Proteome Mapping of Cervical Mucus and Its Potential as a Source of Biomarkers in Female Tract Disorders
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
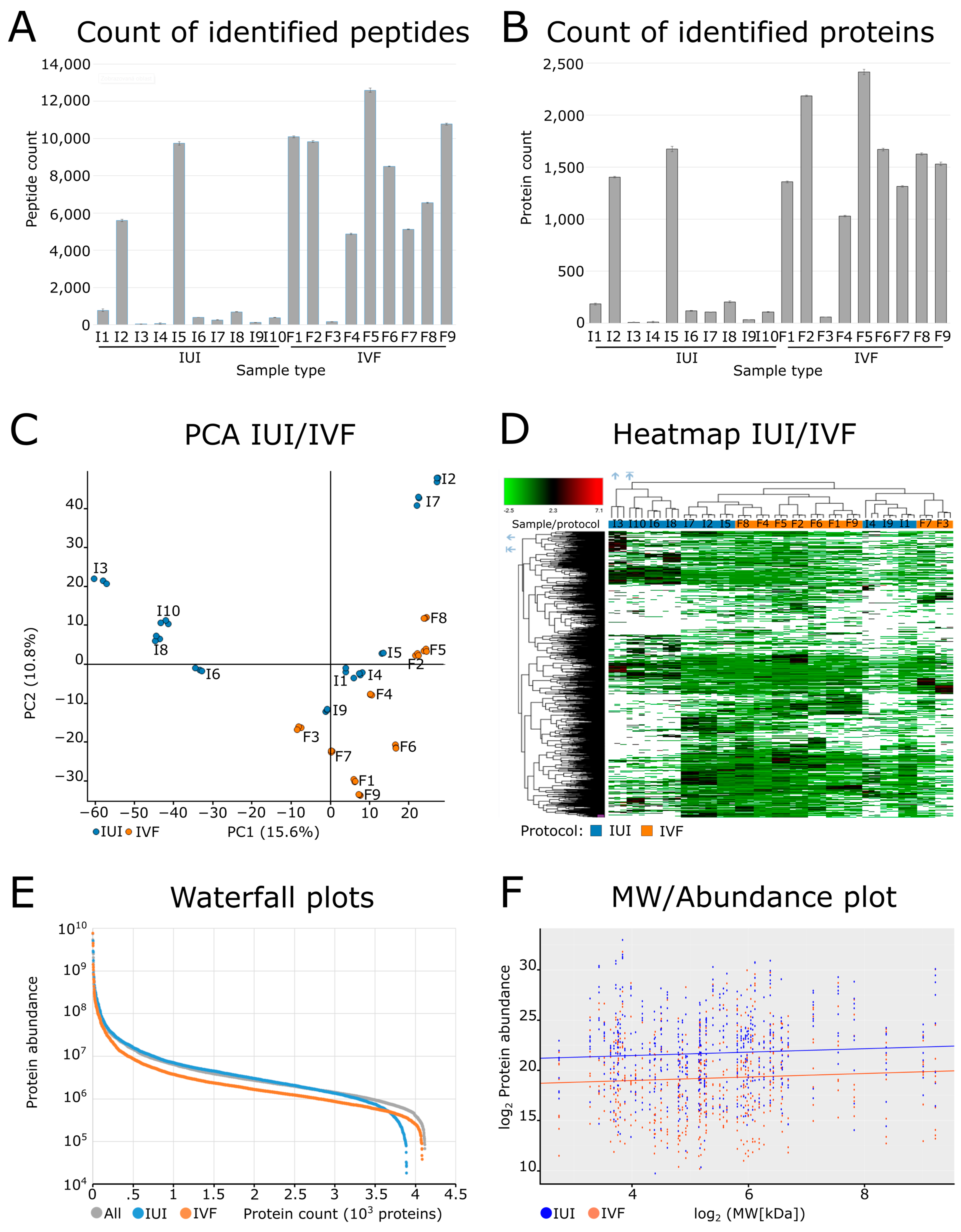
2.2. Quality Control of the Dataset
2.3. Proteomic Characterization of the Cervical Mucus
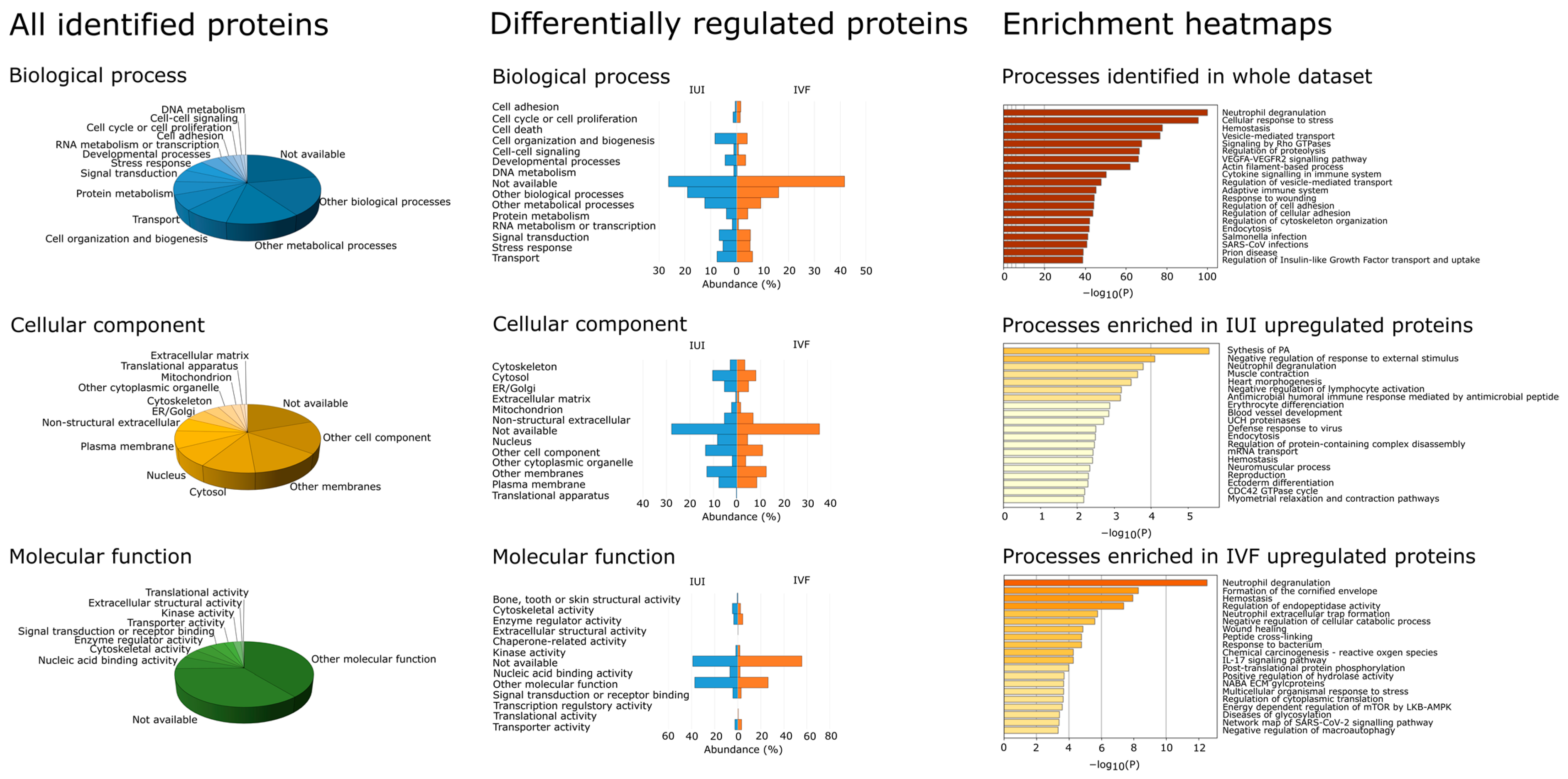
2.4. Protein Annotation of Identified Proteins
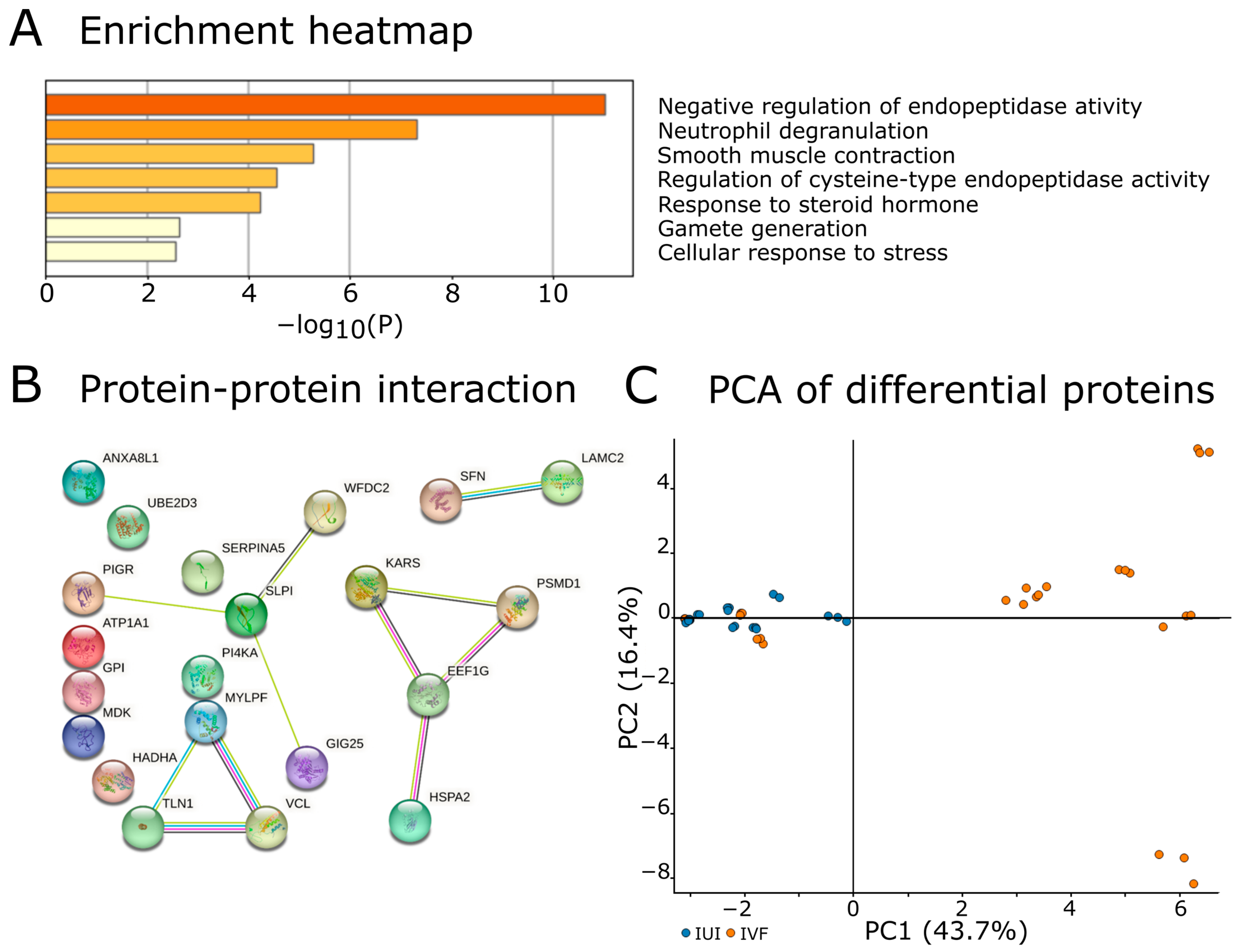
2.5. Proteins Discriminating IUI and IVF Groups
3. Discussion
3.1. Model System for Cervical Mucus Evaluation
3.2. Cervical Mucus Sampling
3.3. Proteomic Approaches in Cervical Mucus Analysis
3.4. Cervical Mucus Proteome
3.5. Differences between IUI and IVF
4. Materials and Methods
4.1. Patient Criteria
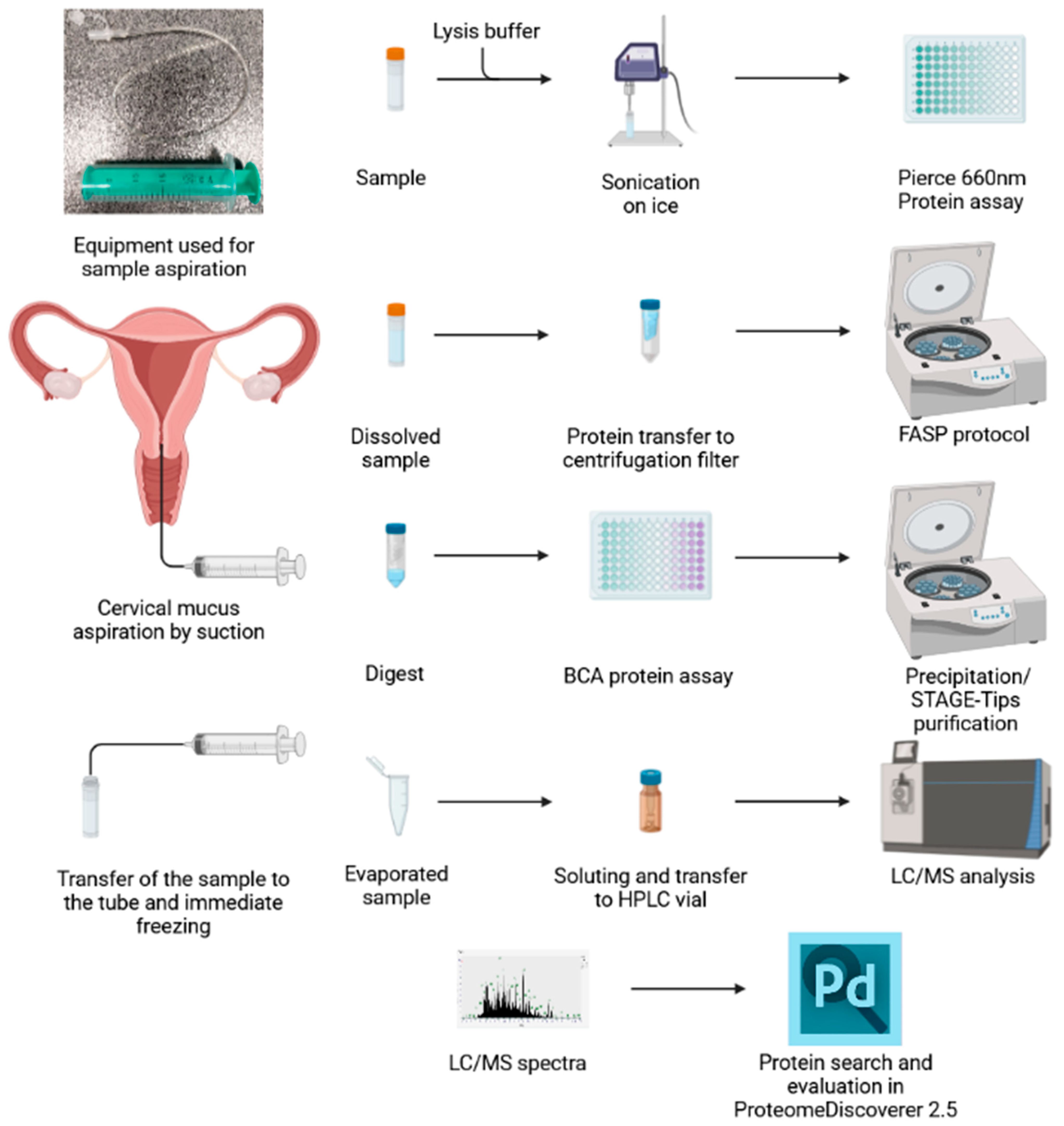
4.2. Sample Aspiration
4.3. Sample Dilution
4.4. Protein Digestion
4.5. Peptide Purification
4.6. LC/MS Analysis
4.7. Protein Search
4.8. Quality Control Metrics
4.9. Statistical Analysis
4.10. Bioinformatics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Katz, D.F. Human Cervical Mucus: Research Update. Am. J. Obstet. Gynecol. 1991, 165, 1984–1986. [Google Scholar] [CrossRef] [PubMed]
- Insler, V.; Melmed, H.; Eichenbrenner, I.; Serr, D.M.; Lunenfeld, B. The Cervical Score. Int. J. Gynecol. Obstet. 1972, 10, 223–228. [Google Scholar] [CrossRef]
- Van Kooij, R.J.; Roelofs, H.J.; Kathmann, G.A.; Kramer, M.F. Human Cervical Mucus and Its Mucous Glycoprotein during the Menstrual Cycle. Fertil. Steril. 1980, 34, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Bigelow, J.L.; Dunson, D.B.; Stanford, J.B.; Ecochard, R.; Gnoth, C.; Colombo, B. Mucus Observations in the Fertile Window: A Better Predictor of Conception than Timing of Intercourse. Hum. Reprod. Oxf. Engl. 2004, 19, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Kota, V.; Sundaram, C.S.; Deendayal, M.; Shivaji, S. Proteome of Human Endometrium: Identification of Differentially Expressed Proteins in Proliferative and Secretory Phase Endometrium. Proteom. Clin. Appl. 2010, 4, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Salamonsen, L.A.; Edgell, T.; Rombauts, L.J.F.; Stephens, A.N.; Robertson, D.M.; Rainczuk, A.; Nie, G.; Hannan, N.J. Proteomics of the Human Endometrium and Uterine Fluid: A Pathway to Biomarker Discovery. Fertil. Steril. 2013, 99, 1086–1092. [Google Scholar] [CrossRef]
- Sadler, T.W. Langman’s Medical Embryology; Williams & Wilkins: Baltimore, MD, USA, 1995; ISBN 978-0-683-07489-5. [Google Scholar]
- Andersch-Björkman, Y.; Thomsson, K.A.; Holmén Larsson, J.M.; Ekerhovd, E.; Hansson, G.C. Large Scale Identification of Proteins, Mucins, and Their O-Glycosylation in the Endocervical Mucus during the Menstrual Cycle. Mol. Cell. Proteom. MCP 2007, 6, 708–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panicker, G.; Lee, D.R.; Unger, E.R. Optimization of SELDI-TOF Protein Profiling for Analysis of Cervical Mucous. J. Proteom. 2009, 71, 637–646. [Google Scholar] [CrossRef]
- Grande, G.; Milardi, D.; Vincenzoni, F.; Pompa, G.; Biscione, A.; Astorri, A.L.; Fruscella, E.; De Luca, A.; Messana, I.; Castagnola, M.; et al. Proteomic Characterization of the Qualitative and Quantitative Differences in Cervical Mucus Composition during the Menstrual Cycle. Mol. Biosyst. 2015, 11, 1717–1725. [Google Scholar] [CrossRef]
- Grande, G.; Vincenzoni, F.; Milardi, D.; Pompa, G.; Ricciardi, D.; Fruscella, E.; Mancini, F.; Pontecorvi, A.; Castagnola, M.; Marana, R. Cervical Mucus Proteome in Endometriosis. Clin. Proteom. 2017, 14, 7. [Google Scholar] [CrossRef]
- Otani, S.; Fujii, T.; Kukimoto, I.; Yamamoto, N.; Tsukamoto, T.; Ichikawa, R.; Nishio, E.; Iwata, A. Cytokine Expression Profiles in Cervical Mucus from Patients with Cervical Cancer and Its Precursor Lesions. Cytokine 2019, 120, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Rocconi, R.P.; Wilhite, A.M.; Schambeau, L.; Scalici, J.; Pannell, L.; Finan, M.A. A Novel Proteomic-Based Screening Method for Ovarian Cancer Using Cervicovaginal Fluids: A Window into the Abdomen. Gynecol. Oncol. 2022, 164, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Finan, M.; Pannell, L.; Billheimer, D.; Schambeau, L.; Blandford, J.; Rocconi, R. A Novel Method of Screening for Endometrial Cancer. Gynecol. Oncol. 2012, 125, S165. [Google Scholar] [CrossRef]
- Simsek, E.; Haydardedeoglu, B.; Hacivelioglu, S.O.; Cok, T.; Parlakgumus, A.; Bagis, T. Effect of Cervical Mucus Aspiration before Intrauterine Insemination. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 2008, 103, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Massai, M.R.; de Ziegler, D.; Lesobre, V.; Bergeron, C.; Frydman, R.; Bouchard, P. Clomiphene Citrate Affects Cervical Mucus and Endometrial Morphology Independently of the Changes in Plasma Hormonal Levels Induced by Multiple Follicular Recruitment. Fertil. Steril. 1993, 59, 1179–1186. [Google Scholar] [CrossRef]
- Marchini, M.; Dorta, M.; Bombelli, F.; Ruspa, M.; Campana, A.; Dolcetta, G.; Radici, E. Effects of Clomiphene Citrate on Cervical Mucus: Analysis of Some Influencing Factors. Int. J. Fertil. 1989, 34, 154–159. [Google Scholar]
- Palomba, A.; Abbondio, M.; Fiorito, G.; Uzzau, S.; Pagnozzi, D.; Tanca, A. Comparative Evaluation of MaxQuant and Proteome Discoverer MS1-Based Protein Quantification Tools. J. Proteome Res. 2021, 20, 3497–3507. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A Subcellular Map of the Human Proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Shaw, J.L.V.; Smith, C.R.; Diamandis, E.P. Proteomic Analysis of Human Cervico-Vaginal Fluid. J. Proteome Res. 2007, 6, 2859–2865. [Google Scholar] [CrossRef]
- Tang, L.-J.; De Seta, F.; Odreman, F.; Venge, P.; Piva, C.; Guaschino, S.; Garcia, R.C. Proteomic Analysis of Human Cervical-Vaginal Fluids. J. Proteome Res. 2007, 6, 2874–2883. [Google Scholar] [CrossRef]
- Ma, Z.; Chen, J.; Luan, T.; Chu, C.; Wu, W.; Zhu, Y.; Gu, Y. Proteomic Analysis of Human Cervical Adenocarcinoma Mucus to Identify Potential Protein Biomarkers. PeerJ 2020, 8, e9527. [Google Scholar] [CrossRef] [PubMed]
- Maddison, J.W.; Rickard, J.P.; Bernecic, N.C.; Tsikis, G.; Soleilhavoup, C.; Labas, V.; Combes-Soia, L.; Harichaux, G.; Druart, X.; Leahy, T.; et al. Oestrus Synchronisation and Superovulation Alter the Cervicovaginal Mucus Proteome of the Ewe. J. Proteom. 2017, 155, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-C.; Hassan, S.S.; Romero, R.; Tarca, A.L.; Bhatti, G.; Gervasi, M.T.; Caruso, J.A.; Stemmer, P.M.; Kim, C.J.; Hansen, L.K.; et al. Protein Profiling Underscores Immunological Functions of Uterine Cervical Mucus Plug in Human Pregnancy. J. Proteom. 2011, 74, 817–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, L.; Park, D.; Reddy, A.; Wilmarth, P.A.; Jensen, J.T. Comparing Endocervical Mucus Proteome of Humans and Rhesus Macaques. Proteom. Clin. Appl. 2021, 15, e2100023. [Google Scholar] [CrossRef] [PubMed]
- Comparetto, C.; Borruto, F. Cervical Cancer Screening: A Never-Ending Developing Program. World J. Clin. Cases 2015, 3, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Rylova, G.; Ozdian, T.; Varanasi, L.; Soural, M.; Hlavac, J.; Holub, D.; Dzubak, P.; Hajduch, M. Affinity-Based Methods in Drug-Target Discovery. Curr. Drug Targets 2015, 16, 60–76. [Google Scholar] [CrossRef]
- Antharavally, B.S.; Mallia, K.A.; Rangaraj, P.; Haney, P.; Bell, P.A. Quantitation of Proteins Using a Dye-Metal-Based Colorimetric Protein Assay. Anal. Biochem. 2009, 385, 342–345. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal Sample Preparation Method for Proteome Analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Ozdian, T.; Holub, D.; Maceckova, Z.; Varanasi, L.; Rylova, G.; Rehulka, J.; Vaclavkova, J.; Slavik, H.; Moudry, P.; Znojek, P.; et al. Proteomic Profiling Reveals DNA Damage, Nucleolar and Ribosomal Stress Are the Main Responses to Oxaliplatin Treatment in Cancer Cells. J. Proteom. 2017, 162, 73–85. [Google Scholar] [CrossRef]
- Wiśniewski, J.R. Quantitative Evaluation of Filter Aided Sample Preparation (FASP) and Multienzyme Digestion FASP Protocols. Anal. Chem. 2016, 88, 5438–5443. [Google Scholar] [CrossRef]
- Erde, J.; Loo, R.R.O.; Loo, J.A. Enhanced FASP (EFASP) to Increase Proteome Coverage and Sample Recovery for Quantitative Proteomic Experiments. J. Proteome Res. 2014, 13, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Odeblad, E. The Discovery of Different Types of Cervical Mucus and the Billings Ovulation Method®. Bull. Ovul. Method Res. Ref. Cent. Aust. 1994, 21, 3–35. [Google Scholar]
- Barrios De Tomasi, J.; Opata, M.M.; Mowa, C.N. Immunity in the Cervix: Interphase between Immune and Cervical Epithelial Cells. J. Immunol. Res. 2019, 2019, 7693183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-Y.; Kannan, A.; Nunn, K.L.; Murphy, M.A.; Subramani, D.B.; Moench, T.; Cone, R.; Lai, S.K. IgG in Cervicovaginal Mucus Traps HSV and Prevents Vaginal Herpes Infections. Mucosal Immunol. 2014, 7, 1036–1044. [Google Scholar] [CrossRef] [Green Version]
- Hein, M.; Petersen, A.C.; Helmig, R.B.; Uldbjerg, N.; Reinholdt, J. Immunoglobulin Levels and Phagocytes in the Cervical Mucus Plug at Term of Pregnancy. Acta Obstet. Gynecol. Scand. 2005, 84, 734–742. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Z.; Xiong, M.; Luo, T.; Dong, X.; Huang, B.; Zhang, H.; Ai, J. Hormonal Replacement Treatment Improves Clinical Pregnancy in Frozen-Thawed Embryos Transfer Cycles: A Retrospective Cohort Study. Am. J. Transl. Res. 2013, 6, 85–90. [Google Scholar]
- De Sutter, P.; Veldeman, L.; Kok, P.; Szymczak, N.; Van der Elst, J.; Dhont, M. Comparison of Outcome of Pregnancy after Intra-Uterine Insemination (IUI) and IVF. Hum. Reprod. 2005, 20, 1642–1646. [Google Scholar] [CrossRef] [Green Version]
- Gekka, Y.; Nakagawa, K.; Watanabe, H.; Kuroda, K.; Horikawa, T.; Takamizawa, S.; Sugiyama, R. Comparison of Pregnancy Outcomes between Fresh Embryo Transfer in a Natural IVF Cycle and IUI Cycle Among Infertile Young Women. J. Reprod. Infertil. 2022, 23, 93–99. [Google Scholar] [CrossRef]
- von Wolff, M.; Fäh, M.; Roumet, M.; Mitter, V.; Stute, P.; Griesinger, G.; Kohl Schwartz, A. Thin Endometrium Is Also Associated with Lower Clinical Pregnancy Rate in Unstimulated Menstrual Cycles: A Study Based on Natural Cycle IVF. Front. Endocrinol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Bourgain, C.; Devroey, P. The Endometrium in Stimulated Cycles for IVF. Hum. Reprod. Update 2003, 9, 515–522. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.M.; Terry, V.; Hosie, M.J.; Gayer, N.; Murphy, C.R. Endometrial Response to IVF Hormonal Manipulation: Comparative Analysis of Menopausal, down Regulated and Natural Cycles. Reprod. Biol. Endocrinol. RBE 2004, 2, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.-Q.; Jin, L.-P. Ovarian Stimulation for in Vitro Fertilization Alters the Protein Profile Expression in Endometrial Secretion. Int. J. Clin. Exp. Pathol. 2013, 6, 1964. [Google Scholar]
- Han, L.; Padua, E.; Hart, K.D.; Edelman, A.; Jensen, J.T. Comparing Cervical Mucus Changes in Response to an Oral Progestin or Oestrogen Withdrawal in Ovarian-Suppressed Women: A Clinical Pilot. Eur. J. Contracept. Reprod. Health Care Off. J. Eur. Soc. Contracept. 2019, 24, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Natavio, M.F.; Taylor, D.; Lewis, R.A.; Blumenthal, P.; Felix, J.C.; Melamed, A.; Gentzschein, E.; Stanczyk, F.Z.; Mishell, D.R. Temporal Changes in Cervical Mucus after Insertion of the Levonorgestrel-Releasing Intrauterine System. Contraception 2013, 87, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Steward, R.; Melamed, A.; Granat, A.; Mishell, D.R. Comparison of Cervical Mucus of 24/4 vs. 21/7 Combined Oral Contraceptives. Contraception 2012, 86, 710–715. [Google Scholar] [CrossRef] [PubMed]
- Wira, C.R.; Rodriguez-Garcia, M.; Patel, M.V. The Role of Sex Hormones in Immune Protection of the Female Reproductive Tract. Nat. Rev. Immunol. 2015, 15, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Boomsma, C.M.; Kavelaars, A.; Eijkemans, M.J.C.; Amarouchi, K.; Teklenburg, G.; Gutknecht, D.; Fauser, B.J.C.M.; Heijnen, C.J.; Macklon, N.S. Cytokine Profiling in Endometrial Secretions: A Non-Invasive Window on Endometrial Receptivity. Reprod. Biomed. Online 2009, 18, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Devroey, P.; Aboulghar, M.; Garcia-Velasco, J.; Griesinger, G.; Humaidan, P.; Kolibianakis, E.; Ledger, W.; Tomás, C.; Fauser, B.C.J.M. Improving the Patient’s Experience of IVF/ICSI: A Proposal for an Ovarian Stimulation Protocol with GnRH Antagonist Co-Treatment. Hum. Reprod. Oxf. Engl. 2009, 24, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Filicori, M.; Cognigni, G.E.; Arnone, R.; Carbone, F.; Falbo, A.; Tabarelli, C.; Ciampaglia, W.; Casadio, P.; Spettoli, D.; Pecorari, R. Role of Different GnRH Agonist Regimens in Pituitary Suppression and the Outcome of Controlled Ovarian Hyperstimulation. Hum. Reprod. Oxf. Engl. 1996, 11 (Suppl. 3), 123–132. [Google Scholar] [CrossRef] [Green Version]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and Go Extraction Tips for Matrix-Assisted Laser Desorption/Ionization, Nanoelectrospray, and LC/MS Sample Pretreatment in Proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- The, M.; MacCoss, M.J.; Noble, W.S.; Käll, L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 2016, 27, 1719–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An Open Source Document Editor for Creating and Analyzing Targeted Proteomics Experiments. Bioinforma. Oxf. Engl. 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein-Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE Database and Related Tools and Resources in 2019: Improving Support for Quantification Data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
| Parameters | All Patients | IUI Group | IVF Group |
|---|---|---|---|
| Count of patients in the group | 19 | 10 | 9 |
| Age (years) | 32.1 (25–43) | 33.8 (26–43) | 30.2 (25–36) |
| BMI (kg/m2) | 25.1 (19–46) | 25.7 (19–46) | 24.5 (20–33) |
| AMH (ng/mL) | 3.8 (0.8–8.6) | 3.5 (0.8–6.8) | 4.2 (2.0–8.6) |
| FSH on the third day (IU/l) | 5.7 (2.7–10.9) | 6.9 (2.7–10.9) | 4.8 (4.1–6.5) |
| Infertility duration (years) | 3.0 (0–7) | 3.25 (1–6) | 2.6 (0–7) |
| Number of deliveries | 0.4 (0–2) | 0.6 (0–2) | 0.2 (0–1) |
| Number of abortions | 0 | 0 | 0 |
| Smoker (%) | 21 | 30 | 11 |
| Endometrium on the CM collection day (mm) | 9.4 (6–15) | 8.6 (6–15) | 10.3 (8–14) |
| Number of retrieved oocytes | 12.9 (3–23)/NA 1 | NA | 12.9 (3–23) |
| Pregnancy rate (%) | 26 | 10 | 44 |
| Protein | Uniprot ID | Gene Symbol | IUI | IVF | Log 2 Fold Change | Wilcox Test p-Value | Tissue Expression |
|---|---|---|---|---|---|---|---|
| WAP four-disulfide core domain protein 2 | Q14508 | WFDC2 | 22.23 (21.61–23.18) | 28.58 (28.07–30.04) | −6.35 | 0.00009 | Tissue enhanced (cervix, salivary gland) |
| Enoyl-CoA hydratase | H0YFD6 | HADHA | 27.4 (26.34–28.32) | 22.5 (21.77–24.36) | 4.9 | 0.00015 | Tissue enhanced (skeletal muscle) |
| Dopamine receptor interacting protein 4 | Q4W4Y1 | DRIP4 | 24.65 (23.99–29.22) | 21.67 (21.64–22.17) | 2.98 | 0.00026 | N/A |
| 26S proteasome non-ATPase regulatory subunit 1 | Q99460 | PSMD1 | 20.81 (20.02–21.45) | 23.23 (22.66–23.39) | −2.42 | 0.00097 | Low tissue specificity |
| Serpin family A member 3 | A0A024R6P0 | SERPINA3 | 23.81 (22.91–24.6) | 26.63 (25.33–26.82) | −2.82 | 0.00097 | Group enriched (liver, pancreas) |
| Connective tissue growth factor | Q5M8T4 | CTGF | 24.38 (22.83–25.49) | 20.38 (20.24–20.76) | 4 | 0.00145 | N/A |
| Glucose-6-phosphate isomerase | P06744 | GPI | 24.68 (23.75–31.52) | 22.16 (21.69–22.44) | 2.52 | 0.00145 | Low tissue specificity |
| Laminin subunit gamma-2 | Q13753 | LAMC2 | 25.02 (23.24–25.35) | 21.37 (21.04–21.54) | 3.65 | 0.00210 | Tissue enhanced (urinary bladder) |
| Serpin family A member 5 | P05154 | SERPINA5 | 23.34 (22.86–24.34) | 26.11 (24.62–26.71) | −2.77 | 0.00299 | Tissue enhanced (adrenal gland, liver, testis) |
| Phosphatidylinositol 4-kinase alpha | P42356 | PI4KA | 22.81 (21.58–23.46) | 20.85 (20.43–21.29) | 1.96 | 0.00299 | Low tissue specificity |
| Antileukoproteinase | P03973 | SLPI | 29.09 (28.37–30.85) | 31.87 (31.66–32.18) | −2.78 | 0.00567 | Group enriched (cervix, salivary gland) |
| Stratifin | P31947 | SFN | 22.25 (20.94–23) | 24.7 (23.61–25.3) | −2.45 | 0.00567 | Group enriched (esophagus, skin, vagina) |
| Polymeric immunoglobulin receptor | P01833 | PIGR | 26.09 (25.25–27.66) | 29.5 (28.89–29.86) | −3.41 | 0.00567 | Tissue enhanced (intestine, salivary gland) |
| Midkine | P21741 | MDK | 21.56 (21.19–23.06) | 24.15 (23.88–24.52) | −2.59 | 0.00762 | Tissue enhanced (ovary) |
| Complement factor D | Q6FHW3 | DF | 22.67 (21.52–23.63) | 27.12 (23.95–27.79) | −4.45 | 0.01013 | N/A |
| Heat shock-related 70 kDa protein 2 | P54652 | HSPA2 | 22.49 (21.58–23.44) | 20.81 (20.43–21.49) | 1.68 | 0.01013 | Tissue enhanced (brain, skeletal muscle) |
| Elongation factor 1-gamma | P26641 | EEF1G | 25.04 (23.62–25.65) | 21.2 (19.37–23.49) | 3.84 | 0.01013 | Low tissue specificity |
| FLJ00385 protein | Q8NF17 | FLJ00385 | 28.94 (26.87–29.81) | 26.31 (24.04–26.89) | 2.63 | 0.01013 | N/A |
| Cystatin C | A0A0K0K1J1 | CSTS3 | 24.16 (23.61–25.99) | 28.73 (27.02–28.83) | −4.57 | 0.01327 | Tissue enhanced (brain) |
| Ubiquitin-conjugating enzyme E2D 3 | A0A024RDH2 | UBE2D3 | 24.12 (23.36–25.01) | 21.1 (19.86–22.89) | 3.02 | 0.01327 | Low tissue specificity |
| Immunoglobulin delta heavy chain | P0DOX3 | N/A | 23.74 (22.24–27) | 21.19 (20.28–21.82) | 2.55 | 0.01327 | N/A |
| Lysine--tRNA ligase | Q15046 | KARS1 | 21.5 (20.19–22.1) | 23.81 (21.76–24.82) | −2.31 | 0.01721 | Low tissue specificity |
| Talin-1 | Q9Y490 | TLN1 | 22.05 (21.48–23.09) | 23.87 (22.72–24.8) | −1.82 | 0.02202 | Low tissue specificity |
| Myosin regulatory light chain 11 | Q96A32 | MYL11 | 22.61 (21–23.08) | 20.21 (19.59–21.23) | 2.4 | 0.02202 | Group enriched (skeletal muscle, tongue) |
| Dermcidin | P81605-2 | DCD | 24.36 (22.79–27.5) | 22.47 (21.57–23.03) | 1.89 | 0.02202 | Tissue enriched (skin) |
| Annexin A8-like protein 1 | Q5VT79 | ANXA8L1 | 22.27 (21.4–25.36) | 20.01 (19.56–21.26) | 2.26 | 0.02793 | Group enriched (esophagus, skin, vagina) |
| Sodium/potassium-transporting ATPase subunit alpha-1 | P05023 | ATP1A1 | 22.94 (22.24–23.32) | 21.3 (21.09–22.28) | 1.64 | 0.03499 | Tissue enhanced (parathyroid gland) |
| Vinculin | A0A024QZN4 | VCL | 22.02 (21.77–22.82) | 23.63 (22.4–24.42) | −1.61 | 0.04347 | Low tissue specificity |
| Keratin 13 | A1A4E9 | KRT13 | 24.18 (23.3–24.74) | 27.66 (26.01–28.76) | −3.48 | 0.04347 | Tissue enhanced (esophagus, vagina) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oždian, T.; Vodička, J.; Dostál, J.; Holub, D.; Václavková, J.; Ješeta, M.; Hamerníková, B.; Kouřilová, P.; Malchar, O.; Dvořák, V.; et al. Proteome Mapping of Cervical Mucus and Its Potential as a Source of Biomarkers in Female Tract Disorders. Int. J. Mol. Sci. 2023, 24, 1038. https://doi.org/10.3390/ijms24021038
Oždian T, Vodička J, Dostál J, Holub D, Václavková J, Ješeta M, Hamerníková B, Kouřilová P, Malchar O, Dvořák V, et al. Proteome Mapping of Cervical Mucus and Its Potential as a Source of Biomarkers in Female Tract Disorders. International Journal of Molecular Sciences. 2023; 24(2):1038. https://doi.org/10.3390/ijms24021038
Chicago/Turabian StyleOždian, Tomáš, Jan Vodička, Jiří Dostál, Dušan Holub, Jana Václavková, Michal Ješeta, Barbora Hamerníková, Pavla Kouřilová, Ondřej Malchar, Vladimír Dvořák, and et al. 2023. "Proteome Mapping of Cervical Mucus and Its Potential as a Source of Biomarkers in Female Tract Disorders" International Journal of Molecular Sciences 24, no. 2: 1038. https://doi.org/10.3390/ijms24021038





