Gene Therapy in ALS and SMA: Advances, Challenges and Perspectives
Abstract
1. Introduction
2. Spinal Muscular Atrophy
2.1. Nusinersen
2.2. Gene Replacement Therapy
2.3. Risdiplam
2.4. Challenges and Future Directions
3. Amyotrophic Lateral Sclerosis
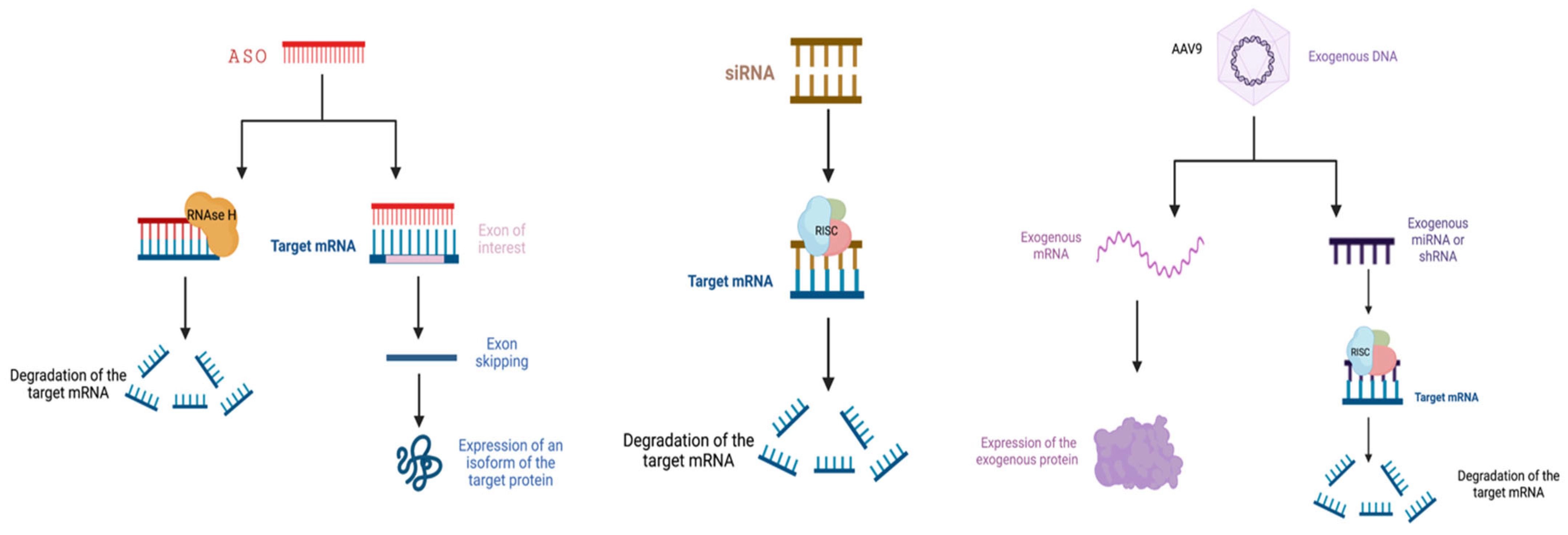
3.1. RNA Interference
3.2. Antisense Oligonucleotides
3.3. Viral Vectors in ALS
3.3.1. Lentiviral Vectors
3.3.2. Adeno-Associated Virus Vectors
3.4. Genome Editing
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tosolini, A.P.; Sleigh, J.N. Motor Neuron Gene Therapy: Lessons from Spinal Muscular Atrophy for Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2017, 10, 405. [Google Scholar] [CrossRef]
- Zakharova, M. Modern Approaches in Gene Therapy of Motor Neuron Diseases. Med. Res. Rev. 2021, 41, 2634–2655. [Google Scholar] [CrossRef]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, Biodistribution and Cell Uptake of Antisense Oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef]
- Sardone, V.; Zhou, H.; Muntoni, F.; Ferlini, A.; Falzarano, M.S. Antisense Oligonucleotide-Based Therapy for Neuromuscular Disease. Molecules 2017, 22, 563. [Google Scholar] [CrossRef]
- Chi, X.; Gatti, P.; Papoian, T. Safety of Antisense Oligonucleotide and SiRNA-Based Therapeutics. Drug Discov. Today 2017, 22, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hu, Y.; Ju, D. Gene Therapy for Neurodegenerative Disorders: Advances, Insights and Prospects. Acta Pharm. Sin. B 2020, 10, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Liu, Y.; Noor, A.; Tran, J.; Li, R. Characteristics and Advantages of Adeno-Associated Virus Vector-Mediated Gene Therapy for Neurodegenerative Diseases. Neural Regen. Res. 2019, 14, 931. [Google Scholar] [CrossRef] [PubMed]
- Kolb, S.J.; Kissel, J.T. Spinal Muscular Atrophy. Neurol. Clin. 2015, 33, 831–846. [Google Scholar] [CrossRef]
- Burghes, A.H.M.; Beattie, C.E. Spinal Muscular Atrophy: Why Do Low Levels of Survival Motor Neuron Protein Make Motor Neurons Sick? Nat. Rev. Neurosci. 2009, 10, 597–609. [Google Scholar] [CrossRef]
- Rossoll, W.; Bassell, G.J. Spinal Muscular Atrophy and a Model for Survival of Motor Neuron Protein Function in Axonal Ribonucleoprotein Complexes. Results Probl. Cell Differ. 2009, 48, 289–326. [Google Scholar] [CrossRef]
- Anderton, R.S.; Meloni, B.P.; Mastaglia, F.L.; Boulos, S. Spinal Muscular Atrophy and the Antiapoptotic Role of Survival of Motor Neuron (SMN) Protein. Mol. Neurobiol. 2013, 47, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Jędrzejowska, M. Rdzeniowy Zanik Mięśni—Deficyt Białka SMN [Spinal Muscular Atrophy: SMN Protein Deficiency]. Neurol. Neurochir. Pol. 2001, 35, 289–297. [Google Scholar] [PubMed]
- Hensel, N.; Brickwedde, H.; Tsaknakis, K.; Grages, A.; Braunschweig, L.; Lüders, K.A.; Lorenz, H.M.; Lippross, S.; Walter, L.M.; Tavassol, F.; et al. Altered Bone Development with Impaired Cartilage Formation Precedes Neuromuscular Symptoms in Spinal Muscular Atrophy. Hum. Mol. Genet. 2020, 29, 2662–2673. [Google Scholar] [CrossRef]
- Jędrzejowska, M.; Milewski, M.; Zimowski, J.; Borkowska, J.; Kostera-Pruszczyk, A.; Sielska, D.; Jurek, M.; Hausmanowa-Petrusewicz, I. Phenotype Modifiers of Spinal Muscular Atrophy: The Number of SMN2 Gene Copies, Deletion in the NAIP Gene and Probably Gender Influence the Course of the Disease. Acta Biochim. Pol. 2009, 56, 103–108. [Google Scholar] [CrossRef]
- Chen, T.H. New and Developing Therapies in Spinal Muscular Atrophy: From Genotype to Phenotype to Treatment and Where Do We Stand? Int. J. Mol. Sci. 2020, 21, 3297. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, N.N. Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes. Adv. Neurobiol. 2018, 20, 31–61. [Google Scholar] [CrossRef] [PubMed]
- Lejman, J.; Zieliński, G.; Gawda, P.; Lejman, M. Alternative Splicing Role in New Therapies of Spinal Muscular Atrophy. Genes 2021, 12, 1346. [Google Scholar] [CrossRef]
- Somers, E.; Lees, R.D.; Hoban, K.; Sleigh, J.N.; Zhou, H.; Muntoni, F.; Talbot, K.; Gillingwater, T.H.; Parson, S.H. Vascular Defects and Spinal Cord Hypoxia in Spinal Muscular Atrophy. Ann. Neurol. 2016, 79, 217–230. [Google Scholar] [CrossRef]
- Pedrotti, S.; Bielli, P.; Paronetto, M.P.; Ciccosanti, F.; Fimia, G.M.; Stamm, S.; Manley, J.L.; Sette, C. The Splicing Regulator Sam68 Binds to a Novel Exonic Splicing Silencer and Functions in SMN2 Alternative Splicing in Spinal Muscular Atrophy. EMBO J. 2010, 29, 1235–1247. [Google Scholar] [CrossRef]
- Wee, C.D.; Havens, M.A.; Jodelka, F.M.; Hastings, M.L. Targeting SR Proteins Improves SMN Expression in Spinal Muscular Atrophy Cells. PLoS ONE 2014, 9, e115205. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chang, J.G.; Liu, T.Y.; Jong, Y.J.; Cheng, W.L.; Yuo, C.Y. Securinine Enhances SMN2 Exon 7 Inclusion in Spinal Muscular Atrophy Cells. Biomed. Pharmacother. 2017, 88, 708–714. [Google Scholar] [CrossRef]
- Garcia-Lopez, A.; Tessaro, F.; Jonker, H.R.A.; Wacker, A.; Richter, C.; Comte, A.; Berntenis, N.; Schmucki, R.; Hatje, K.; Petermann, O.; et al. Targeting RNA Structure in SMN2 Reverses Spinal Muscular Atrophy Molecular Phenotypes. Nat. Commun. 2018, 9, 2032. [Google Scholar] [CrossRef] [PubMed]
- Claborn, M.K.; Stevens, D.L.; Walker, C.K.; Gildon, B.L. Nusinersen: A Treatment for Spinal Muscular Atrophy. Ann. Pharm. 2019, 53, 61–69. [Google Scholar] [CrossRef]
- Mercuri, E.; Lucibello, S.; Perulli, M.; Coratti, G.; de Sanctis, R.; Pera, M.C.; Pane, M.; Montes, J.; de Vivo, D.C.; Darras, B.T.; et al. Longitudinal Natural History of Type i Spinal Muscular Atrophy: A Critical Review. Orphanet. J. Rare Dis. 2020, 15, 84. [Google Scholar] [CrossRef]
- Bérard, C.; Payan, C.; Hodgkinson, I.; Fermanian, J. A Motor Function Measure for Neuromuscular Diseases. Construction and Validation Study. Neuromuscul. Disord. 2005, 15, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, D.; Scoto, M.; Mayhew, A.; Main, M.; Mazzone, E.S.; Montes, J.; de Sanctis, R.; Young, S.D.; Salazar, R.; Glanzman, A.M.; et al. Revised Hammersmith Scale for Spinal Muscular Atrophy: A SMA Specific Clinical Outcome Assessment Tool. PLoS ONE 2017, 12, e0172346. [Google Scholar] [CrossRef]
- Frederiksen, S.B.; Holm, L.L.; Larsen, M.R.; Doktor, T.K.; Andersen, H.S.; Hastings, M.L.; Hua, Y.; Krainer, A.R.; Andresen, B.S. Identification of SRSF10 as a Regulator of SMN2 ISS-N1. Hum. Mutat. 2021, 42, 246–260. [Google Scholar] [CrossRef]
- Messina, S.; Sframeli, M. New Treatments in Spinal Muscular Atrophy: Positive Results and New Challenges. J. Clin. Med. 2020, 9, 2222. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.K.; Kapp, D.; Schroth, M. Gene Therapy for Spinal Muscular Atrophy: An Emerging Treatment Option for a Devastating Disease. J. Manag. Care Spec. Pharm. 2018, 24, S3–S16. [Google Scholar] [CrossRef]
- Finkel, R.S.; Chiriboga, C.A.; Vajsar, J.; Day, J.W.; Montes, J.; de Vivo, D.C.; Yamashita, M.; Rigo, F.; Hung, G.; Schneider, E.; et al. Treatment of Infantile-Onset Spinal Muscular Atrophy with Nusinersen: A Phase 2, Open-Label, Dose-Escalation Study. Lancet 2016, 388, 3017–3026. [Google Scholar] [CrossRef] [PubMed]
- A Study of Multiple Doses of Nusinersen (ISIS 396443) Delivered to Infants with Genetically Diagnosed and Presymptomatic Spinal Muscular Atrophy. Available online: https://clinicaltrials.gov/ct2/show/NCT02386553 (accessed on 20 December 2022).
- Finkel, R.S.; Mercuri, E.; Darras, B.T.; Connolly, A.M.; Kuntz, N.L.; Kirschner, J.; Chiriboga, C.A.; Saito, K.; Servais, L.; Tizzano, E.; et al. Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, E.S.; Mayhew, A.; Montes, J.; Ramsey, D.; Fanelli, L.; Young, S.D.; Salazar, R.; de Sanctis, R.; Pasternak, A.; Glanzman, A.; et al. Revised Upper Limb Module for Spinal Muscular Atrophy: Development of a New Module. Muscle Nerve 2017, 55, 869–874. [Google Scholar] [CrossRef]
- Bonanno, S.; Marcuzzo, S.; Malacarne, C.; Giagnorio, E.; Masson, R.; Zanin, R.; Arnoldi, M.T.; Andreetta, F.; Simoncini, O.; Venerando, A.; et al. Circulating MyomiRs as Potential Biomarkers to Monitor Response to Nusinersen in Pediatric SMA Patients. Biomedicines 2020, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Bonanno, S.; Cavalcante, P.; Salvi, E.; Giagnorio, E.; Malacarne, C.; Cattaneo, M.; Andreetta, F.; Venerando, A.; Pensato, V.; Gellera, C.; et al. Identification of a Cytokine Profile in Serum and Cerebrospinal Fluid of Pediatric and Adult Spinal Muscular Atrophy Patients and Its Modulation upon Nusinersen Treatment. Front. Cell. Neurosci. 2022, 16, 437. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Samulski, R.J.; Muzyczka, N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu. Rev. Virol. 2014, 1, 427–451. [Google Scholar] [CrossRef]
- Blessing, D.; Déglon, N. Adeno-Associated Virus and Lentivirus Vectors: A Refined Toolkit for the Central Nervous System. Curr. Opin. Virol. 2016, 21, 61–66. [Google Scholar] [CrossRef]
- Lykken, E.A.; Shyng, C.; Edwards, R.J.; Rozenberg, A.; Gray, S.J. Recent Progress and Considerations for AAV Gene Therapies Targeting the Central Nervous System. J. Neurodev. Disord. 2018, 10, 16. [Google Scholar] [CrossRef]
- Gray, S.J.; Woodard, K.T.; Samulski, R.J. Viral Vectors and Delivery Strategies for CNS Gene Therapy. Ther. Deliv. 2010, 1, 517–534. [Google Scholar] [CrossRef]
- Shevtsova, Z.; Malik, J.M.I.; Michel, U.; Bähr, M.; Kügler, S. Promoters and Serotypes: Targeting of Adeno-Associated Virus Vectors for Gene Transfer in the Rat Central Nervous System in Vitro and in Vivo. Exp. Physiol. 2005, 90, 53–59. [Google Scholar] [CrossRef]
- Jara, J.H.; Stanford, M.J.; Zhu, Y.; Tu, M.; Hauswirth, W.W.; Bohn, M.C.; Devries, S.H.; Özdinler, P.H. Healthy and Diseased Corticospinal Motor Neurons Are Selectively Transduced upon Direct AAV2-2 Injection into the Motor Cortex. Gene Ther. 2016, 23, 272–282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Colella, P.; Ronzitti, G.; Mingozzi, F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018, 8, 87–104. [Google Scholar] [CrossRef] [PubMed]
- Pena, S.A.; Iyengar, R.; Eshraghi, R.S.; Bencie, N.; Mittal, J.; Aljohani, A.; Mittal, R.; Eshraghi, A.A. Gene Therapy for Neurological Disorders: Challenges and Recent Advancements. J. Drug Target. 2020, 28, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R. Onasemnogene Abeparvovec for Spinal Muscular Atrophy: The Costlier Drug Ever. Int. J. Appl. Basic Med. Res. 2019, 9, 127. [Google Scholar] [CrossRef]
- Day, J.W.; Finkel, R.S.; Chiriboga, C.A.; Connolly, A.M.; Crawford, T.O.; Darras, B.T.; Iannaccone, S.T.; Kuntz, N.L.; Peña, L.D.M.; Shieh, P.B.; et al. Onasemnogene Abeparvovec Gene Therapy for Symptomatic Infantile-Onset Spinal Muscular Atrophy in Patients with Two Copies of SMN2 (STR1VE): An Open-Label, Single-Arm, Multicentre, Phase 3 Trial. Lancet Neurol. 2021, 20, 284–293. [Google Scholar] [CrossRef]
- Al-Zaidy, S.A.; Mendell, J.R. From Clinical Trials to Clinical Practice: Practical Considerations for Gene Replacement Therapy in SMA Type 1. Pediatr. Neurol. 2019, 100, 3–11. [Google Scholar] [CrossRef]
- Meyer, K.; Ferraiuolo, L.; Schmelzer, L.; Braun, L.; McGovern, V.; Likhite, S.; Michels, O.; Govoni, A.; Fitzgerald, J.; Morales, P.; et al. Improving Single Injection CSF Delivery of AAV9-Mediated Gene Therapy for SMA: A Dose-Response Study in Mice and Nonhuman Primates. Mol. Ther. 2015, 23, 477–487. [Google Scholar] [CrossRef]
- Friese, J.; Geitmann, S.; Holzwarth, D.; Müller, N.; Sassen, R.; Baur, U.; Adler, K.; Kirschner, J. Safety Monitoring of Gene Therapy for Spinal Muscular Atrophy with Onasemnogene Abeparvovec—A Single Centre Experience. J. Neuromuscul. Dis. 2021, 8, 209–216. [Google Scholar] [CrossRef]
- van Alstyne, M.; Tattoli, I.; Delestrée, N.; Recinos, Y.; Workman, E.; Shihabuddin, L.S.; Zhang, C.; Mentis, G.Z.; Pellizzoni, L. Gain of Toxic Function by Long-Term AAV9-Mediated SMN Overexpression in the Sensorimotor Circuit. Nat. Neurosci. 2021, 24, 930–940. [Google Scholar] [CrossRef]
- Mendell, J.R.; Al-Zaidy, S.A.; Lehman, K.J.; McColly, M.; Lowes, L.P.; Alfano, L.N.; Reash, N.F.; Iammarino, M.A.; Church, K.R.; Kleyn, A.; et al. Five-Year Extension Results of the Phase 1 START Trial of Onasemnogene Abeparvovec in Spinal Muscular Atrophy. JAMA Neurol. 2021, 78, 834–841. [Google Scholar] [CrossRef]
- Dhillon, S. Risdiplam: First Approval. Drugs 2020, 80, 1853–1858. [Google Scholar] [CrossRef] [PubMed]
- FDA; CDER. Highlights of Prescribing Information: EVRYSDI (Risdisplam). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213535s003s005lbl.pdf (accessed on 24 August 2022).
- Sivaramakrishnan, M.; McCarthy, K.D.; Campagne, S.; Huber, S.; Meier, S.; Augustin, A.; Heckel, T.; Meistermann, H.; Hug, M.N.; Birrer, P.; et al. Binding to SMN2 Pre-MRNA-Protein Complex Elicits Specificity for Small Molecule Splicing Modifiers. Nat. Commun. 2017, 8, 1476. [Google Scholar] [CrossRef]
- FDA Genentech Inc. EvrysdiTM (Risdiplam): US Prescribing Information. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/213535s003s005lbl.pdf (accessed on 5 January 2023).
- Wang, J.; Schultz, P.G.; Johnson, K.A. Mechanistic Studies of a Small-Molecule Modulator of SMN2 Splicing. Proc. Natl. Acad. Sci. USA 2018, 115, E4604–E4612. [Google Scholar] [CrossRef]
- Poirier, A.; Weetall, M.; Heinig, K.; Bucheli, F.; Schoenlein, K.; Alsenz, J.; Bassett, S.; Ullah, M.; Senn, C.; Ratni, H.; et al. Risdiplam Distributes and Increases SMN Protein in Both the Central Nervous System and Peripheral Organs. Pharmacol. Res. Perspect. 2018, 6, e00447. [Google Scholar] [CrossRef] [PubMed]
- Baranello, G.; Servais, L.; Day, J.; Deconinck, N.; Mercuri, E.; Klein, A.; Darras, B.; Masson, R.; Kletzl, H.; Cleary, Y.; et al. P.353FIREFISH Part 1: 16-Month Safety and Exploratory Outcomes of Risdiplam (RG7916) Treatment in Infants with Type 1 Spinal Muscular Atrophy. Neuromuscul. Disord. 2019, 29, S184. [Google Scholar] [CrossRef]
- Kolb, S.J.; Coffey, C.S.; Yankey, J.W.; Krosschell, K.; Arnold, W.D.; Rutkove, S.B.; Swoboda, K.J.; Reyna, S.P.; Sakonju, A.; Darras, B.T.; et al. Natural History of Infantile-Onset Spinal Muscular Atrophy. Ann. Neurol. 2017, 82, 883–891. [Google Scholar] [CrossRef]
- Servais, L.; Baranello, G.; Masson, R.; Mazurkiewicz-Bełdzińska, M.; Rose, K.; Vlodavets, D.; Xiong, H.; Zanoteli, E.; El-Khairi, M.; Fuerst-Recktenwald, S.; et al. FIREFISH Part 2: Efficacy and Safety of Risdiplam (RG7916) in Infants with Type 1 Spinal Muscular Atrophy (SMA). Eur. Respir. J. 2020, 56 (Suppl. S64), 117. [Google Scholar] [CrossRef]
- PTC Therapeutics Announces 2-Year Data from Part 1 of SUNFISH and New Data from JEWELFISH Trials for Risdiplam in Patients with Spinal Muscular Atrophy|PTC Therapeutics, Inc. Available online: https://ir.ptcbio.com/news-releases/news-release-details/ptc-therapeutics-announces-2-year-data-part-1-sunfish-and-new (accessed on 22 December 2022).
- Mercuri, E.; Barisic, N.; Boespflug-Tanguy, O.; Deconinck, N.; Kostera-Pruszczyk, A.; Masson, R.; Mazzone, E.; Nascimento, A.; Saito, K.; Vlodavets, D.; et al. SUNFISH Part 2: Efficacy and Safety of Risdiplam (RG7916) in Patients with Type 2 or Non-Ambulant Type 3 Spinal Muscular Atrophy (SMA) (1260). Neurology. 2020, 94 (Suppl. S15). Available online: https://n.neurology.org/content/94/15_Supplement/1260 (accessed on 24 August 2022).
- Chiriboga, C.A.; Mercuri, E.; Fischer, D.; Kraus, D.; Yeung, W.Y.; Kletzl, H.; Gerber, M.; Cleary, Y.; Gorni, K.; Khwaja, O. JEWELFISH: Risdiplam (RG7916) Increases SMN Protein in Non-Naïve Patients with SMA. Available online: https://medically.roche.com/content/dam/pdmahub/non-restricted/neurology/wms-2018/WMS_2018_JEWELFISH_risdiplam_poster_Chiriboga.pdf (accessed on 24 August 2022).
- Finkel, R.S.; Al-Muhaizea, M.; Farrar, M.A.; Nelson, L.; Prufer, A.; Servais, L.; Wang, Y.; Zanoteli, E.; Palfreeman, L.; El-Khairi, M.; et al. RAINBOWFISH: A Study of Risdiplam in Newborns with Presymptomatic Spinal Muscular Atrophy (SMA). In Proceedings of the MDA Clinical & Scientific Conference, Dallas, TX, USA, 19–22 March 2023; Available online: https://www.mdaconference.org/abstract-library/rainbowfish-a-study-of-risdiplam-in-newborns-with-presymptomatic-spinal-muscular-atrophy-sma/ (accessed on 22 December 2022).
- Erdos, J.; Wild, C. Mid- and long-term (at least 12 months) follow-up of patients with spinal muscular atrophy (SMA) treated with nusinersen, onasemnogene abeparvovec, risdiplam or combination therapies: A systematic review of real-world study data. Eur. J. Paediatr. Neurol. 2022, 39, 1–10. [Google Scholar] [CrossRef]
- Calder, A.N.; Androphy, E.J.; Hodgetts, K.J. Small Molecules in Development for the Treatment of Spinal Muscular Atrophy. J. Med. Chem. 2016, 59, 10067–10083. [Google Scholar] [CrossRef]
- Peters, T.; Deconinck, N.; Valentin, M.; Kieloch, A.; Theil, D.; de Pommerol, J.H.P. 273Serum Neurofilament Light Chain in Type 1 Spinal Muscular Atrophy: 30 Months Data from First Part of a Branaplam Phase II Study. Neuromuscul. Disord. 2019, 29, S147. [Google Scholar] [CrossRef]
- d’Ydewalle, C.; Ramos, D.M.; Pyles, N.J.; Ng, S.Y.; Gorz, M.; Pilato, C.M.; Ling, K.; Kong, L.; Ward, A.J.; Rubin, L.L.; et al. The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy. Neuron 2017, 93, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Hazell, G.; Shabanpoor, F.; Saleh, A.F.; Bowerman, M.; Sleigh, J.N.; Meijboom, K.E.; Zhou, H.; Muntoni, F.; Talbot, K.; et al. Systemic Peptide-Mediated Oligonucleotide Therapy Improves Long-Term Survival in Spinal Muscular Atrophy. Proc. Natl. Acad. Sci. USA 2016, 113, 10962–10967. [Google Scholar] [CrossRef] [PubMed]
- Torres-Benito, L.; Schneider, S.; Rombo, R.; Ling, K.K.; Grysko, V.; Upadhyay, A.; Kononenko, N.L.; Rigo, F.; Bennett, C.F.; Wirth, B. NCALD Antisense Oligonucleotide Therapy in Addition to Nusinersen Further Ameliorates Spinal Muscular Atrophy in Mice. Am. J. Hum. Genet. 2019, 105, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, B.; Kröber, S.; Torres-Benito, L.; Borgmann, A.; Peters, M.; Hosseini Barkooie, S.M.; Tejero, R.; Jakubik, M.; Schreml, J.; Milbradt, J.; et al. Plastin 3 Ameliorates Spinal Muscular Atrophy via Delayed Axon Pruning and Improves Neuromuscular Junction Functionality. Hum. Mol. Genet. 2013, 22, 1328–1347. [Google Scholar] [CrossRef] [PubMed]
- Oprea, G.E.; Kröber, S.; McWhorter, M.L.; Rossoll, W.; Müller, S.; Krawczak, M.; Bassell, G.J.; Beattie, C.E.; Wirth, B. Plastin 3 Is a Protective Modifier of Autosomal Recessive Spinal Muscular Atrophy. Science 2008, 320, 524–527. [Google Scholar] [CrossRef]
- Ahmad, S.; Wang, Y.; Shaik, G.M.; Burghes, A.H.; Gangwani, L. The Zinc Finger Protein ZPR1 Is a Potential Modifier of Spinal Muscular Atrophy. Hum. Mol. Genet. 2012, 21, 2745–2758. [Google Scholar] [CrossRef]
- Mehta, P.; Kaye, W.; Raymond, J.; Wu, R.; Larson, T.; Punjani, R.; Heller, D.; Cohen, J.; Peters, T.; Muravov, O.; et al. Prevalence of Amyotrophic Lateral Sclerosis—United States, 2014. MMWR Morb. Mortal. Wkly. Rep. 2019, 67, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Marin, B.; Boumédiene, F.; Logroscino, G.; Couratier, P.; Babron, M.C.; Leutenegger, A.L.; Copetti, M.; Preux, P.M.; Beghi, E. Variation in Worldwide Incidence of Amyotrophic Lateral Sclerosis: A Meta-Analysis. Int. J. Epidemiol. 2017, 46, 57–74. [Google Scholar] [CrossRef]
- Hulisz, D. Amyotrophic Lateral Sclerosis: Disease State Overview. Am. J. Manag. Care 2018, 24, S320–S326. [Google Scholar]
- Jaiswal, M.K. Riluzole and Edaravone: A Tale of Two Amyotrophic Lateral Sclerosis Drugs. Med. Res. Rev. 2019, 39, 733–748. [Google Scholar] [CrossRef]
- Niedermeyer, S.; Murn, M.; Choi, P.J. Respiratory Failure in Amyotrophic Lateral Sclerosis. Chest 2019, 155, 401–408. [Google Scholar] [CrossRef]
- Cappella, M.; Ciotti, C.; Cohen-Tannoudji, M.; Biferi, M.G. Gene Therapy for ALS—A Perspective. Int. J. Mol. Sci. 2019, 20, 4388. [Google Scholar] [CrossRef] [PubMed]
- FDA; CDER. Highlights of Prescribing Information: RILUTEK. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2016/020599s017lbl.pdf (accessed on 24 August 2022).
- Chen, J.J. Overview of Current and Emerging Therapies for Amyotrophic Lateral Sclerosis. Am. J. Manag. Care 2020, 26, S191–S197. [Google Scholar] [CrossRef]
- Thakor, K.; Naud, S.; Howard, D.; Tandan, R.; Waheed, W. Effect of Riluzole on Weight in Short-Term and Long-Term Survivors of Amyotrophic Lateral Sclerosis. Amyotroph. Lateral Scler. Front. Degener. 2021, 22, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Safety Study of Oral Edaravone Administered in Subjects with ALS. Available online: https://clinicaltrials.gov/ct2/show/NCT04165824 (accessed on 22 December 2022).
- Yoshino, H.; Kimura, A. Investigation of the Therapeutic Effects of Edaravone, a Free Radical Scavenger, on Amyotrophic Lateral Sclerosis (Phase II Study). Amyotroph. Lateral Scler. 2006, 7, 247–251. [Google Scholar] [CrossRef]
- Genge, A.; Pattee, G.L.; Sobue, G.; Aoki, M.; Yoshino, H.; Couratier, P.; Lunetta, C.; Petri, S.; Selness, D.; Bidani, S.; et al. Oral Edaravone Demonstrated a Favorable Safety Profile in Patients with Amyotrophic Lateral Sclerosis after 48 Weeks of Treatment. Muscle Nerve 2022. [CrossRef]
- Zhao, M.; Kim, J.R.; van Bruggen, R.; Park, J. RNA-Binding Proteins in Amyotrophic Lateral Sclerosis. Moleucles Cells 2018, 41, 818–829. [Google Scholar] [CrossRef]
- Huai, J.; Zhang, Z. Structural Properties and Interaction Partners of Familial ALS-Associated SOD1 Mutants. Front. Neurol. 2019, 10, 527. [Google Scholar] [CrossRef]
- Chen, L.X.; Xu, H.F.; Wang, P.S.; Yang, X.X.; Wu, Z.Y.; Li, H.F. SOD1 Mutation Spectrum and Natural History of ALS Patients in a 15-Year Cohort in Southeastern China. Front. Genet. 2021, 12, 1891. [Google Scholar] [CrossRef]
- Hayashi, Y.; Homma, K.; Ichijo, H. SOD1 in Neurotoxicity and Its Controversial Roles in SOD1 Mutation-Negative ALS. Adv. Biol. Regul. 2016, 60, 95–104. [Google Scholar] [CrossRef]
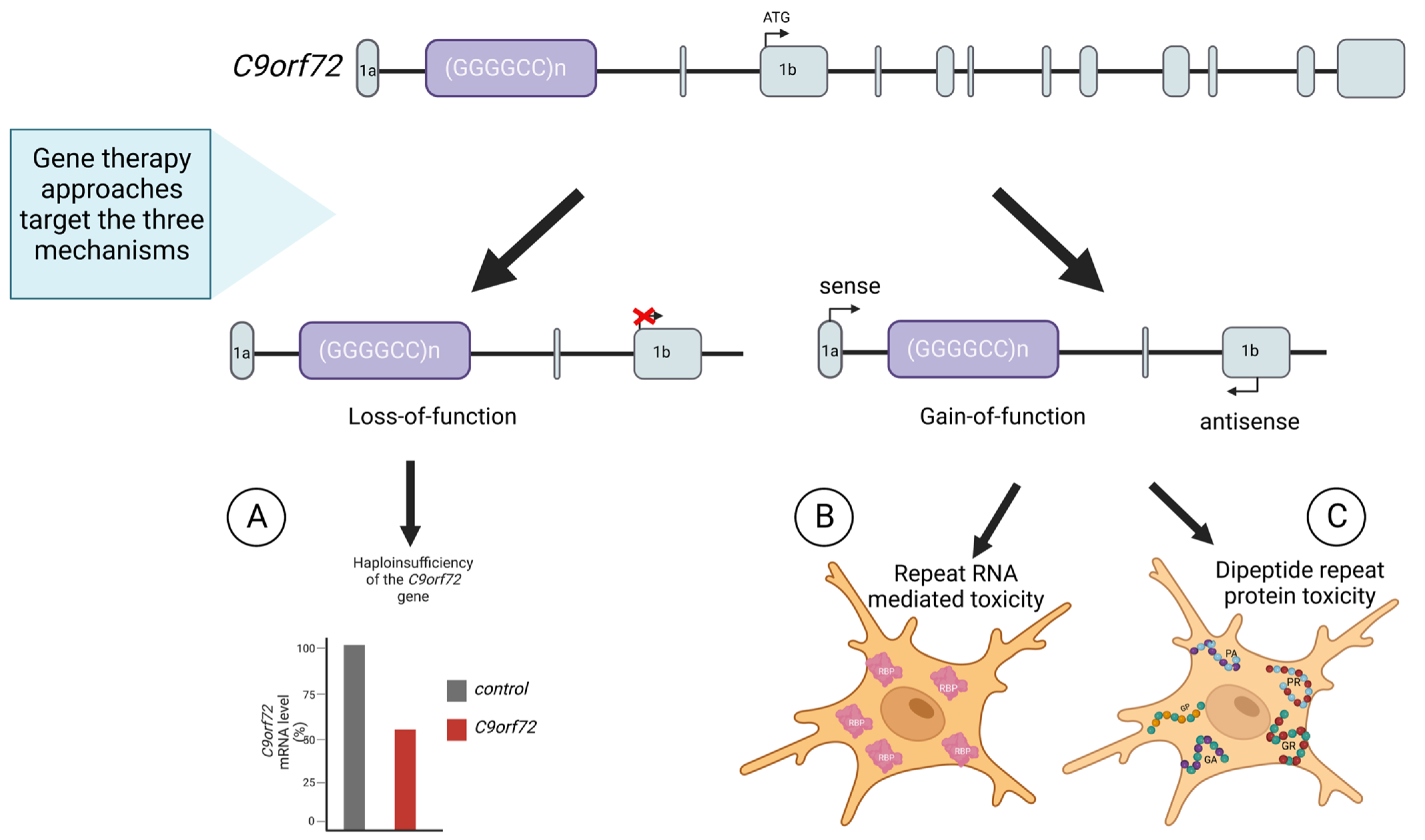
- Dafinca, R.; Barbagallo, P.; Talbot, K. The Role of Mitochondrial Dysfunction and ER Stress in TDP-43 and C9ORF72 ALS. Front. Cell Neurosci. 2021, 15, 97. [Google Scholar] [CrossRef]
- Pang, W.; Hu, F. Cellular and Physiological Functions of C9ORF72 and Implications for ALS/FTD. J. Neurochem. 2021, 157, 334–350. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.C.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Foggin, S.; Mesquita-Ribeiro, R.; Dajas-Bailador, F.; Layfield, R. Biological Significance of MicroRNA Biomarkers in ALS-Innocent Bystanders or Disease Culprits? Front. Neurol. 2019, 10, 578. [Google Scholar] [CrossRef]
- Miller, T.M.; Kaspar, B.K.; Kops, G.J.; Yamanaka, K.; Christian, L.J.; Gage, F.H.; Cleveland, D.W. Virus-Delivered Small RNA Silencing Sustains Strength in Amyotrophic Lateral Sclerosis. Ann. Neurol. 2005, 57, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Ralph, G.S.; Radcliffe, P.A.; Day, D.M.; Carthy, J.M.; Leroux, M.A.; Lee, D.C.P.; Wong, L.F.; Bilsland, L.G.; Greensmith, L.; Kingsman, S.M.; et al. Silencing Mutant SOD1 Using RNAi Protects against Neurodegeneration and Extends Survival in an ALS Model. Nat. Med. 2005, 11, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Raoul, C.; Abbas-Terki, T.; Bensadoun, J.C.; Guillot, S.; Haase, G.; Szulc, J.; Henderson, C.E.; Aebischer, P. Lentiviral-Mediated Silencing of SOD1 through RNA Interference Retards Disease Onset and Progression in a Mouse Model of ALS. Nat. Med. 2005, 11, 423–428. [Google Scholar] [CrossRef]
- Foust, K.D.; Salazar, D.L.; Likhite, S.; Ferraiuolo, L.; Ditsworth, D.; Ilieva, H.; Meyer, K.; Schmelzer, L.; Braun, L.; Cleveland, D.W.; et al. Therapeutic AAV9-Mediated Suppression of Mutant SOD1 Slows Disease Progression and Extends Survival in Models of Inherited ALS. Mol. Ther. 2013, 21, 2148–2159. [Google Scholar] [CrossRef]
- Wang, H.; Yang, B.; Qiu, L.; Yang, C.; Kramer, J.; Su, Q.; Guo, Y.; Brown, R.H.; Gao, G.; Xu, Z. Widespread Spinal Cord Transduction by Intrathecal Injection of RAAV Delivers Efficacious RNAi Therapy for Amyotrophic Lateral Sclerosis. Hum. Mol. Genet. 2014, 23, 668–681. [Google Scholar] [CrossRef]
- Rizvanov, A.A.; Mukhamedyarov, M.A.; Palotás, A.; Islamov, R.R. Retrogradely Transported SiRNA Silences Human Mutant SOD1 in Spinal Cord Motor Neurons. Exp. Brain Res. 2009, 195, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Borel, F.; Gernoux, G.; Cardozo, B.; Metterville, J.P.; Toro Cabreja, G.C.; Song, L.; Su, Q.; Gao, G.P.; ElMallah, M.K.; Brown, R.H.; et al. Therapeutic RAAVrh10 Mediated SOD1 Silencing in Adult SOD1(G93A) Mice and Nonhuman Primates. Hum. Gene Ther. 2016, 27, 19–31. [Google Scholar] [CrossRef]
- Biferi, M.G.; Cohen-Tannoudji, M.; Cappelletto, A.; Giroux, B.; Roda, M.; Astord, S.; Marais, T.; Bos, C.; Voit, T.; Ferry, A.; et al. A New AAV10-U7-Mediated Gene Therapy Prolongs Survival and Restores Function in an ALS Mouse Model. Mol. Ther. 2017, 25, 2038–2052. [Google Scholar] [CrossRef]
- Ediriweera, G.R.; Chen, L.; Yerbury, J.J.; Thurecht, K.J.; Vine, K.L. Non-Viral Vector-Mediated Gene Therapy for ALS: Challenges and Future Perspectives. Mol. Pharm. 2021, 18, 2142–2160. [Google Scholar] [CrossRef]
- Smith, R.A.; Miller, T.M.; Yamanaka, K.; Monia, B.P.; Condon, T.P.; Hung, G.; Lobsiger, C.S.; Ward, C.M.; McAlonis-Downes, M.; Wei, H.; et al. Antisense Oligonucleotide Therapy for Neurodegenerative Disease. J. Clin. Investig. 2006, 116, 2290–2296. [Google Scholar] [CrossRef]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K.; et al. An Antisense Oligonucleotide against SOD1 Delivered Intrathecally for Patients with SOD1 Familial Amyotrophic Lateral Sclerosis: A Phase 1, Randomised, First-in-Man Study. Lancet Neurol. 2013, 12, 435–442. [Google Scholar] [CrossRef]
- Safety, Tolerability, and Activity Study of ISIS SOD1Rx to Treat Familial Amyotrophic Lateral Sclerosis (ALS) Caused by SOD1 Gene Mutations. Available online: https://www.clinicaltrials.gov/ct2/show/NCT01041222 (accessed on 22 December 2022).
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1–2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.M.; Cudkowicz, M.E.; Genge, A.; Shaw, P.J.; Sobue, G.; Bucelli, R.C.; Chiò, A.; van Damme, P.; Ludolph, A.C.; Glass, J.D.; et al. Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2022, 387, 1099–1110. [Google Scholar] [CrossRef]
- Biogen and Ionis Announce Topline Phase 1 Study Results of Investigational Drug in C9orf72 Amyotrophic Lateral Sclerosis | Biogen. Available online: https://investors.biogen.com/news-releases/news-release-details/biogen-and-ionis-announce-topline-phase-1-study-results (accessed on 22 December 2022).
- An Efficacy, Safety, Tolerability, Pharmacokinetics and Pharmacodynamics Study of BIIB067 in Adults with Inherited Amyotrophic Lateral Sclerosis (ALS). Available online: https://clinicaltrials.gov/ct2/show/NCT02623699 (accessed on 22 December 2022).
- Donnelly, C.J.; Zhang, P.W.; Pham, J.T.; Heusler, A.R.; Mistry, N.A.; Vidensky, S.; Daley, E.L.; Poth, E.M.; Hoover, B.; Fines, D.M.; et al. RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron 2013, 80, 415–428. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Baughn, M.; Rigo, F.; Sun, S.; Liu, P.; Li, H.R.; Jiang, J.; Watt, A.T.; Chun, S.; Katz, M.; et al. Targeted Degradation of Sense and Antisense C9orf72 RNA Foci as Therapy for ALS and Frontotemporal Degeneration. Proc. Natl. Acad. Sci. USA 2013, 110, E4530–E4539. [Google Scholar] [CrossRef] [PubMed]
- Sareen, D.; O’Rourke, J.G.; Meera, P.; Muhammad, A.K.M.G.; Grant, S.; Simpkinson, M.; Bell, S.; Carmona, S.; Ornelas, L.; Sahabian, A.; et al. Targeting RNA Foci in IPSC-Derived Motor Neurons from ALS Patients with a C9ORF72 Repeat Expansion. Sci. Transl. Med. 2013, 5, 208ra149. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhu, Q.; Gendron, T.F.; Saberi, S.; McAlonis-Downes, M.; Seelman, A.; Stauffer, J.E.; Jafar-nejad, P.; Drenner, K.; Schulte, D.; et al. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron 2016, 90, 535–550. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M.; Cleary, J.D.; Guo, S.; Ranum, L.P.W. Therapeutic Strategies for C9orf72 Amyotrophic Lateral Sclerosis and Frontotemporal Dementia. Curr. Opin. Neurol. 2021, 34, 748–755. [Google Scholar] [CrossRef]
- Liu, Y.; Dodart, J.C.; Tran, H.; Berkovitch, S.; Braun, M.; Byrne, M.; Durbin, A.F.; Hu, X.S.; Iwamoto, N.; Jang, H.G.; et al. Variant-Selective Stereopure Oligonucleotides Protect against Pathologies Associated with C9orf72-Repeat Expansion in Preclinical Models. Nat. Commun. 2021, 12, 847. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C. Tailored Treatment for ALS Poised to Move Ahead. Nat. Med. 2019. [CrossRef]
- A Study to Evaluate the Efficacy, Safety, Pharmacokinetics and Pharmacodynamics of ION363 in Amyotrophic Lateral Sclerosis Participants with Fused in Sarcoma Mutations (FUS-ALS). Available online: https://clinicaltrials.gov/ct2/show/NCT04768972 (accessed on 22 December 2022).
- Codron, P.; Cassereau, J.; Vourc’h, P. InFUSing Antisense Oligonucleotides for Treating ALS. Trends Mol. Med. 2022, 28, 253–254. [Google Scholar] [CrossRef]
- Ionis Pipeline: Antisense Drugs for a Broad Range of Diseases. Available online: https://www.ionispharma.com/ionis-innovation/pipeline/ (accessed on 22 December 2022).
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic Reduction of Ataxin-2 Extends Lifespan and Reduces Pathology in TDP-43 Mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef]
- Duque, S.; Joussemet, B.; Riviere, C.; Marais, T.; Dubreil, L.; Douar, A.M.; Fyfe, J.; Moullier, P.; Colle, M.A.; Barkats, M. Intravenous Administration of Self-Complementary AAV9 Enables Transgene Delivery to Adult Motor Neurons. Mol. Ther. 2009, 17, 1187–1196. [Google Scholar] [CrossRef]
- O’Connor, D.M.; Boulis, N.M. Gene Therapy for Neurodegenerative Diseases. Trends Mol. Med. 2015, 21, 504–512. [Google Scholar] [CrossRef]
- Azzouz, M. Gene Therapy for ALS: Progress and Prospects. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2006, 1762, 1122–1127. [Google Scholar] [CrossRef]
- Escors, D.; Breckpot, K. Lentiviral Vectors in Gene Therapy: Their Current Status and Future Potential. Arch. Immunol. Ther. Exp. 2010, 58, 107–119. [Google Scholar] [CrossRef]
- Trabalza, A.; Eleftheriadou, I.; Sgourou, A.; Liao, T.-Y.; Patsali, P.; Lee, H.; Mazarakis, N.D. Enhanced Central Nervous System Transduction with Lentiviral Vectors Pseudotyped with RVG/HIV-1gp41 Chimeric Envelope Glycoproteins. J. Virol. 2014, 88, 2877–2890. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kato, S.; Kobayashi, K. Genetic Manipulation of Specific Neural Circuits by Use of a Viral Vector System. J. Neural. Transm. 2018, 125, 67–75. [Google Scholar] [CrossRef]
- Choudhury, S.R.; Hudry, E.; Maguire, C.A.; Sena-Esteves, M.; Breakefield, X.O.; Grandi, P. Viral Vectors for Therapy of Neurologic Diseases. Neuropharmacology 2017, 120, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Svendsen, C.N. Ex Vivo Gene Therapy Using Human Mesenchymal Stem Cells to Deliver Growth Factors in the Skeletal Muscle of a Familial ALS Rat Model. Methods Mol. Biol. 2016, 1382, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Iannitti, T.; Scarrott, J.M.; Likhite, S.; Coldicott, I.R.P.; Lewis, K.E.; Heath, P.R.; Higginbottom, A.; Myszczynska, M.A.; Milo, M.; Hautbergue, G.M.; et al. Translating SOD1 Gene Silencing toward the Clinic: A Highly Efficacious, Off-Target-Free, and Biomarker-Supported Strategy for FALS. Mol. Ther. Nucleic Acids 2018, 12, 75–88. [Google Scholar] [CrossRef]
- AveXis, ALS Gene Therapy AVXS-301 to Be Bought by Novartis for $8.7B. Available online: https://alsnewstoday.com/news/avexis-and-als-therapy-avxs-301-to-be-acquired-novartis/ (accessed on 22 December 2022).
- Statement from Novartis Gene Therapies: OAV301 Program for Familial ALS Caused by SOD1 Mutation—Les Turner ALS Foundation. Available online: https://lesturnerals.org/oav301-program-for-familial-als-caused-by-sod1-mutation-statement-from-novartis-gene-therapies/ (accessed on 22 December 2022).
- Apic Bio Receives FDA Fast Track Designation for APB-102 | Apic Bio. Available online: https://apic-bio.com/apic-bio-receives-fda-fast-track-designation-for-apb-102-for-the-treatment-of-patients-with-sod1-als/ (accessed on 22 December 2022).
- APB-102 Gene Therapy for SOD1 ALS Wins FDA Fast Track Status. Available online: https://alsnewstoday.com/news/apb-102-gene-therapy-sod1-als-wins-fda-fast-track/ (accessed on 22 December 2022).
- Mueller, C.; Berry, J.D.; McKenna-Yasek, D.M.; Gernoux, G.; Owegi, M.A.; Pothier, L.M.; Douthwright, C.L.; Gelevski, D.; Luppino, S.D.; Blackwood, M.; et al. SOD1 Suppression with Adeno-Associated Virus and MicroRNA in Familial ALS. N. Engl. J. Med. 2020, 383, 151–158. [Google Scholar] [CrossRef]
- Miyoshi, S.; Tezuka, T.; Arimura, S.; Tomono, T.; Okada, T.; Yamanashi, Y. DOK7 Gene Therapy Enhances Motor Activity and Life Span in ALS Model Mice. EMBO Mol. Med. 2017, 9, 880–889. [Google Scholar] [CrossRef] [PubMed]
- Bartus, R.T.; Baumann, T.L.; Brown, L.; Kruegel, B.R.; Ostrove, J.M.; Herzog, C.D. Advancing Neurotrophic Factors as Treatments for Age-Related Neurodegenerative Diseases: Developing and Demonstrating “Clinical Proof-of-Concept” for AAV-Neurturin (CERE-120) in Parkinson’s Disease. Neurobiol. Aging 2013, 34, 35–61. [Google Scholar] [CrossRef]
- Tovar-y-Romo, L.B.; Ramírez-Jarquín, U.N.; Lazo-Gómez, R.; Tapia, R. Trophic Factors as Modulators of Motor Neuron Physiology and Survival: Implications for ALS Therapy. Front. Cell. Neurosci. 2014, 8, 61. [Google Scholar] [CrossRef]
- Gross, S.K.; Shim, B.S.; Bartus, R.T.; Deaver, D.; McEachin, Z.; Bétourné, A.; Boulis, N.M.; Maragakis, N.J. Focal and Dose-Dependent Neuroprotection in ALS Mice Following AAV2-Neurturin Delivery. Exp. Neurol. 2020, 323, 113091. [Google Scholar] [CrossRef]
- Lin, H.Q.; Hu, H.J.; Duan, W.S.; Liu, Y.L.; Tan, G.J.; Li, Z.Y.; Liu, Y.K.; bin Deng, B.; Song, X.Q.; Wang, W.; et al. Intramuscular Delivery of ScAAV9-HIGF1 Prolongs Survival in the HSOD1G93A ALS Mouse Model via Upregulation of D-Amino Acid Oxidase. Mol. Neurobiol. 2018, 55, 682–695. [Google Scholar] [CrossRef] [PubMed]
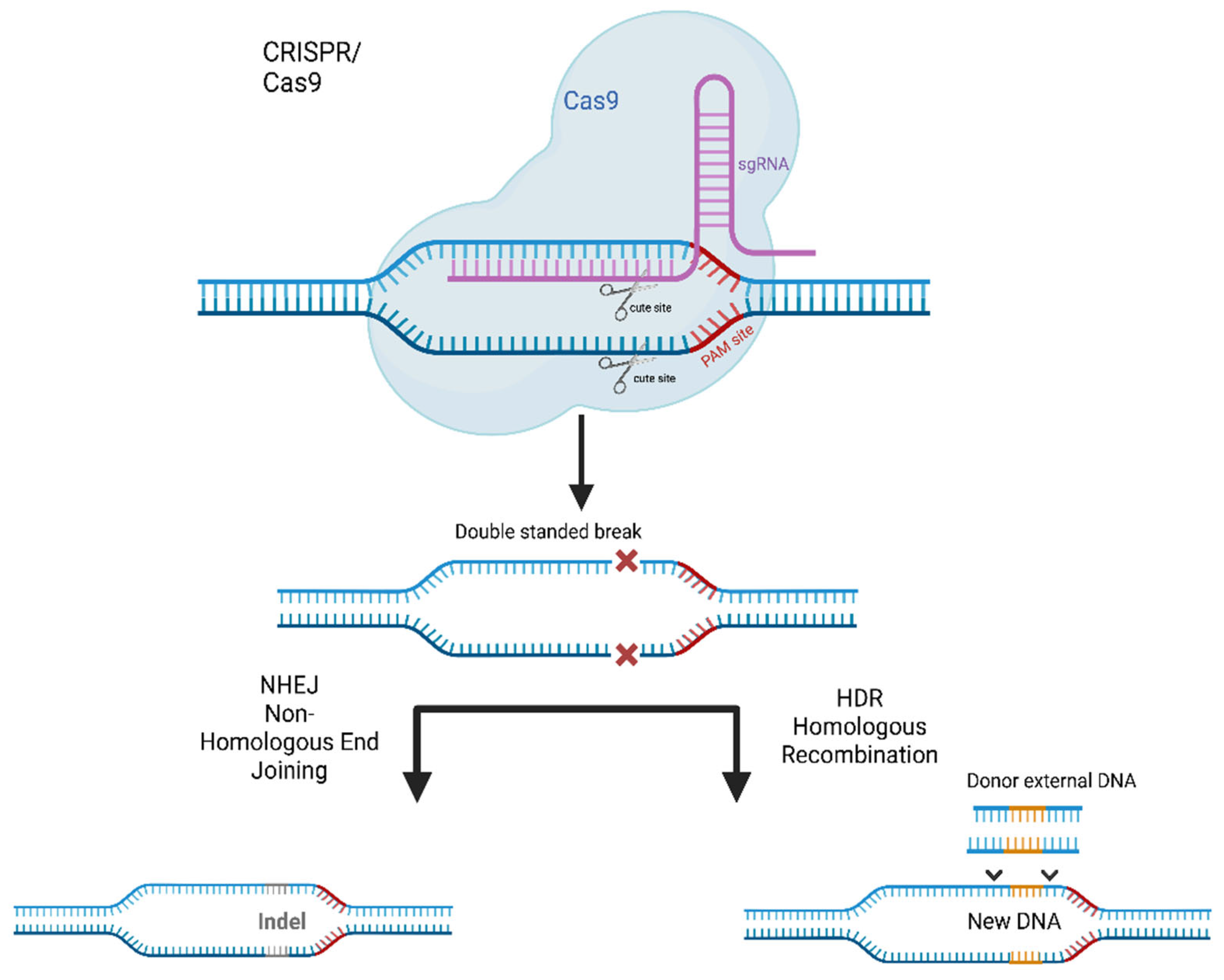
- Cai, L.; Fisher, A.L.; Huang, H.; Xie, Z. CRISPR-Mediated Genome Editing and Human Diseases. Genes Dis. 2016, 3, 244–251. [Google Scholar] [CrossRef]
- Cribbs, A.P.; Perera, S.M.W. Focus: Genome Editing: Science and Bioethics of CRISPR-Cas9 Gene Editing: An Analysis Towards Separating Facts and Fiction. Yale J. Biol. Med. 2017, 90, 625. [Google Scholar] [PubMed]
- Karpe, Y.; Chen, Z.; Li, X.J. Stem Cell Models and Gene Targeting for Human Motor Neuron Diseases. Pharmaceuticals 2021, 14, 565. [Google Scholar] [CrossRef]
- Yun, Y.; Ha, Y. CRISPR/Cas9-Mediated Gene Correction to Understand ALS. Int. J. Mol. Sci. 2020, 21, 3801. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-Homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Amado, D.A.; Davidson, B.L. Gene Therapy for ALS: A Review. Mol. Ther. 2021, 29, 3345–3358. [Google Scholar] [CrossRef]
- Miccio, A.; Antoniou, P.; Ciura, S.; Kabashi, E. Novel Genome-Editing-Based Approaches to Treat Motor Neuron Diseases: Promises and Challenges. Mol. Ther. 2022, 30, 47–53. [Google Scholar] [CrossRef]
- Nakade, S.; Yamamoto, T.; Sakuma, T. Cas9, Cpf1 and C2c1/2/3―What’s Next? Bioengineered 2017, 8, 265–273. [Google Scholar] [CrossRef]
- Gaj, T.; Ojala, D.S.; Ekman, F.K.; Byrne, L.C.; Limsirichai, P.; Schaffer, D.V. In Vivo Genome Editing Improves Motor Function and Extends Survival in a Mouse Model of ALS. Sci. Adv. 2017, 3, eaar3952. [Google Scholar] [CrossRef]
- Duan, W.; Guo, M.; Yi, L.; Liu, Y.; Li, Z.; Ma, Y.; Zhang, G.; Liu, Y.; Bu, H.; Song, X.; et al. The Deletion of Mutant SOD1 via CRISPR/Cas9/SgRNA Prolongs Survival in an Amyotrophic Lateral Sclerosis Mouse Model. Gene Ther. 2019, 27, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.X.; Zhai, H.; Shi, Y.; Liu, G.; Lowry, J.; Liu, B.; Ryan, É.B.; Yan, J.; Yang, Y.; Zhang, N.; et al. Efficacy and Long-Term Safety of CRISPR/Cas9 Genome Editing in the SOD1-Linked Mouse Models of ALS. Commun. Biol. 2021, 4, 396. [Google Scholar] [CrossRef] [PubMed]
- Pribadi, M.; Yang, Z.; Kim, T.S.; Swartz, E.W.; Huang, A.Y.; Chen, J.A.; Dokuru, D.; Baek, J.; Gao, F.; Fua, A.T.; et al. CRISPR-Cas9 Targeted Deletion of the C9orf72 Repeat Expansion Mutation Corrects Cellular Phenotypes in Patient-Derived IPS Cells. bioRxiv 2016, 051193. [Google Scholar] [CrossRef]
- Lopez-Gonzalez, R.; Yang, D.; Pribadi, M.; Kim, T.S.; Krishnan, G.; Choi, S.Y.; Lee, S.; Coppola, G.; Gao, F.B. Partial Inhibition of the Overactivated Ku80-Dependent DNA Repair Pathway Rescues Neurodegeneration in C9ORF72-ALS/FTD. Proc. Natl. Acad. Sci. USA 2019, 116, 9628–9633. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 Systems Have off-Target Activity with Insertions or Deletions between Target DNA and Guide RNA Sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef]
- Kruminis-Kaszkiel, E.; Juranek, J.; Maksymowicz, W.; Wojtkiewicz, J. CRISPR/Cas9 Technology as an Emerging Tool for Targeting Amyotrophic Lateral Sclerosis (ALS). Int. J. Mol. Sci. 2018, 19, 906. [Google Scholar] [CrossRef]
| Drug Name | Type | Mechanism of Action | Clinical Trials | Status |
|---|---|---|---|---|
| Nusinersen (Spinraza) | Antisense oligonucleotide | Binding SMN2 mRNA to modify splicing to increase SMN protein production | ENDEAR (Phase III) NURTURE (Phase II) CHERISH (Phase III) DEVOTE (Phase II/III) ONWARD (Phase III) | Approved by FDA in 2016 |
| Onasemnogene abeparvovec (Zolgensma) | Gene replacement therapy | SMN1 gene delivery via adenovirus vector (AAV9) | START (Phase I) STR1VE (Phase III) NCT05089656 (Phase III) | Approved by FDA in 2019 |
| Risdiplam (Evrysdi) | Small molecule | Binding directly to ESE2 of SMN2 transcript | FIREFISH (Phase II, III) SUNFISH (Phase II, III) JEWELFISH (Phase II) RAINBOWFISH (Phase II) | Approved by FDA in 2020 |
| Gene Therapy | Agent | Phase | Citation | Date |
|---|---|---|---|---|
| SOD1 | I | Miller et al. [104] | 2013 | |
| Tofersen (BIIB067) | I/II | Miller et al. [106] | 2020 | |
| III | NCT02623699 | current | ||
| C9orf72 | BIIB078 | I | NCT03626012 | 2021 |
| FUS | Jacifusen (ION363) | III | NCT04768972 | current |
| ATXN2 | BIIB100 | I | NCT03945279 | current |
| Gene Therapy | Model | Study | Major Findings | References |
|---|---|---|---|---|
| AAV-RNAi | ||||
| SOD1 | SOD1G93A mouse | Raoul et al., 2005 | Delayed disease onset preserved motor neurons and axons, and improved motor function. | [96] |
| SOD1G93A mouse | Wang et al., 2008 | It would improve the survival and function of motor neurons. | [89,130] | |
| SOD1G93A mouse; SOD1G37R mouse | Foust et al., 2013 | Improved motor function, increased muscle mass and extended life expectancy in mice. Injections of neutral SOD1 mRNA reductions in the spinal cord of non-human primates. | [97] | |
| SOD1G93A mouse | Borel et al., 2016 | They obtained an extension of the median life expectancy of SOD1-G93A mice by 21% and preserved motor and respiratory function. | [100] | |
| SOD1G93A mouse | Biferi et al., 2017 | Increased survival in the tested mice and prevented the deterioration of neuromuscular function in them. | [101] | |
| Human SOD1-ALS phase I | Mueller et al., 2020 | In 1 patient, clinical stabilization reduced the level of SOD1 in the spinal cord. Patient 2 without clinical benefits. | [134] | |
| Neuromuscular junction modulators | ||||
| DOK7 | SOD1G93A mouse | Miyoshi et al., 2017 | Improvement in motor activity and an extension of the life of the tested mice but no effect on motor neuron counts. | [135] |
| Neurotrophic support | ||||
| IGF | SOD1G93A mouse | Lin et al., 2018 | Significantly reduced the loss of the anterior lumbar spinal cord motor neurons and delayed muscle wasting in the tested mice. It delayed the onset of the disease and prolonged the lifespan of ALS mice. | [139] |
| AAV-CRISPR | ||||
| SOD1 | SOD1G93A mouse | Gaj et al., 2017 | Reduced the mutant SOD1 protein > 2.5-fold in the thoracic and lumbar spine. In the final stage, the mice tested showed about 50% more motor neurons, and about 37% of the disease was delayed. | [148] |
| SOD1G93A mouse | Duan et al., 2020 | Deletion of the SOD1 gene and the improvement in the life expectancy of the mice tested by 54.6%. | [149] | |
| Human SOD1G93A transgenic mouse | Deng et al., 2021 | No evidence of other diseases beyond the age of two in genome-edited mice. | [150] | |
| C9orf72 | Human FTD/ALS patient-derived iPSCs | Pribadi et al., 2016 | Prevented RNA foci formation and promoter hypermethylation, two phenotypes of the C9orf72 mutation, but this did not significantly alter the expression of C9orf72 at the mRNA or protein level. | [151] |
| Drosophila C9; human iPSC C9-ALS | Lopez-Gonzalez et al., 2019 | Decreased activation of the apoptotic pathway and reduced nuclear foci. | [152] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lejman, J.; Panuciak, K.; Nowicka, E.; Mastalerczyk, A.; Wojciechowska, K.; Lejman, M. Gene Therapy in ALS and SMA: Advances, Challenges and Perspectives. Int. J. Mol. Sci. 2023, 24, 1130. https://doi.org/10.3390/ijms24021130
Lejman J, Panuciak K, Nowicka E, Mastalerczyk A, Wojciechowska K, Lejman M. Gene Therapy in ALS and SMA: Advances, Challenges and Perspectives. International Journal of Molecular Sciences. 2023; 24(2):1130. https://doi.org/10.3390/ijms24021130
Chicago/Turabian StyleLejman, Jan, Kinga Panuciak, Emilia Nowicka, Angelika Mastalerczyk, Katarzyna Wojciechowska, and Monika Lejman. 2023. "Gene Therapy in ALS and SMA: Advances, Challenges and Perspectives" International Journal of Molecular Sciences 24, no. 2: 1130. https://doi.org/10.3390/ijms24021130
APA StyleLejman, J., Panuciak, K., Nowicka, E., Mastalerczyk, A., Wojciechowska, K., & Lejman, M. (2023). Gene Therapy in ALS and SMA: Advances, Challenges and Perspectives. International Journal of Molecular Sciences, 24(2), 1130. https://doi.org/10.3390/ijms24021130





