2022, 11, 439. [Google Scholar] [CrossRef]
- Wei, X.; Pu, A.; Liu, Q.; Hou, Q.; Zhang, Y.; An, X.; Long, Y.; Jiang, Y.; Dong, Z.; Wu, S.; et al. The Bibliometric Landscape of Gene Editing Innovation and Regulation in the Worldwide. Cells 2022, 11, 2682. [Google Scholar] [CrossRef]
- Jiang, Y.; An, X.; Li, Z.; Yan, T.; Zhu, T.; Xie, K.; Liu, S.; Hou, Q.; Zhao, L.; Wu, S.; et al. CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 2021, 19, 1769–1784. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, X. Breeding with dominant genic male-sterility genes to boost crop grain yield in the post-heterosis utilization era. Mol. Plant 2021, 14, 531–534. [Google Scholar] [CrossRef]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
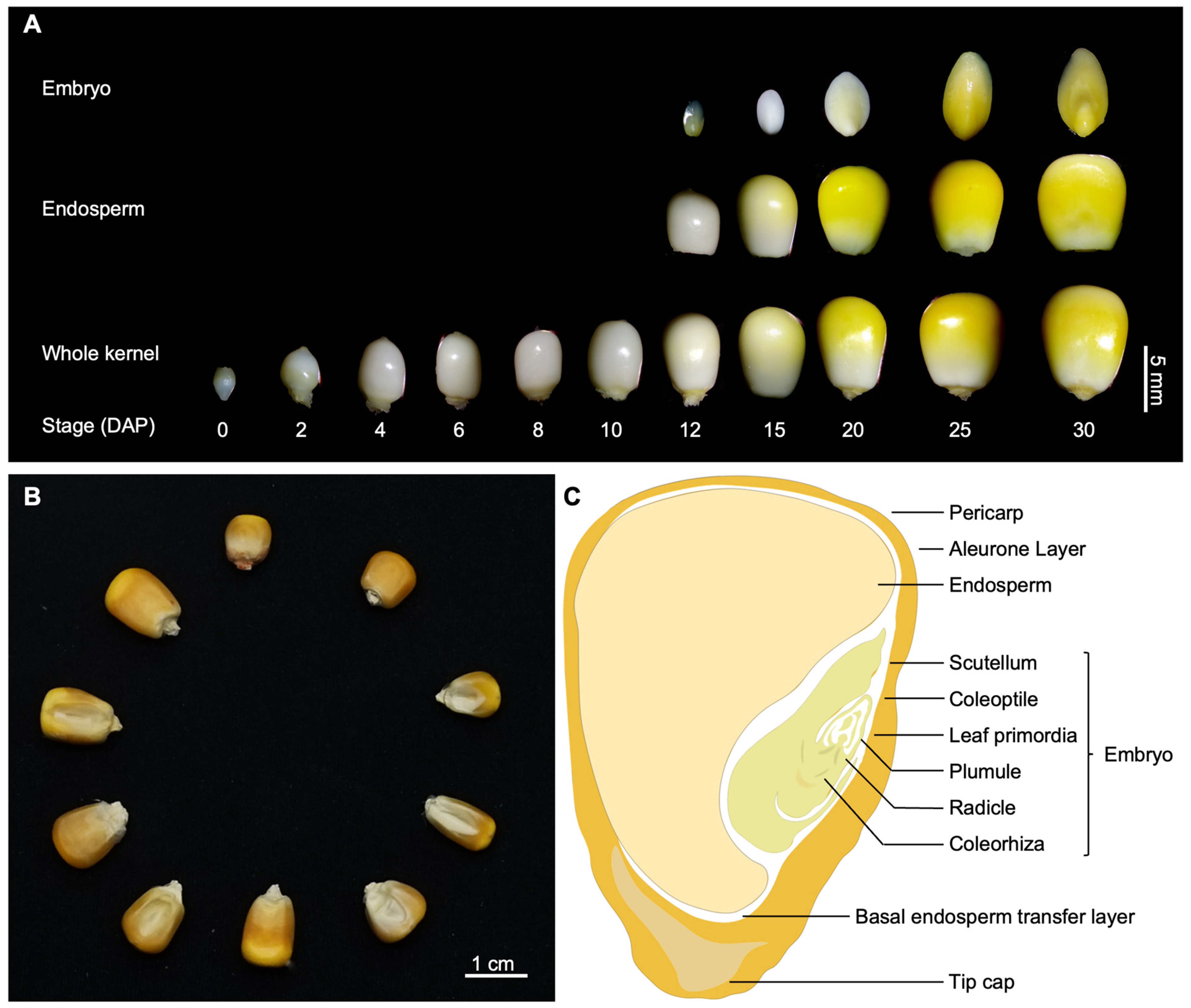
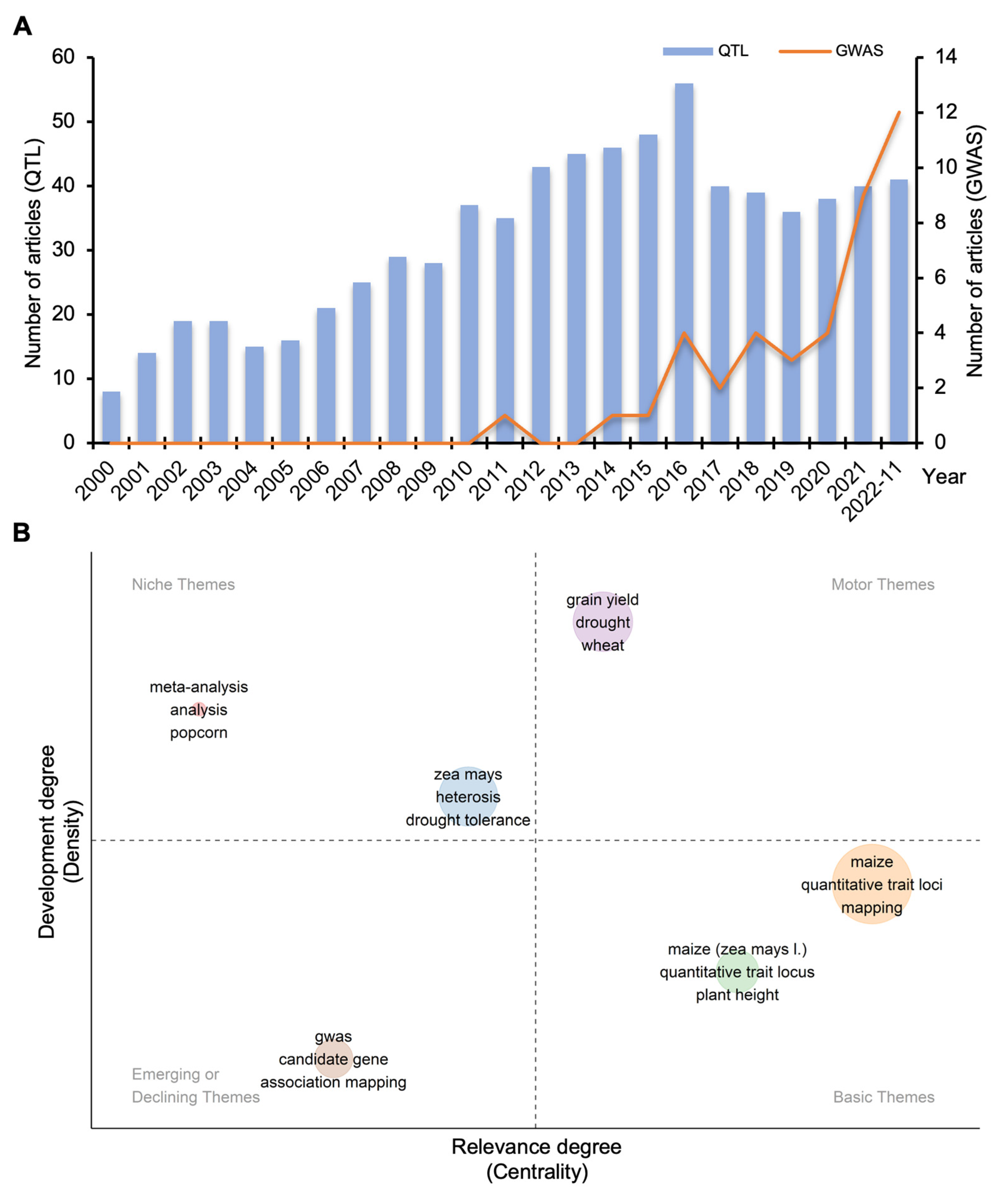
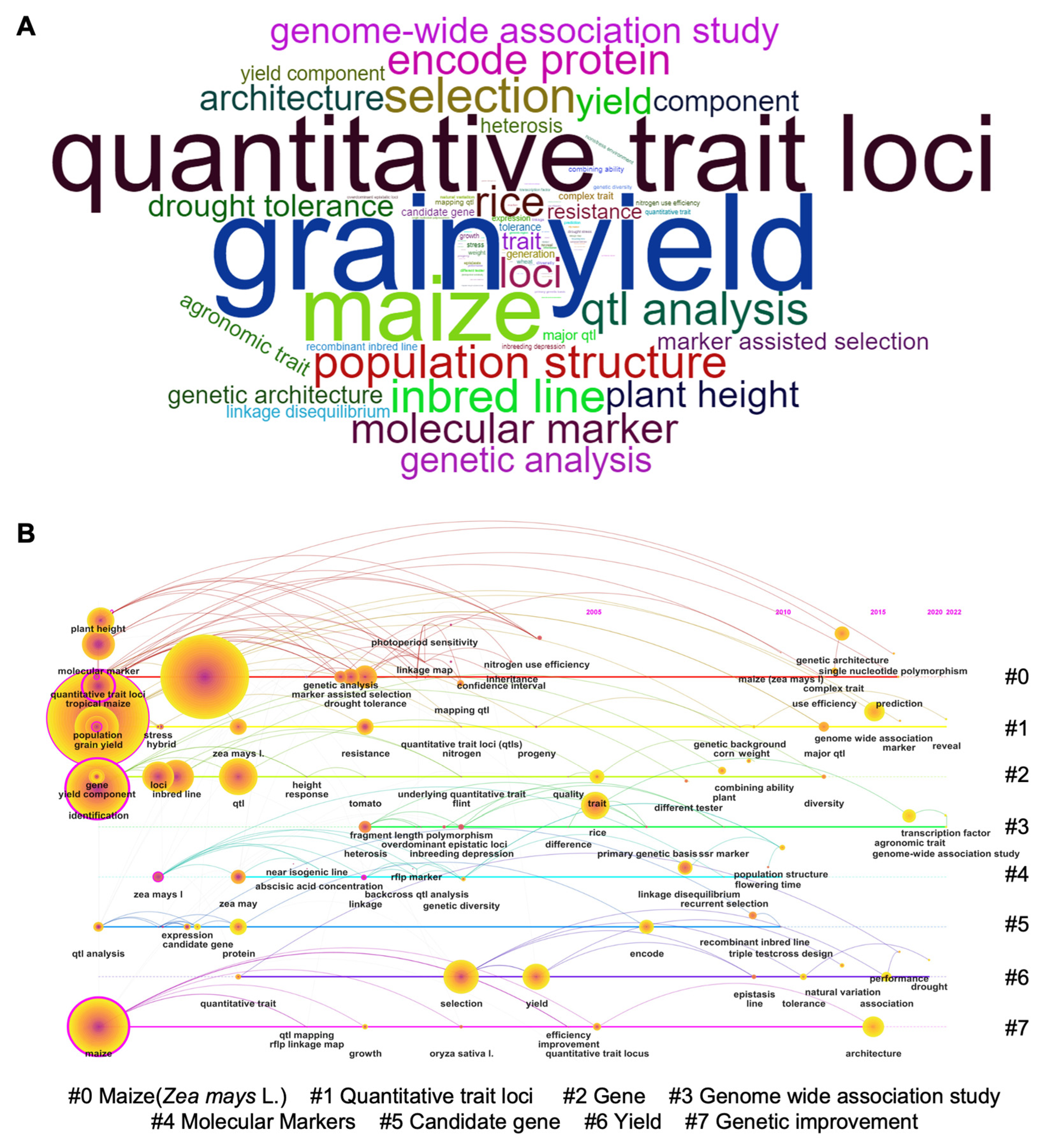
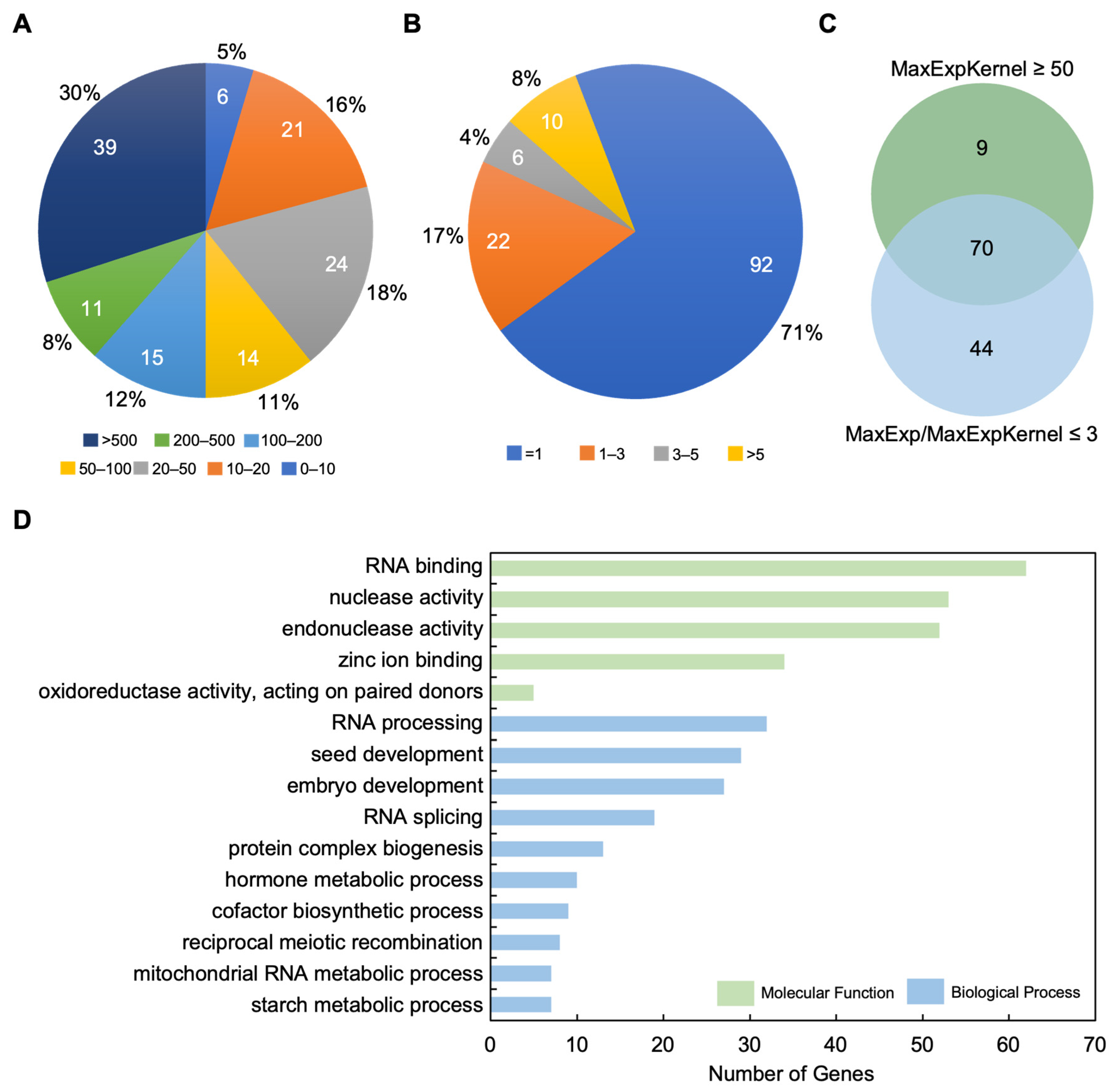

| Phenotype a | Gene Name | Gene ID (B73_V4) | Chr | Gene Annotation | Reference |
|---|---|---|---|---|---|
| Dek | Arm4 | Zm00001d053964 | 4 | ARM repeat protein, unknown pathway | [35] |
| Dsc1 | Zm00001d049871 | 4 | ARF-GTPases, vesicular material transport | [36] | |
| Ehd1 | Zm00001d053858 | 4 | EHD proteins, regulation of auxin homeostasis | [37] | |
| Qpt1 | Zm00001d006311 | 2 | Quinolinate phosphoribosyltransferase, vitamin B biosynthesis | [38] | |
| Rgh3 | Zm00001d016836 | 5 | U12 splicing factor, U12-type intron splicing | [39] | |
| Skus1 | Zm00001d043090 | 3 | Multi-copper oxidase, regulation of redox homeostasis | [40] | |
| Dek53 | Zm00001d041326 | 3 | PPR protein, RNA editing | [41] | |
| Nzp1 | Zm00001d010994 | 8 | Mitochondrial 50S ribosomal protein L10, protein body formation, mitochondrial complex assembly | [42] | |
| Reas1 | Zm00001d038475 | 6 | Ribosome export associated1, ribosome biosynthesis | [43] | |
| Dek47 | Zm00001d021372 | 7 | RCC1 domain-containing protein RUG3, RNA splicing | [44] | |
| Dek48 | Zm00001d002539 | 2 | PPR protein, RNA editing | [45] | |
| Dek504 | Zm00001d022394 | 7 | PPR protein, RNA editing | [46] | |
| Dek55 | Zm00001d014471 | 5 | PPR protein, RNA splicing and editing | [47] | |
| Dek1 | Zm00001d028818 | 1 | Membrane protein, plant signal transduction | [48] | |
| Dek10 | Zm00001d053802 | 4 | PPR protein, RNA editing | [49] | |
| Dek15 | Zm00001d052197 | 4 | Cohesin-loading complex subunit SCC4, ensuring proper chromosome segregation | [50] | |
| Dek19 | Zm00001d038257 | 6 | PPR protein, unknown pathway | [51] | |
| Dek2 | Zm00001d034882 | 1 | PPR protein, RNA splicing | [52] | |
| Dek33 | Zm00001d016475 | 5 | Pyrimidine reductase, riboflavin biosynthesis | [53] | |
| Dek35 | Zm00001d033749 | 1 | PPR protein, RNA splicing | [54] | |
| Dek36 | Zm00001d013136 | 5 | PPR protein, RNA editing | [55] | |
| Dek37 | Zm00001d003543 | 2 | PPR protein, RNA splicing | [56] | |
| Dek38 | Zm00001d014595 | 5 | Tel2-interacting protein 2, promoting early seed development through the action of PIKKs | [57] | |
| Dek39 | Zm00001d047013 | 9 | PPR protein, RNA editing | [58] | |
| Dek40 | Zm00001d011478 | 8 | PBAC4 Protein, ubiquitin-20S Proteasome Biogenesis | [59] | |
| Dek41 | Zm00001d021053 | 7 | PPR protein, RNA splicing | [60] | |
| Rbm48 | Zm00001d054077 | 4 | RNA-binding protein, pre-mRNA spliceosome formation | [61] | |
| Dek44 | Zm00001d052865 | 4 | Mitochondrial ribosomal protein L9, well-functioning in oxidative phosphorylation | [62] | |
| Dek45 | Zm00001d023331 | 10 | PPR protein, RNA editing | [63] | |
| Dek46 | Zm00001d043107 | 3 | PPR protein, RNA editing | [64] | |
| Dek5 | Zm00001d039612 | 3 | E. coli TamB homologous, chloroplast envelope biogenesis; | [65] | |
| Dek605 | Zm00001d016798 | 5 | PPR protein, RNA editing | [66] | |
| MPPR6 | Zm00001d034111 | 1 | PPR protein, facilitating translation initiation | [67] | |
| Nkd1 | Zm00001d002654 | 2 | IDD transcription factors, central regulators of gene expression in endosperm development | [68] | |
| Nkd2 | Zm00001d026113 | 10 | IDD transcription factors, central regulators of gene expression in endosperm development | [68] | |
| Shai1 | Zm00001d002661 | 2 | RWP-RK transcription factor, embryo polarity establishment, polar transport of IAA | [69] | |
| Dof3 | Zm00001d035651 | 6 | Dof-type transcription factor, starch accumulation and aleurone layer development | [70] | |
| Emb | Bige1 | Zm00001d012883 | 5 | MATE-type transporter, CYP78A pathway (transport of growth factors) | [71] |
| Emb12 | Zm00001d018366 | 5 | Translation initiation factor 3, plastid protein synthesis | [72] | |
| Emb14 | Zm00001d054079 | 4 | Plastid-targeted cGTPase, ribosome formation in plastid | [73] | |
| Why1 | Zm00001d036148 | 6 | DNA/RNA binding protein, genome stabilization and ribosome formation in plastids | [74] | |
| Emb-7L | Zm00001d021871 | 7 | Plastid PPR protein, RNA splicing | [75] | |
| Lem1 | Zm00001d034192 | 1 | Plastid 30S ribosomal protein S9, maintenance of plastid stability and ribosome formation | [76] | |
| PPR8522 | Zm00001d034962 | 1 | Plastid PPR protein, chloroplast transcription | [77] | |
| PRPL35-1 | Zm00001d046555 | 9 | Plastid ribosomal L35 subunit, translation | [78] | |
| Emp | Emp2441 | Zm00001d036689 | 6 | Nuclear-encoded maturase 3 protein, RNA splicing | [79] |
| Ppr14 | Zm00001d002157 | 2 | PPR protein, RNA splicing | [80] | |
| Ppr22 | Zm00001d028422 | 1 | PPR protein, RNA editing | [81] | |
| Ppr166 | Zm00001d040222 | 3 | PPR protein, RNA editing | [82] | |
| Mcsf1 | Zm00001d024429 | 10 | CRM domain-containing protein, interaction with PPR protein | [83] | |
| Ppr-smr | Zm00001d002345 | 2 | PPR protein, RNA splicing | [83] | |
| Emp25 | Zm00001d022184 | 7 | PPR protein, RNA splicing | [84] | |
| Emp603 | Zm00001d012528 | 8 | PPR protein, RNA splicing | [85] | |
| Emp80 | Zm00001d009677 | 8 | PPR protein, RNA editing | [86] | |
| Emp11 | Zm00001d052450 | 4 | PPR protein, RNA splicing | [87] | |
| Emp12 | Zm00001d002098 | 2 | PPR protein, RNA splicing | [88] | |
| Emp16 | Zm00001d011559 | 8 | PPR protein, RNA splicing | [89] | |
| Emp18 | Zm00001d034253 | 1 | PPR protein, RNA editing | [90] | |
| Emp10 | Zm00001d033992 | 1 | PPR protein, RNA splicing | [91] | |
| Emp2 | Zm00001d005675 | 2 | Heat shock binding protein 1, heat shock response | [92] | |
| Emp21 | Zm00001d033495 | 1 | PPR protein, RNA editing | [93] | |
| Emp32 | Zm00001d040363 | 3 | PPR protein, RNA splicing | [94] | |
| Emp4 | Zm00001d033869 | 1 | PPR protein, correct expression of mitochondrial transcripts | [95] | |
| Emp6 | Zm00001d005959 | 2 | PORR protein, mitochondrial intron splicing | [96] | |
| Emp602 | Zm00001d028046 | 1 | PPR protein, RNA splicing | [97] | |
| Emp7 | Zm00001d008298 | 8 | PPR protein, RNA editing | [98] | |
| Emp8 | Zm00001d049796 | 4 | PPR protein, RNA splicing | [99] | |
| Emp9 | Zm00001d022480 | 7 | PPR protein, RNA editing | [100] | |
| Ppr101 | Zm00001d010942 | 8 | PPR protein, RNA splicing | [101] | |
| Ppr27 | Zm00001d029061 | 1 | PPR protein, RNA editing | [82] | |
| Ppr18 | Zm00001d007927 | 2 | PPR protein, RNA splicing | [102] | |
| Ppr20 | Zm00001d039548 | 3 | PPR protein, RNA splicing | [103] | |
| Emp5 | Zm00001d042039 | 3 | PPR protein, RNA editing | [104] | |
| Sal1 | Zm00001d046599 | 9 | Lass E vacuolar sorting protein, aleurone layer differentiation | [105] | |
| SWEET4c | Zm00001d015912 | 5 | Bidirectional sugar transporter SWEET4-like, hexose transport | [106] | |
| End | Cesa5 | Zm00001d034553 | 1 | Cellulose synthase 5, flange cell wall ingrowths formation | [107] |
| Mn6 | Zm00001d037926 | 6 | ER SPases I, signal cleavage | [13] | |
| Cr4 | Zm00001d023425 | 10 | Receptor-like kinase, cell differentiation | [108] | |
| De18 | Zm00001d023718 | 10 | Endosperm-specific YUCCA1 protein, IAA biosynthesis | [109] | |
| Mdh4 | Zm00001d032695 | 1 | Cytosolic malate dehydrogenase 4, interconversion between malic acid and oxaloacetic acid (OAA) | [110] | |
| O11 | Zm00001d003677 | 2 | bHLH transcription factor, important regulators of endosperm development and metabolism | [111] | |
| opaque/floury | Ocd1 | Zm00001d008739 | 8 | Oxalyl-CoA decarboxylase, oxalate degradation | [112] |
| Fl1 | Zm00001d003398 | 2 | Endoplasmic reticulum protein, protein body assembly | [113] | |
| Fl2 | Zm00001d049243 | 4 | 22-kD a-zein protein, zein biosynthesis | [114] | |
| Fl3 | Zm00001d009292 | 8 | PLATZ protein, tRNA and 5S rRNA transcription | [115] | |
| Fl4 | Zm00001d048851 | 4 | 19-kD a-zein z1A-6, protein body assembly | [116] | |
| Mc | Zm00001d005793 | 2 | 16-kD-γ-zein, zein biosynthesis | [117] | |
| O1 | Zm00001d052110 | 4 | Myosin XI motor protein, morphology and movement of the endoplasmic reticulum, protein body formation | [118] | |
| O10 | Zm00001d033654 | 1 | Novel cereal-specific protein, regulation of protein distribution | [119] | |
| O2 | Zm00001d018971 | 7 | bZIP transcription factor, multiple biological process regulators in the endosperm | [15] | |
| O5 | Zm00001d020537 | 7 | Monogalactosyldiacylglycerol synthase, MGDG biosynthesis | [120] | |
| O6/Pro1 | Zm00001d010056 | 8 | P5CS, proline biosynthesis | [121] | |
| O7 | Zm00001d026649 | 10 | Acyl-activating enzyme, zein biosynthesis | [122] | |
| Pbf1 | Zm00001d005100 | 2 | Prolamin-box binding factor, regulation of zein expression | [123] | |
| Pdk1 | Zm00001d038163 | 6 | Pyruvate phosphate dikinase, energy production and metabolism | [124] | |
| Pdk2 | Zm00001d010321 | 8 | Pyruvate phosphate dikinase, energy production and metabolism | [124] | |
| Smu2 | Zm00001d023239 | 10 | RNA-splicing factor, rRNA processing and protein synthesis | [125] | |
| shrunken | Ae1 | Zm00001d016684 | 5 | Starch-branching enzyme IIb, starch biosynthesis | [126] |
| Bt2 | Zm00001d050032 | 4 | ADP-glucose pyrophosphorylase, starch biosynthesis | [127] | |
| Se1 | Zm00001d007657 | 2 | FAF domain protein, starch biosynthesis | [128] | |
| Sh1 | Zm00001d045042 | 9 | Sucrose synthase, starch biosynthesis | [16] | |
| Sh2 | Zm00001d044129 | 3 | AGPase subunit, starch biosynthesis | [129] | |
| Su1 | Zm00001d049753 | 4 | Isoamylase, starch biosynthesis | [130] | |
| NAC128 | Zm00001d040189 | 3 | NAC transcription factor, starch and zein biosynthesis | [131] | |
| NAC130 | Zm00001d008403 | 8 | NAC transcription factor, starch and zein biosynthesis | [131] | |
| Smk | Chao2 | Zm00001d011819 | 8 | Chlorophyll a oxygenase 1, chlorophyll B synthesis | [132] |
| Drg10 | Zm00001d003349 | 2 | Cytochrome P450 protein, brassinosteroid biosynthesis | [133] | |
| Expb14 | Zm00001d045792 | 9 | Expansin protein, miR164 pathway, participating in kernel expansion | [134] | |
| Expb15 | Zm00001d045861 | 9 | Expansin protein, miR164 pathway, participating in kernel expansion | [134] | |
| qKW9 | Zm00001d048451 | 9 | Plastid PPR protein, RNA editing | [135] | |
| Ppr78 | Zm00001d034428 | 1 | PPR protein, nad5 mature and mRNA stabilization | [136] | |
| Ppr278 | Zm00001d015156 | 5 | PPR protein, RNA splicing and editing | [137] | |
| Smk1 | Zm00001d007100 | 2 | PPR protein, RNA editing | [138] | |
| Smk10 | Zm00001d001803 | 2 | Choline transporter-like protein, choline transport pathway | [139] | |
| Smk501 | Zm00001d008256 | 8 | RUBylation activating enzyme E1 subunit ECR1, ubiquitin-related RUB pathway | [140] | |
| Vks1 | Zm00001d018624 | 7 | Kinesin-14 motor protein, regulation of mitosis and cytokinesis | [141] | |
| Mn2 | Zm00001d019294 | 7 | Nitrate transporter, bidirectional transport of nitrate | [142] | |
| Incw1 | Zm00001d016708 | 5 | Cell wall invertases 1, sucrose cleavage and transport | [143] | |
| Hda101 | Zm00001d053595 | 4 | Histone deacetylase, maintenance of histone acetylation | [144] | |
| Mn1 | Zm00001d003776 | 2 | Cell wall isozymes 2, sucrose cleavage and transport | [11] | |
| MRP-1 | Zm00001d010889 | 8 | Transfer cell-specific transcriptional activator, regulator of the differentiation of transfer cells | [145] | |
| Ppr231 | Zm00001d018219 | 5 | PPR protein, RNA splicing | [101] | |
| Ppr2263 | Zm00001d045089 | 9 | PPR protein, RNA editing | [146] | |
| VPS29 | Zm00001d053371 | 4 | Retromer complex subunit, regulation of IAA homeostasis | [147] | |
| Smk2 | Zm00001d053981 | 4 | Glutaminase, vitamin B6 Biosynthesis | [148] | |
| Smk3 | Zm00001d041537 | 3 | Mitochondrial transcription termination factor, intron splicing and complex assembly | [149] | |
| Smk4 | Zm00001d049196 | 4 | PPR protein, RNA editing | [150] | |
| Smk6 | Zm00001d025446 | 10 | PPR protein, RNA editing | [151] | |
| Smk7 | Zm00001d035960 | 6 | RNA polymerase III subunit, transcriptional regulation of tRNA and 5s rRNA | [152] | |
| Ubl1 | Zm00001d017432 | 5 | Putative RNA exonuclease, pre-mRNA splicing | [153] | |
| Urb2 | Zm00001d028096 | 1 | Urb2 domain-containing protein, pre-ribosomal RNA processing | [154] | |
| Ysl2 | Zm00001d017427 | 5 | Iron-nicotianamine transporter, Fe stabilization and storage | [155] |
| ID | Chr | Physical Position (B73_V4, nt) | Overlapped Cluster | Cloned Gene | ||
|---|---|---|---|---|---|---|
| Number | Detail | Number | Detail | |||
| HS01 | 1 | 12,622,245–17,191,112 | 3 | KW-gCL1-1, KW-qCL1-1, HKW-qCL1-1 | 0 | |
| HS02 | 1 | 20,505,000–52,520,534 | 14 | KW-qCL1-2, HKW-qCL1-2, KT-qCL1-1, KL-gCL1-1, KT-gCL1-1, KT-qCL1-2, KT-qCL1-3, HKW-gCL1-1, KW-gCL1-2, KT-qCL1-4, KT-gCL1-2, KT-qCL1-5, KT-qCL1-6, KT-qCL1-7 | 4 | Emp602, Urb2, Ppr22, Dek1 |
| HS03 | 1 | 53,029,436–59,415,031 | 3 | KW-gCL1-3, KL-qCL1-1, HKW-qCL1-2 | 1 | Ppr27 |
| HS04 | 1 | 211,711,329–221,020,719 | 3 | KL-gCL1-2, KT-qCL1-9, HKW-qCL1-4 | 0 | |
| HS05 | 1 | 238,107,995–242,396,593 | 4 | KL-gCL1-3, KL-qCL1-2, KT-qCL1-11, KW-qCL1-5 | 0 | |
| HS06 | 1 | 244,038,128–252,279,335 | 6 | KW-qCL1-5, HKW-qCL1-5, KL-qCL1-3, KW-gCL1-5, KW-qCL1-6, HKW-qCL1-6 | 0 | |
| HS07 | 1 | 270,393,381–288,290,428 | 6 | KL-qCL1-3, KT-gCL1-3, KW-gCL1-6, KT-qCL1-15, KT-qCL1-16, HKW-gCL1-3 | 6 | Dek35, Emp4, Emp10, MPPR6, Lem1, Emp18 |
| HS08 | 2 | 1,645,703–3,317,858 | 3 | KL-qCL2-1, HKW-qCL2-1, KT-qCL2-1 | 0 | |
| HS09 | 2 | 19,436,743–33,434,209 | 5 | KW-qCL2-1, HKW-qCL2-7, KT-qCL2-3, KW-qCL2-2, KW-qCL2-3 | 0 | |
| HS10 | 2 | 193,515,365–196,000,000 | 3 | KW-qCL2-5, KL-qCL2-4, HKW-qCL2-11 | 1 | Emp6 |
| HS11 | 3 | 1,325,039–3,615,750 | 8 | KW-gCL3-1, KT-gCL3-1, KL-gCL3-1, KW-qCL3-1, KL-qCL3-1, KL-qCL3-2, KW-qCL3-2, HKW-qCL3-1 | 0 | |
| HS12 | 3 | 4,388,286–6,099,395 | 5 | KL-gCL3-1, KW-qCL3-3, HKW-qCL3-2, KT-qCL3-1, KW-qCL3-4 | 0 | |
| HS13 | 4 | 3,561,183–6,010,197 | 3 | KL-qCL4-1, HKW-gCL4-1, KW-gCL4-1 | 0 | |
| HS14 | 4 | 158,189,789–163,298,733 | 4 | KW-gCL4-4, HKW-gCL4-4, KW-qCL4-2, KL-gCL4-3 | 0 | |
| HS15 | 4 | 175,915,845–193,659,742 | 8 | KT-gCL4-1, HKW-gCL4-5, HKW-qCL4-3, KL-gCL4-4, KW-qCL4-3, KL-qCL4-3, KW-qCL4-4, KW-gCL4-5 | 3 | O1, Dek15, Emp11 |
| HS16 | 4 | 196,013,048–201,338,547 | 3 | HKW-qCL4-4, KW-qCL4-6, KW-gCL4-5 | 0 | |
| HS17 | 4 | 237,957,206–240,000,000 | 3 | KW-qCL4-7, KW-gCL4-6, HKW-qCL4-6 | 0 | |
| HS18 | 5 | 14,156,572–16,289,485 | 3 | KW-qCL5-1, HKW-qCL5-2, KL-gCL5-1 | 0 | |
| HS19 | 5 | 34,324,375–65,639,673 | 4 | KW-qCL5-2, KW-gCL5-2, KW-gCL5-3, HKW-qCL5-2 | 2 | Dek36, Dek55 |
| HS20 | 5 | 188,839,192–195,089,478 | 5 | HKW-qCL5-6, HKW-qCL5-5, KW-gCL5-5, KT-gCL5-2, KL-gCL5-4 | 0 | |
| HS21 | 5 | 196,011,766–199,615,219 | 3 | KW-qCL5-3, HKW-qCL5-7, KL-qCL5-2 | 0 | |
| HS22 | 5 | 202,544,328–209,785,812 | 5 | HKW-qCL5-11, HKW-qCL5-10, KL-gCL5-5, HKW-qCL5-9, KW-qCL5-4 | 0 | |
| HS23 | 5 | 211,000,000–212,583,743 | 3 | HKW-qCL5-12, KT-gCL5-3, KL-gCL5-5 | 0 | |
| HS24 | 7 | 112,866,130–118,353,636 | 3 | HKW-qCL7-3, KL-qCL7-1, KL-gCL7-1 | 0 | |
| HS25 | 7 | 125,132,474–129,166,008 | 4 | HKW-qCL7-3, KL-gCL7-2, KW-qCL7-1, KW-qCL7-2 | 0 | |
| HS26 | 7 | 132,311,932–156,631,694 | 11 | HKW-gCL7-1, KW-qCL7-4, KL-gCL7-3, KL-qCL7-2, KL-qCL7-3, KL-qCL7-4, KW-qCL7-5, KW-gCL7-1, KW-qCL7-6, KW-qCL7-7, HKW-qCL7-3 | 2 | Dek41, Dek47 |
| HS27 | 7 | 172,547,374–174,900,000 | 4 | HKW-qCL7-5, KT-gCL7-2, KL-gCL7-4, KL-qCL7-8 | 0 | |
| HS28 | 8 | 167,000,000–170,036,314 | 3 | KW-qCL8-2, KL-qCL8-3, HKW-qCL8-2 | 0 | |
| HS29 | 9 | 149,400,961–157,000,000 | 9 | HKW-qCL9-2, KL-gCL9-1, KW-qCL9-2, HKW-gCL9-2, KW-qCL9-3, KT-gCL9-1, KL-qCL9-8, KW-qCL9-4, KW-qCL9-5 | 1 | qKW9 |
| HS30 | 10 | 109,814,884–122,563,176 | 3 | HKW-gCL10-1, KW-gCL10-2, HKW-qCL10-1 | 1 | Smk6 |
| HS31 | 10 | 130,739,025–149,279,019 | 10 | HKW-qCL10-2, KW-gCL10-3, HKW-gCL10-2, KL-gCL10-1, KT-gCL10-1, HKW-qCL10-3, KW-qCL10-1, HKW-qCL10-4, KL-qCL10-1, HKW-qCL10-5 | 1 | Nkd2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Li, H.; Long, Y.; Dong, Z.; Wang, J.; Liu, C.; Wei, X.; Wan, X. A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize. Int. J. Mol. Sci. 2023, 24, 1025. https://doi.org/10.3390/ijms24021025
Wang C, Li H, Long Y, Dong Z, Wang J, Liu C, Wei X, Wan X. A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize. International Journal of Molecular Sciences. 2023; 24(2):1025. https://doi.org/10.3390/ijms24021025
Chicago/Turabian StyleWang, Cheng, Huangai Li, Yan Long, Zhenying Dong, Jianhui Wang, Chang Liu, Xun Wei, and Xiangyuan Wan. 2023. "A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize" International Journal of Molecular Sciences 24, no. 2: 1025. https://doi.org/10.3390/ijms24021025
APA StyleWang, C., Li, H., Long, Y., Dong, Z., Wang, J., Liu, C., Wei, X., & Wan, X. (2023). A Systemic Investigation of Genetic Architecture and Gene Resources Controlling Kernel Size-Related Traits in Maize. International Journal of Molecular Sciences, 24(2), 1025. https://doi.org/10.3390/ijms24021025






