A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize
Abstract
1. Introduction
2. Results
2.1. The Nomenclature and Characterization of Maize LTP Genes
2.2. Phylogenetic Analysis and Classification of LTPs in Maize
2.3. Gene Ontology Analysis of LTPs in Maize
2.4. Functions of LTP Transporters in Rice and Arabidopsis and Their Implications in Maize
2.4.1. Anther and Pollen Development
| No. | Gene name | Gene ID | Gene Name in This Paper | Maize Orthologs | Expression Organs | Biological Functions | References |
|---|---|---|---|---|---|---|---|
| I. Anther development and male fertility | |||||||
| 1 | AtLTP5 | At3g51600 | AtLTP1.8 | ZmLTP1.8-1 | Root, Shoot, Leaf, Pistil | Vegetative and pollen tube growth | [44,45] |
| 2 | AtLTP12 | At3g51590 | AtLTP1.7 | ZmLTP1.8-1 | Pollen | Restore the fertility of proline-deficient microspores | [43] |
| 3 | AtLTPc1 | At5g07230 | AtLTPc1 | ZmLTPc1 | Anther | Transport exine precursors from tapetal ER to microspore surface | [14] |
| 4 | AtLTPc3 | At5g62080 | AtLTPc3 | ZmLTPc1 | Anther | Transport exine precursors from tapetal ER to microspore surface | [14] |
| 5 | OsC4 | Os08g0546300 | OsLTPc1 | ZmLTPc1 | Anther | Related to tapetal PCD of anther | [39,40] |
| 6 | ZmMs44 | Zm00001d052736 | ZmLTPc1 | – | Anther | Works as signal peptide and facilitates the secretion of lipids from tapetal cells into the locule | [32] |
| 7 | AtLTPG3 | At1g18280 | AtLTPg3 | ZmLTPg15 | Anther | Pollen grain development | [17] |
| 8 | AtLTPG4 | At1g27950 | AtLTPg4 | ZmLTPg4/ ZmTLPg14 | Anther | Pollen grain development | [17] |
| 9 | OsC6 | Os11g0582500 | OsLTPg25 | ZmLTPx1 | Anther | Male reproductive development | [16] |
| 10 | OsLTP47 | Os01g0607100 | OsLTPg28 | ZmLTPg26 | Anther | Pollen wall development | [31] |
| 11 | OsDIL/ OsLTP6 | Os10g0148000 | OsLTPd11 | ZmLTPg15/ ZmTLP16 | Leaf, Lemma, Palea, Anther, Pistil | Drought stress, tapetal and anther sacs development | [41,42] |
| 12 | OsLTPL94/ OsEPAD1 | Os03g0663900 | OsLTPg29 | ZmLTPg11/ ZmLTPx2 | Anther | Pollen exine formation | [5,15] |
| 13 | ZmLTPg11 | Zm00001d003600 | ZmLTPg11 | – | Anther | The orthologs of TaMs1, but zmltpg11zmltpx2 mutant is normal in pollen development | [33] |
| 14 | ZmLTPx2 | Zm00001d025467 | ZmLTPx2 | – | Anther | ||
| 15 | MZm3-3 | Zm00001d021226 | ZmLTPc2 | – | Anther | Contribute to pollen coat formation | [34] |
| II. Vegetative organ and seed development | |||||||
| 1 | AtLTP1 | At2g38540 | AtLTP1.5 | ZmLTP1.8-1 | Stem, leaf, and root | Export of lipids to the plant surface | [21,25,59] |
| 2 | AtLTP2 | At2g38530 | AtLTP1.4 | ZmLTP1.8-1 | Epidermal Cells | Play major structural roles | [60] |
| 3 | AtLTP7 | At2g15050 | AtLTP1.1 | ZmLTP1.8-1 | Leaf | – | [45] |
| 4 | AtLTP6 | At3g08770 | AtLTP1.6 | ZmLTP1.1 | Leaf and root | – | [45] |
| 5 | OsPSD1/ OsPTD1 | Os01g0822900 | OsLTP1.2 | ZmLTP1.1 | Stem, Leaf | Cell development, plant height, sensitivity to temperature conditions | [22,24] |
| 6 | AtLTPG2 | At3g43720 | AtLTPg21 | ZmLTPg2 | Stem | Alkane and wax transport | [23] |
| 7 | AtLTPG5 | At1g36150 | AtLTPg5 | ZmLTPg25 | Seed | Seed coat permeability | [17] |
| 8 | AtLTPG6 | At1g55260 | AtLTPg6 | ZmLTPg9 | Seed | Cuticle development and seed coat suberization | [17] |
| 9 | AtLTPG15 | At2g48130 | AtLTPG15 | ZmLTPg5 | Root, seed | Seed coat permeability | [61] |
| 10 | AtLTPG23 | At4g08670 | AtLTPg23 | ZmLTPg5 | - | Related to suberin biosynthesis | [3] |
| 11 | AtLTPG26 | At4g14815 | AtLTPg26 | ZmLTPg7 | - | Related to suberin biosynthesis | [3] |
| 12 | OsLTPL36 | Os03g0369100 | OsLTPd13 | ZmLTPd6 | Seed | Seed quality, seed development and germination | [20] |
| 13 | ZmBETL-9 | Zm00001d041822 | ZmLTPd6 | - | Endosperm | Transcribed in outer surface of developing endosperm | [35] |
| 14 | AtEND1 | At1g32280 | AtLTPd9 | ZmLTPg15/ ZmTLPg16 | Root, Leaf, Stem, Flower, Seed. | Gametophytic tissues and developing endosperm | [62] |
| 15 | AtLSR1 | At1g62500 | AtLSR1 | – | Leaf | Regulate leaf senescence | [63] |
| III. Biotic and abiotic stress response | |||||||
| 1 | AtDIR1 | At5g48485 | AtLTPd1 | ZmLTPd5 | Leaf | Transmission of mobile signal(s) during systemic acquired resistance | [64] |
| 2 | AtDIR1-like | At5g48490 | AtLTPd2 | ZmLTPd5 | Leaf | Transmission of mobile signal(s) during systemic acquired resistance | [65] |
| 3 | AtLTP3 | At5g59320 | AtLTP1.12 | ZmLTP1.8-1 | Leaf, Flower, Silique, Root | Contributes to disease susceptibility | [19,66,67] |
| 4 | AtLTP4 | At5g59310 | AtLTP1.11 | ZmLTP1.8-1 | Shoot apex, Leaf, Root | Reduced susceptibility to Pseudomonas and down-regulation of ABA biosynthesis genes in ltp3/ltp4 mutant | [45,66] |
| 5 | AtLTPG1 | At1g03103 | AtLTPg1 | ZmLTPg7/ ZmTLPg13 | Stem, Leaf | Alter cuticular lipid composition, increase plastoglobulus, enhance susceptibility to infection by Fungal Pathogen | [27] |
| 6 | OsLTP5 | Os11g0115400 | OsLTP1.22 | ZmLTP1.2-2/ ZmLTP1.6 | Stem, Flower | Response to ABA, salicylic acid, and 16-hydroxypalmitic acid | [26] |
| 7 | Zm-LTP | Zm00001d044686 | ZmLTP1.2 | – | – | Binds to calmodulin (CaM) in a Ca2+-independent manner | [36] |
| 8 | OsLTPL159 | Os10g0505500 | OsLTP2.11 | ZmLTP2.4 | Seeding, root, node, Leaf, Sheath, Spikelet | Involved in cold tolerance at early seedling stage in rice | [30] |
| 9 | OsLTP110 | Os11g0115100 | OsLTP1.9 | ZmLTP1.7 | – | Inhibit germination of Pyricularia oryzae spores, resistance to biotic stresses | [68] |
| 10 | AtDRN1 | At2g45180 | – | – | Leaf | Response to avirulent bacterial phytopathogen Pst DC3000 | [69] |
| 11 | AtAZI1 | At4g12470 | – | – | Root, Leaf | Salt stress tolerance, regulate systemic acquired resistance | [29,38] |
| 12 | OsLTP1 | CAX20937 | – | – | Leaf, Root, Lemma, Palea, Anther | Structural barriers and organ protection against mechanical disruption and pathogen attack | [70,71] |
| 13 | ZmLTP3 | Zm00001d043049 | ZmLTP1.1 | – | Root, Coleoptile, Leaf, Silk, Ovary | Improve plant survival under salt and drought stresses | [37] |
2.4.2. Vegetative and Female Organ Development
2.4.3. Biotic and Abiotic Stress Response
2.5. Functional Prediction of LTP Genes Based on Bioinformatics Analysis
2.6. Substrate Identification Strategies of Plant LTP Transporters
3. Discussion
4. Materials and Methods
4.1. Identification of LTP Genes in Maize
4.2. Phylogenetic and GO Analysis of Maize LTP Genes
4.3. RNA Extraction, qPCR, and RNA-Seq Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef]
- Kader, J.C. Lipid-Transfer Proteins in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 627–654. [Google Scholar] [CrossRef]
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997. [Google Scholar] [CrossRef]
- Edstam, M.M.; Viitanen, L.; Salminen, T.A.; Edqvist, J. Evolutionary history of the non-specific lipid transfer proteins. Mol. Plant 2011, 4, 947–964. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kim, Y.J.; Yang, L.; Liu, Z.; Zhang, J.; Shi, H.; Huang, G.; Persson, S.; Zhang, D.; Liang, W. Grass-specific EPAD1 is essential for pollen exine patterning in rice. Plant Cell 2020, 32, 3961–3977. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Gatta, A.T.; Levine, T.P. Lipid transfer proteins: The lipid commute via shuttles, bridges and tubes. Nat. Rev. Mol. Cell Biol. 2019, 20, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Copic, A.; Levine, T.P. Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 2017, 42, 516–530. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K. Lipid transfer proteins rectify inter-organelle flux and accurately deliver lipids at membrane contact sites. J. Lipid Res. 2018, 59, 1341–1366. [Google Scholar] [CrossRef]
- Holthuis, J.C.; Menon, A.K. Lipid landscapes and pipelines in membrane homeostasis. Nature 2014, 510, 48–57. [Google Scholar] [CrossRef]
- Julke, S.; Ludwig-Muller, J. Response of Arabidopsis thaliana roots with altered lipid transfer protein (LTP) gene expression to the clubroot disease and salt stress. Plants 2015, 5, 2. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Mineev, K.S.; Finkina, E.I.; Arseniev, A.S.; Ovchinnikova, T.V. A novel lipid transfer protein from the dill Anethum graveolens L.: Isolation, structure, heterologous expression, and functional characteristics. J. Pept. Sci. 2016, 22, 59–66. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Zhong, X. Non-specific lipid transfer proteins in maize. BMC Plant Biol. 2014, 14, 281. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.D.; Chen, T.L.; Huang, A.H. Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiol. 2013, 163, 1218–1229. [Google Scholar] [CrossRef]
- Tao, Y.; Zou, T.; Zhang, X.; Liu, R.; Chen, H.; Yuan, G.; Zhou, D.; Xiong, P.; He, Z.; Li, G.; et al. Secretory lipid transfer protein OsLTPL94 acts as a target of EAT1 and is required for rice pollen wall development. Plant J. 2021, 108, 358–377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liang, W.; Yin, C.; Zong, J.; Gu, F.; Zhang, D. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010, 154, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant 2014, 152, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Kreis, M.; Forde, B.G.; Rahman, S.; Miflin, B.J.; Shewry, P.R. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985, 183, 499–502. [Google Scholar] [CrossRef]
- Pagnussat, L.A.; Oyarburo, N.; Cimmino, C.; Pinedo, M.L.; de la Canal, L. On the role of a Lipid-Transfer Protein. Arabidopsis ltp3 mutant is compromised in germination and seedling growth. Plant Signal. Behav. 2015, 10, e1105417. [Google Scholar] [CrossRef]
- Wang, X.; Zhou, W.; Lu, Z.; Ouyang, Y.; O, C.S.; Yao, J. A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 2015, 239, 200–208. [Google Scholar] [CrossRef]
- Deeken, R.; Saupe, S.; Klinkenberg, J.; Riedel, M.; Leide, J.; Hedrich, R.; Mueller, T.D. The nonspecific lipid transfer protein AtLtpI-4 is involved in suberin formation of Arabidopsis thaliana Crown Galls. Plant Physiol. 2016, 172, 1911–1927. [Google Scholar] [CrossRef]
- Deng, W.; Li, R.; Xu, Y.; Mao, R.; Chen, S.; Chen, L.; Chen, L.; Liu, Y.G.; Chen, Y. A lipid transfer protein variant with a mutant eight-cysteine motif causes photoperiod- and thermo-sensitive dwarfism in rice. J. Exp. Bot. 2020, 71, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.B.; Kim, H.J.; Min, M.K.; Hwang, I.; Suh, M.C. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1391–1403. [Google Scholar] [CrossRef]
- Li, R.; Xia, J.; Xu, Y.; Zhao, X.; Liu, Y.G.; Chen, Y. Characterization and genetic mapping of a Photoperiod-sensitive dwarf 1 locus in rice (Oryza sativa L.). Theor. Appl. Genet. 2014, 127, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Potocka, I.; Baldwin, T.C.; Kurczynska, E.U. Distribution of lipid transfer protein 1 (LTP1) epitopes associated with morphogenic events during somatic embryogenesis of Arabidopsis thaliana. Plant Cell Rep. 2012, 31, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Park, J.H.; Kim, M.C.; Cho, S.H. Cutin monomer induces expression of the rice OsLTP5 lipid transfer protein gene. J. Plant Physiol. 2008, 165, 345–349. [Google Scholar] [CrossRef]
- Lee, S.B.; Go, Y.S.; Bae, H.J.; Park, J.H.; Cho, S.H.; Cho, H.J.; Lee, D.S.; Park, O.K.; Hwang, I.; Suh, M.C. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol. 2009, 150, 42–54. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Finkina, E.I.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant Pathogenesis-related proteins binding lipids and other hydrophobic ligands. Russ. J. Bioorg. Chem. 2019, 44, 586–594. [Google Scholar] [CrossRef]
- Pitzschke, A.; Datta, S.; Persak, H. Salt stress in Arabidopsis: Lipid transfer protein AZI1 and its control by mitogen-activated protein kinase MPK3. Mol. Plant 2014, 7, 722–738. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, S.; Qin, J.; Sun, C.; Liu, F. The lipid transfer protein OsLTPL159 is involved in cold tolerance at the early seedling stage in rice. Plant Biotechnol. J. 2020, 18, 756–769. [Google Scholar] [CrossRef]
- Chen, L.; Ji, C.; Zhou, D.; Gou, X.; Tang, J.; Jiang, Y.; Han, J.; Liu, Y.G.; Chen, L.; Xie, Y. OsLTP47 may function in a lipid transfer relay essential for pollen wall development in rice. J. Genet. Genomics 2022, 49, 481–491. [Google Scholar] [CrossRef]
- Fox, T.; DeBruin, J.; Haug Collet, K.; Trimnell, M.; Clapp, J.; Leonard, A.; Li, B.; Scolaro, E.; Collinson, S.; Glassman, K.; et al. A single point mutation in Ms44 results in dominant male sterility and improves nitrogen use efficiency in maize. Plant Biotechnol. J. 2017, 15, 942–952. [Google Scholar] [CrossRef]
- Li, J.; Wang, Z.; Chang, Z.; He, H.; Tang, X.; Ma, L.; Deng, X.W. A functional characterization of TaMs1 orthologs in Poaceae plants. Crop J. 2021, 9, 1291–1300. [Google Scholar] [CrossRef]
- Lauga, B.; Charbonnel-Campaa, L.; Combes, D. Characterization of MZm3-3, a Zea mays tapetum-specific transcript. Plant Sci. 2000, 157, 65–75. [Google Scholar] [CrossRef]
- Royo, J.; Gomez, E.; Sellam, O.; Gerentes, D.; Paul, W.; Hueros, G. Two maize END-1 orthologs, BETL9 and BETL9like, are transcribed in a non-overlapping spatial pattern on the outer surface of the developing endosperm. Front. Plant Sci. 2014, 5, 180. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xie, W.; Bai, W.; Li, Z.; Zhao, Y.; Liu, H. Calmodulin binds to maize lipid transfer protein and modulates its lipids binding ability. FEBS J. 2008, 275, 5298–5308. [Google Scholar] [CrossRef]
- Zou, H.W.; Tian, X.H.; Ma, G.H.; Li, Z.X. Isolation and functional analysis of ZmLTP3, a homologue to Arabidopsis LTP3. Int. J. Mol. Sci. 2013, 14, 5025–5035. [Google Scholar] [CrossRef]
- Yu, K.; Soares, J.M.; Mandal, M.K.; Wang, C.; Chanda, B.; Gifford, A.N.; Fowler, J.S.; Navarre, D.; Kachroo, A.; Kachroo, P. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 2013, 3, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, T.; Toriyama, K.; Ejiri, S.; Hinata, K. Molecular characterization of rice genes specifically expressed in the anther tapetum. Plant Mol. Biol. 1994, 26, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Li, X.; Wang, Z.; Ding, M.; Sun, Y.; Dong, F.; Chen, F.; Liu, L.; Doughty, J.; Li, Y.; et al. Histone H2B monoubiquitination mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is involved in anther development by regulating tapetum degradation-related genes in rice. Plant Physiol. 2015, 168, 1389–1405. [Google Scholar] [CrossRef]
- Guo, C.; Ge, X.; Ma, H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 2013, 82, 239–253. [Google Scholar] [CrossRef]
- Liu, X.; Shangguan, Y.; Zhu, J.; Lu, Y.; Han, B. The rice OsLTP6 gene promoter directs anther-specific expression by a combination of positive and negative regulatory elements. Planta 2013, 238, 845–857. [Google Scholar] [CrossRef]
- Mattioli, R.; Biancucci, M.; El Shall, A.; Mosca, L.; Costantino, P.; Funck, D.; Trovato, M. Proline synthesis in developing microspores is required for pollen development and fertility. BMC Plant Biol. 2018, 18, 356. [Google Scholar] [CrossRef] [PubMed]
- Chae, K.; Kieslich, C.A.; Morikis, D.; Kim, S.C.; Lord, E.M. A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 2009, 21, 3902–3914. [Google Scholar] [CrossRef] [PubMed]
- Chae, K.; Gonong, B.J.; Kim, S.C.; Kieslich, C.A.; Morikis, D.; Balasubramanian, S.; Lord, E.M. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010, 61, 4277–4290. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Q.; Liu, C.; Song, M.; Xu, J.; Zhu, Z. Comprehensive transcriptome analysis of rare carpinus putoensis plants under NO2 stress. Genes 2021, 12, 754. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol Plant 2020, 13, 955–983. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, Z.; Liu, X.; Zhu, T.; Xie, K.; Hou, Q.; Yan, T.; Niu, C.; Zhang, S.; Yang, M.; et al. ZmFAR1 and ZmABCG26 Regulated by microRNA Are Essential for Lipid Metabolism in Maize Anther. Int. J. Mol. Sci. 2021, 22, 7916. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, S.; Niu, C.; Liu, D.; Yan, T.; Xie, K.; Li, Z.; Wang, Y.; Zhao, W.; Dong, Z.; et al. ZmMs25 encoding a plastid-localized fatty acyl reductase is critical for anther and pollen development in maize. J. Exp. Bot. 2021, 72, 4298–4318. [Google Scholar] [CrossRef]
- Xie, K.; Wu, S.; Li, Z.; Zhou, Y.; Zhang, D.; Dong, Z.; An, X.; Zhu, T.; Zhang, S.; Liu, S.; et al. Map-based cloning and characterization of Zea mays male sterility33 (ZmMs33) gene, encoding a glycerol-3-phosphate acyltransferase. Theor. Appl. Genet. 2018, 131, 1363–1378. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, H.; Zhao, Y.; Chen, X.; Huang, Y.; Yan, S.; Li, S.; Liu, M.; Huang, W.; Zhang, X.; et al. Maize male sterile 33 encodes a putative glycerol-3-phosphate acyltransferase that mediates anther cuticle formation and microspore development. BMC Plant Biol. 2018, 18, 318. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Li, Z.; An, X.; Long, Y.; Xue, X.; Xie, K.; Ma, B.; Zhang, D.; Guan, Y.; Niu, C.; et al. Normal structure and function of endothecium chloroplasts maintained by ZmMs33-mediated lipid biosynthesis in tapetal cells are critical for anther development in maize. Mol. Plant 2020, 13, 1624–1643. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wu, S.; Zhang, D.; Li, Z.; Xie, K.; An, X.; Ma, B.; Hou, Q.; Dong, Z.; Tian, Y.; et al. Genome-wide analysis of maize GPAT gene family and cytological characterization and breeding application of ZmMs33/ZmGPAT6 gene. Theor. Appl. Genet. 2019, 132, 2137–2154. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Dong, Z.; Tian, Y.; Xie, K.; Wu, S.; Zhu, T.; Zhang, D.; Zhou, Y.; Niu, C.; Ma, B.; et al. ZmMs30 encoding a novel GDSL lipase is essential for male fertility and valuable for hybrid breeding in maize. Mol. Plant 2019, 12, 343–359. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, Y.; Wu, S.; Wang, J.; Fang, C.; Zhang, S.; Xie, R.; Zhao, L.; An, X.; Wan, X. The ZmMYB84-ZmPKSB regulatory module controls male fertility through modulating anther cuticle-pollen exine trade-off in maize anthers. Plant Biotechnol. J. 2022, 20, 2342–2356. [Google Scholar] [CrossRef]
- Fang, C.; Wu, S.; Niu, C.; Hou, Q.; An, X.; Wei, X.; Zhao, L.; Jiang, Y.; Liu, X.; Wan, X. Triphasic regulation of ZmMs13 encoding an ABCG transporter is sequentially required for callose dissolution, pollen exine and anther cuticle formation in maize. J. Adv. Res. 2022. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Fang, C.; Li, Z.; Wang, Y.; Pan, S.; Wu, Y.; An, X.; Long, Y.; Wan, X. ATP-bnding cassette G transporters and their multiple roles especially for male fertility in Arabidopsis, rice and maize. Int. J. Mol. Sci. 2022, 23, 9304. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; Dong, Z.; An, X.; Ma, B.; Tian, Y.; Li, J. Maize genic male-sterility genes and their applications in hybrid breeding: Progress and perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef]
- Thoma, S.; Kaneko, Y.; Somerville, C. A non-specific lipid transfer protein from Arabidopsis is a cell wall protein. Plant J. 1993, 3, 427–436. [Google Scholar] [CrossRef]
- Jacq, A.; Pernot, C.; Martinez, Y.; Domergue, F.; Payre, B.; Jamet, E.; Burlat, V.; Pacquit, V.B. The Arabidopsis lipid transfer protein 2 (AtLTP2) is involved in cuticle-cell wall interface integrity and in etiolated hypocotyl permeability. Front. Plant Sci. 2017, 8, 263. [Google Scholar] [CrossRef]
- Lee, S.B.; Suh, M.C. Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein 15 affects seed coat permeability in Arabidopsis. Plant J. 2018, 96, 1206–1217. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lopato, S.; Hrmova, M.; Pickering, M.; Shirley, N.; Koltunow, A.M.; Langridge, P. Expression patterns and protein structure of a lipid transfer protein END1 from Arabidopsis. Planta 2014, 240, 1319–1334. [Google Scholar] [CrossRef]
- Feng, G.; Zhong, Y.; Zou, W. Lipid transporter LSR1 positively regulates leaf senescence in Arabidopsis. Plant Signal. Behav. 2022, 17, 2007328. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; von Dahl, C.C.; Park, S.W.; Klessig, D.F. Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and tobacco. Plant Physiol. 2011, 155, 1762–1768. [Google Scholar] [CrossRef]
- Champigny, M.J.; Isaacs, M.; Carella, P.; Faubert, J.; Fobert, P.R.; Cameron, R.K. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 2013, 4, 230. [Google Scholar] [CrossRef]
- Gao, S.; Guo, W.; Feng, W.; Liu, L.; Song, X.; Chen, J.; Hou, W.; Zhu, H.; Tang, S.; Hu, J. LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 2016, 17, 412–426. [Google Scholar] [CrossRef]
- Rojas, M.; Jimenez-Bremont, F.; Villicana, C.; Carreon-Palau, L.; Arredondo-Vega, B.O.; Gomez-Anduro, G. Involvement of OpsLTP1 from Opuntia streptacantha in abiotic stress adaptation and lipid metabolism. Funct. Plant Biol. 2019, 46, 816–829. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Chen, J.; Li, N.; Lin, Y.; Sun, C.; Cao, K. Resistance function of rice lipid transfer protein LTP110. J. Biochem. Mol. Biol. 2003, 36, 603–607. [Google Scholar] [CrossRef]
- Dhar, N.; Caruana, J.; Erdem, I.; Raina, R. An Arabidopsis disease related nonspecific lipid transfer protein 1 is required for resistance against various phytopathogens and tolerance to salt stress. Gene 2020, 753, 144802. [Google Scholar] [CrossRef]
- Vignols, F.; Lund, G.; Pammi, S.; Trémousaygue, D.; Grellet, F.; Kader, J.-C.; Puigdomènech, P.; Delseny, M. Characterization of a rice gene coding for a lipid transfer protein. Gene 1994, 142, 265–270. [Google Scholar] [CrossRef]
- Guiderdoni, E.; Cordero, M.J.; Vignols, F.; Garcia-Garrido, J.M.; Lescot, M.; Tharreau, D.; Meynard, D.; Ferriere, N.; Notteghem, J.L.; Delseny, M. Inducibility by pathogen attack and developmental regulation of the rice Ltp1 gene. Plant Mol. Biol. 2002, 49, 683–699. [Google Scholar] [CrossRef]
- Liu, Z.; Bao, W.; Liang, W.; Yin, J.; Zhang, D. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Biol. 2010, 52, 670–678. [Google Scholar] [CrossRef]
- Aya, K.; Ueguchi-Tanaka, M.; Kondo, M.; Hamada, K.; Yano, K.; Nishimura, M.; Matsuoka, M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 2009, 21, 1453–1472. [Google Scholar] [CrossRef]
- Wu, L.; Jing, X.; Zhang, B.; Chen, S.; Xu, R.; Duan, P.; Zou, D.; Huang, S.; Zhou, T.; An, C.; et al. A natural allele of OsMS1 responds to temperature changes and confers thermosensitive genic male sterility. Nat. Commun. 2022, 13, 2055. [Google Scholar] [CrossRef]
- Xu, J.; Ding, Z.; Vizcay-Barrena, G.; Shi, J.; Liang, W.; Yuan, Z.; Werck-Reichhart, D.; Schreiber, L.; Wilson, Z.A.; Zhang, D. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. Plant Cell 2014, 26, 1544–1556. [Google Scholar] [CrossRef]
- Li, N.; Zhang, D.S.; Liu, H.S.; Yin, C.S.; Li, X.X.; Liang, W.Q.; Yuan, Z.; Xu, B.; Chu, H.W.; Wang, J.; et al. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 2006, 18, 2999–3014. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, C.; Yuan, Z.; Zhang, D.; Gondwe, M.Y.; Ding, Z.; Liang, W.; Zhang, D.; Wilson, Z.A. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. Plant Cell 2010, 22, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Liang, W.; Yang, X.; Jin, W.; Wilson, Z.A.; Hu, J.; Zhang, D. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 2013, 4, 1445. [Google Scholar] [CrossRef] [PubMed]
- Chun, H.J.; Zheng, L.; Ahmad, M.; Wang, J.; Speirs, C.K.; Siegel, R.M.; Dale, J.K.; Puck, J.; Davis, J.; Hall, C.G.; et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 2002, 419, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Z.; Wu, S.; Wan, X. The essential roles of sugar metabolism for pollen development and male fertility in plants. Crop J. 2021, 9, 1223–1236. [Google Scholar] [CrossRef]
- Jiang, Y.; An, X.; Li, Z.; Yan, T.; Zhu, T.; Xie, K.; Liu, S.; Hou, Q.; Zhao, L.; Wu, S.; et al. CRISPR/Cas9-based discovery of maize transcription factors regulating male sterility and their functional conservation in plants. Plant Biotechnol. J. 2021, 19, 1769–1784. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, J.; Chen, S.; Heng, Y.; Chen, Z.; Yang, J.; Zhou, K.; Pei, J.; He, H.; Deng, X.W.; et al. Poaceae-specific MS1 encodes a phospholipid-binding protein for male fertility in bread wheat. Proc. Natl. Acad. Sci. USA 2017, 114, 12614–12619. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, D.; Bogdanov, I.; Ovchinnikova, T.; Finkina, E. Interaction between the lentil lipid transfer protein Lc-LTP2 and its novel signal ligand PI(4,5)P2. Membranes 2020, 10, 357. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Cheng, C.; Zhang, K.; Lou, Q.; Li, J.; Chen, J. Genome-wide analysis of a putative lipid transfer protein LTP_2 gene family reveals CsLTP_2 genes involved in response of cucumber against root-knot nematode (Meloidogyne incognita). Genome 2020, 63, 225–238. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, S.; Jiang, Y.; Yan, T.; Fang, C.; Hou, Q.; Wu, S.; Xie, K.; An, X.; Wan, X. Use of CRISPR/Cas9-based gene editing to simultaneously mutate multiple homologous genes required for pollen development and male fertility in maize. Cells 2022, 11, 439. [Google Scholar] [CrossRef]
- Wei, X.; Pu, A.; Liu, Q.; Hou, Q.; Zhang, Y.; An, X.; Long, Y.; Jiang, Y.; Dong, Z.; Wu, S.; et al. The bibliometric landscape of gene editing innovation and regulation in the worldwide. Cells 2022, 11, 2682. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; An, X.; Xie, K.; Dong, Z.; Zhou, Y.; Xu, L.; Fang, W.; Liu, S.; Liu, S.; et al. Construction of a multicontrol sterility system for a maize male-sterile line and hybrid seed production based on the ZmMs7 gene encoding a PHD-finger transcription factor. Plant Biotechnol. J. 2018, 16, 459–471. [Google Scholar] [CrossRef]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D.; et al. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, X. Breeding with dominant genic male-sterility genes to boost crop grain yield in the post-heterosis utilization era. Mol. Plant 2021, 14, 531–534. [Google Scholar] [CrossRef]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
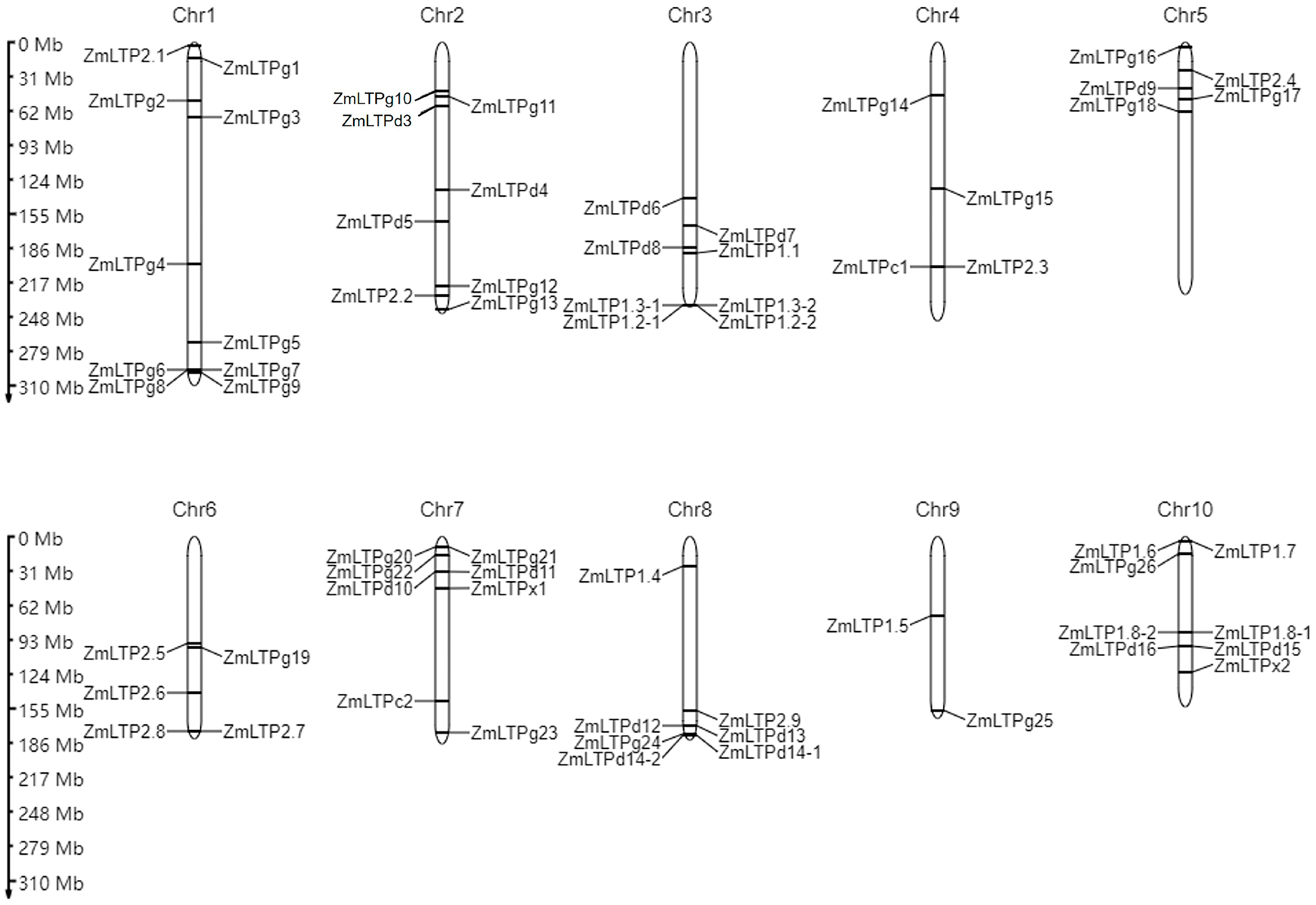
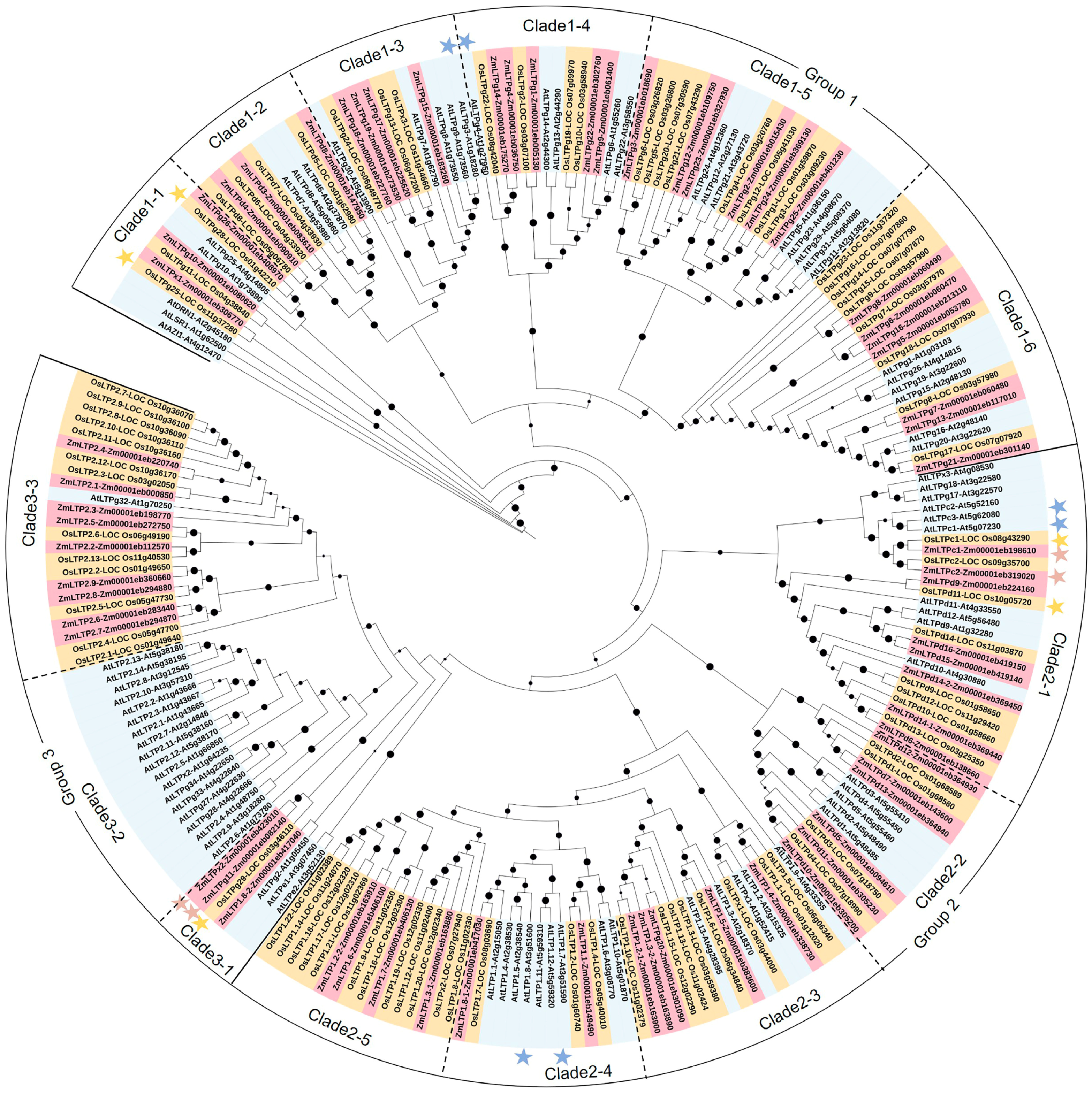
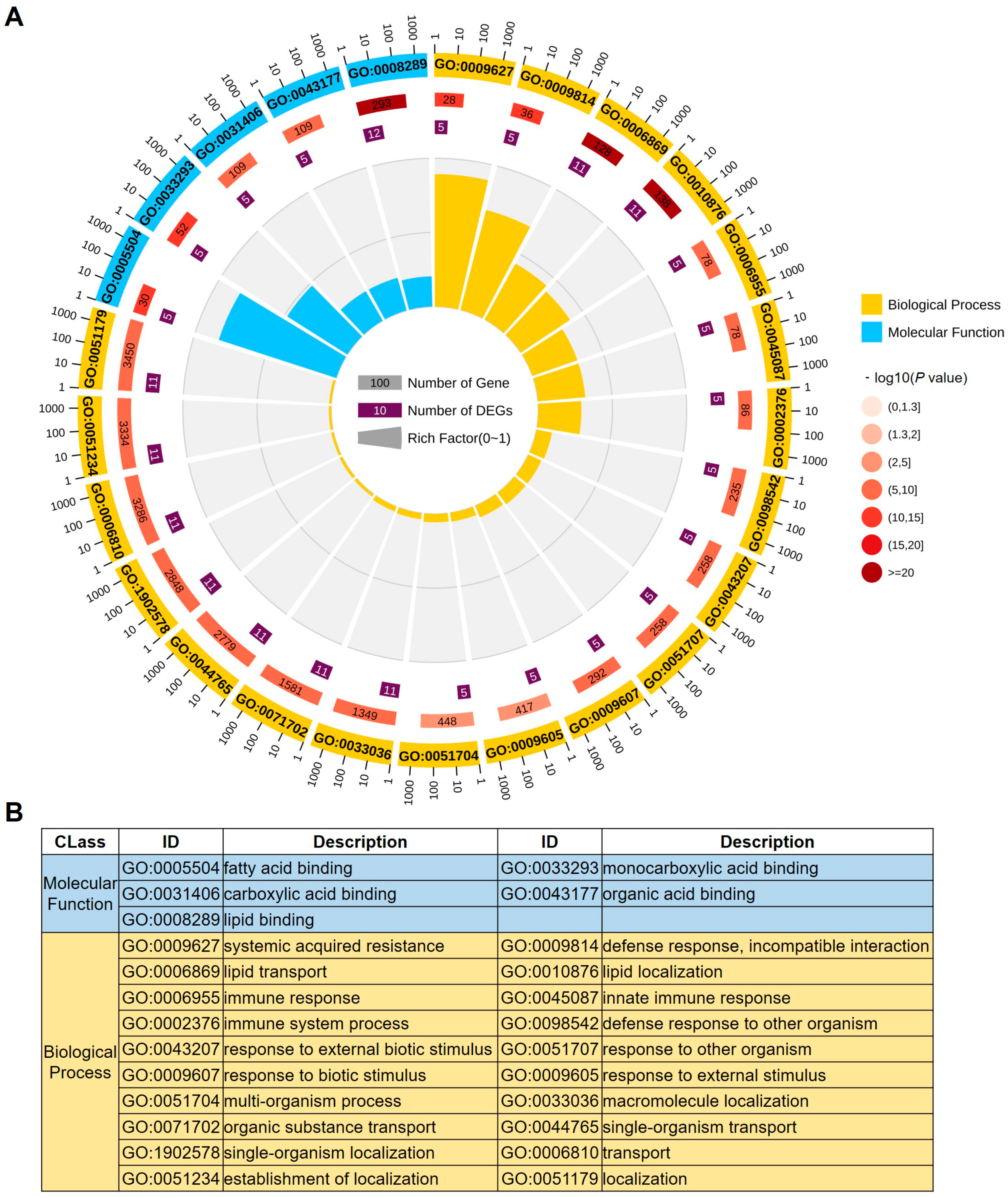

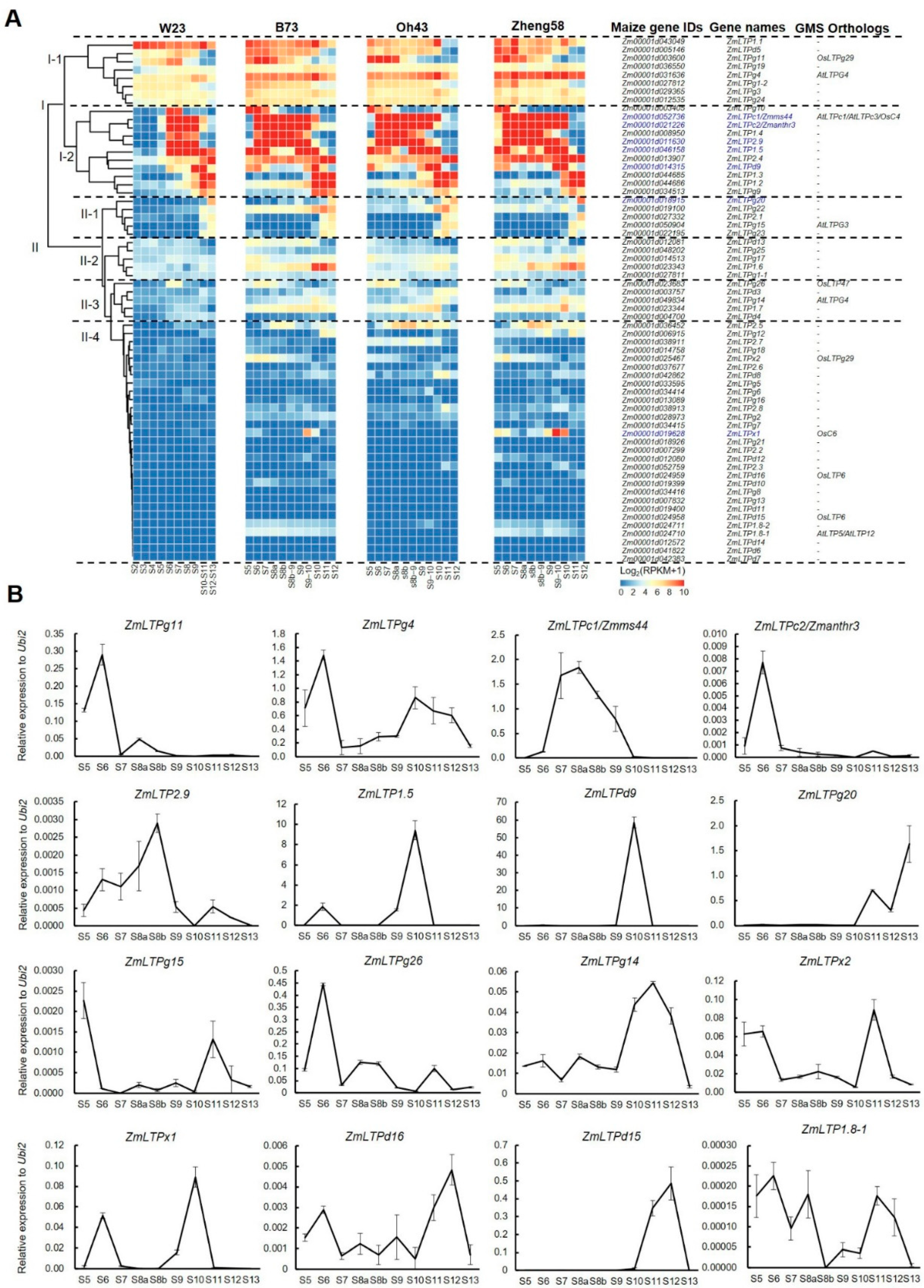
| No. | Name | Gene ID (B73 V5) 1 | Expression Patterns 2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Root | Leaf | Sem and sam | Internode | Tassel | Anther | Silk | Cob | Seed | Endosperm | Enbryo | Pericarp | |||
| 1 | ZmLTP1.1 | Zm00001eb149490 | − | + | + | − | + | − | − | + | + | − | + | − |
| 2 | ZmLTP1.2-1 | Zm00001eb163900 | − | + | − | + | + | − | − | + | + | − | + | − |
| 3 | ZmLTP1.2-2 | Zm00001eb163910 | ||||||||||||
| 4 | ZmLTP1.3-1 | Zm00001eb163880 | − | + | − | + | + | − | + | − | + | − | − | − |
| 5 | ZmLTP1.3-2 | Zm00001eb163890 | ||||||||||||
| 6 | ZmLTP1.4 | Zm00001eb338730 | − | + | − | − | + | − | − | − | − | − | − | − |
| 7 | ZmLTP1.5 | Zm00001eb383600 | − | − | − | − | + | − | − | − | − | − | − | − |
| 8 | ZmLTP1.6 | Zm00001eb406100 | − | + | − | − | − | − | + | − | + | − | + | − |
| 9 | ZmLTP1.7 | Zm00001eb406130 | − | − | + | − | + | − | − | + | + | + | + | − |
| 10 | ZmLTP1.8-1 | Zm00001eb417030 | − | − | − | − | − | − | − | − | + | + | − | − |
| 11 | ZmLTP1.8-2 | Zm00001eb417040 | ||||||||||||
| 12 | ZmLTP2.1 | Zm00001eb000850 | − | + | − | − | − | + | − | − | + | − | − | + |
| 13 | ZmLTP2.2 | Zm00001eb112570 | − | − | − | − | − | − | − | − | + | + | − | + |
| 14 | ZmLTP2.3 | Zm00001eb198770 | − | − | − | − | − | − | − | − | + | + | − | + |
| 15 | ZmLTP2.4 | Zm00001eb220740 | − | + | − | − | − | + | − | − | + | − | + | − |
| 16 | ZmLTP2.5 | Zm00001eb272750 | − | + | − | − | − | − | − | − | − | − | + | − |
| 17 | ZmLTP2.6 | Zm00001eb283440 | + | − | + | − | − | − | − | − | + | − | − | − |
| 18 | ZmLTP2.7 | Zm00001eb294870 | + | + | − | + | − | − | − | − | − | − | − | − |
| 19 | ZmLTP2.8 | Zm00001eb294880 | + | + | − | + | + | − | − | − | + | − | − | − |
| 20 | ZmLTP2.9 | Zm00001eb360660 | − | − | − | − | + | − | − | − | − | − | − | − |
| 21 | ZmLTPc1 | Zm00001eb198610 | − | − | − | − | + | − | − | − | − | − | − | − |
| 22 | ZmLTPc2 | Zm00001eb319020 | − | − | − | − | + | − | − | − | − | − | − | − |
| 23 | ZmLTPd3 | Zm00001eb083610 | − | + | − | − | − | − | − | − | + | − | − | − |
| 24 | ZmLTPd4 | Zm00001eb090910 | + | + | − | + | + | − | − | − | − | − | − | − |
| 25 | ZmLTPd5 | Zm00001eb094610 | + | + | − | + | + | − | − | − | + | − | + | + |
| 26 | ZmLTPd6 | Zm00001eb138660 | − | − | − | − | − | − | − | − | + | + | + | + |
| 27 | ZmLTPd7 | Zm00001eb143600 | + | − | − | − | − | − | − | − | − | − | − | − |
| 28 | ZmLTPd8 | Zm00001eb147950 | + | + | − | + | + | − | − | − | + | − | − | − |
| 29 | ZmLTPd9 | Zm00001eb224160 | − | − | − | − | + | − | − | − | − | − | − | − |
| 30 | ZmLTPd10 | Zm00001eb305200 | + | + | − | + | − | − | − | − | − | − | − | − |
| 31 | ZmLTPd11 | Zm00001eb305230 | + | + | − | + | − | − | − | − | − | − | − | − |
| 32 | ZmLTPd12 | Zm00001eb364930 | + | − | − | + | − | − | − | − | − | − | − | − |
| 33 | ZmLTPd13 | Zm00001eb364940 | + | + | − | + | + | − | − | + | + | + | + | + |
| 34 | ZmLTPd14-1 | Zm00001eb369440 | − | − | − | − | − | − | − | − | + | + | + | + |
| 35 | ZmLTPd14-2 | Zm00001eb369450 | ||||||||||||
| 36 | ZmLTPd15 | Zm00001eb419140 | − | − | − | − | − | − | − | − | + | + | − | + |
| 37 | ZmLTPd16 | Zm00001eb419150 | − | − | − | − | − | − | − | − | + | + | − | + |
| 38 | ZmLTPg1-1 | Zm00001eb005130 | − | + | − | + | + | − | + | − | + | + | + | + |
| 39 | ZmLTPg2 | Zm00001eb015430 | − | + | − | − | − | − | + | + | + | − | − | + |
| 40 | ZmLTPg3 | Zm00001eb018690 | + | + | − | + | − | − | + | + | + | + | + | + |
| 41 | ZmLTPg4 | Zm00001eb036750 | − | + | − | − | + | − | + | + | + | + | + | + |
| 42 | ZmLTPg5 | Zm00001eb053780 | + | + | − | − | − | − | − | − | − | − | − | − |
| 43 | ZmLTPg6 | Zm00001eb060470 | + | + | − | − | + | + | − | − | − | − | − | − |
| 44 | ZmLTPg7 | Zm00001eb060480 | + | + | − | − | + | − | − | − | + | − | − | − |
| 45 | ZmLTPg8 | Zm00001eb060490 | + | + | − | − | + | − | − | − | − | − | − | − |
| 46 | ZmLTPg9 | Zm00001eb061400 | − | + | − | − | + | + | − | − | − | − | − | + |
| 47 | ZmLTPg10 | Zm00001eb080620 | − | + | − | − | + | + | − | − | + | − | − | − |
| 48 | ZmLTPg11 | Zm00001eb082140 | − | − | − | − | + | − | − | − | + | + | − | − |
| 49 | ZmLTPg12 | Zm00001eb109750 | + | − | − | − | − | − | − | − | − | − | − | − |
| 50 | ZmLTPg13 | Zm00001eb117010 | + | + | − | − | + | − | − | − | − | − | − | − |
| 51 | ZmLTPg14 | Zm00001eb175270 | − | − | − | − | + | − | − | − | − | + | + | + |
| 52 | ZmLTPg15 | Zm00001eb183260 | − | + | − | − | + | − | − | − | − | − | + | − |
| 53 | ZmLTPg16 | Zm00001eb213110 | + | + | − | − | + | − | − | − | − | − | − | − |
| 54 | ZmLTPg17 | Zm00001eb225620 | + | + | + | + | + | + | − | + | + | + | + | + |
| 55 | ZmLTPg18 | Zm00001eb227760 | + | + | + | + | − | − | + | − | + | − | − | − |
| 56 | ZmLTPg19 | Zm00001eb273530 | + | + | + | + | + | + | + | + | + | + | + | + |
| 57 | ZmLTPg20 | Zm00001eb301090 | − | − | − | − | − | + | − | − | − | − | − | − |
| 58 | ZmLTPg21 | Zm00001eb301140 | + | + | − | − | + | − | − | − | + | − | − | − |
| 59 | ZmLTPg22 | Zm00001eb302760 | + | + | + | + | + | − | − | − | + | + | − | + |
| 60 | ZmLTPg23 | Zm00001eb327930 | − | + | − | − | − | + | + | − | − | − | − | − |
| 61 | ZmLTPg24 | Zm00001eb369130 | − | + | + | − | + | − | + | + | + | − | − | − |
| 62 | ZmLTPg25 | Zm00001eb401230 | + | + | + | + | + | − | − | − | + | + | − | + |
| 63 | ZmLTPg26 | Zm00001eb408970 | + | − | − | − | + | − | − | − | − | − | + | + |
| 64 | ZmLTPx1 | Zm00001eb306770 | − | − | − | − | + | − | − | − | − | − | − | − |
| 65 | ZmLTPx2 | Zm00001eb423010 | − | − | − | − | − | − | − | − | − | + | − | − |
| No. | LTP Transporters | Substrate(s) | Method of Substrate Identification | References |
|---|---|---|---|---|
| 1 | TaMs1 | Phospholipid | Purification of the fusion proteins of MBP-TaMs1-His in E. coli and protein-lipid overlay assay by PIP lipid strips and membrane lipid strips | [33,83] |
| 2 | OsLTPg29/OsLTPL94/OsEPAD1 | Phospholipid | Purification of the fusion proteins of MBP-OsLTPg29-His in E. coli and protein-lipid overlay assay by PIP lipid strips and membrane lipid strips | [33] |
| 3 | ZmLTPg11 | Phospholipid | Purification of the fusion proteins of MBP-ZmLTPg11-His in E. coli and protein-lipid overlay assay by PIP lipid strips and membrane lipid strips | [33] |
| 4 | AtDIR1 | Acid azelaic acid; phosphorylated sugar derivative glycerol- 3-phosphate | 14C-containing products measured by TLC method | [38] |
| 5 | AtAZI1 | |||
| 6 | OpsLTP1 | 16C and 18C fatty acids, linoleic acid | Quantification of total lipids by spectrophotometric methods | [67] |
| 7 | AtLTP3 | 16C and 18C fatty acids | Quantification of total lipids by spectrophotometric methods | [67] |
| 8 | Lc-LTP2 | FAs (C12-C22) and lysolipids | Molecular modeling, 2-p-toluidinonaphthalene-6-sulphonate (TNS) displacement and liposome leakage experiments | [84] |
| 9 | AtLTP1 | Cuticular wax | Substance analysis by using GC-MS system | [21] |
| 10 | AtLTP2 | Cuticular wax | Substance analysis by using GC-MS system | [60] |
| 11 | AtLTP3 | Fatty acid | Substance analysis by using GC-MS system | [19] |
| 12 | AtLTPG1 | Cuticular wax | Substance analysis by using GC-MS system | [27] |
| 13 | AtLTPG2 | Cuticular wax | Substance analysis by using GC-MS system | [23] |
| 14 | AtLTPG4 | Cuticular wax | Substance analysis by using GC-MS system | [17] |
| 15 | AtLTPG6 | Cuticular wax | Substance analysis by using GC-MS system | [17] |
| 16 | OsLTPL36 | Fatty acid | Substance analysis by using GC-MS system | [20] |
| 17 | OpsLTP1 | Fatty acid | Substance analysis by using GC-MS system | [67] |
| 18 | OsLTP5 | Fatty acid | Substance analysis by using GC-MS system | [26] |
| 19 | AtLTPG15 | Suberin monomer | Substance analysis by using GC-MS system | [61] |
| 20 | OsLTP47 | Fatty acid | Substance analysis by using GC-MS system | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, C.; Wu, S.; Li, Z.; Pan, S.; Wu, Y.; An, X.; Long, Y.; Wei, X.; Wan, X. A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize. Int. J. Mol. Sci. 2023, 24, 1660. https://doi.org/10.3390/ijms24021660
Fang C, Wu S, Li Z, Pan S, Wu Y, An X, Long Y, Wei X, Wan X. A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize. International Journal of Molecular Sciences. 2023; 24(2):1660. https://doi.org/10.3390/ijms24021660
Chicago/Turabian StyleFang, Chaowei, Suowei Wu, Ziwen Li, Shuangshuang Pan, Yuru Wu, Xueli An, Yan Long, Xun Wei, and Xiangyuan Wan. 2023. "A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize" International Journal of Molecular Sciences 24, no. 2: 1660. https://doi.org/10.3390/ijms24021660
APA StyleFang, C., Wu, S., Li, Z., Pan, S., Wu, Y., An, X., Long, Y., Wei, X., & Wan, X. (2023). A Systematic Investigation of Lipid Transfer Proteins Involved in Male Fertility and Other Biological Processes in Maize. International Journal of Molecular Sciences, 24(2), 1660. https://doi.org/10.3390/ijms24021660







