CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber
Abstract
1. Introduction
2. Results
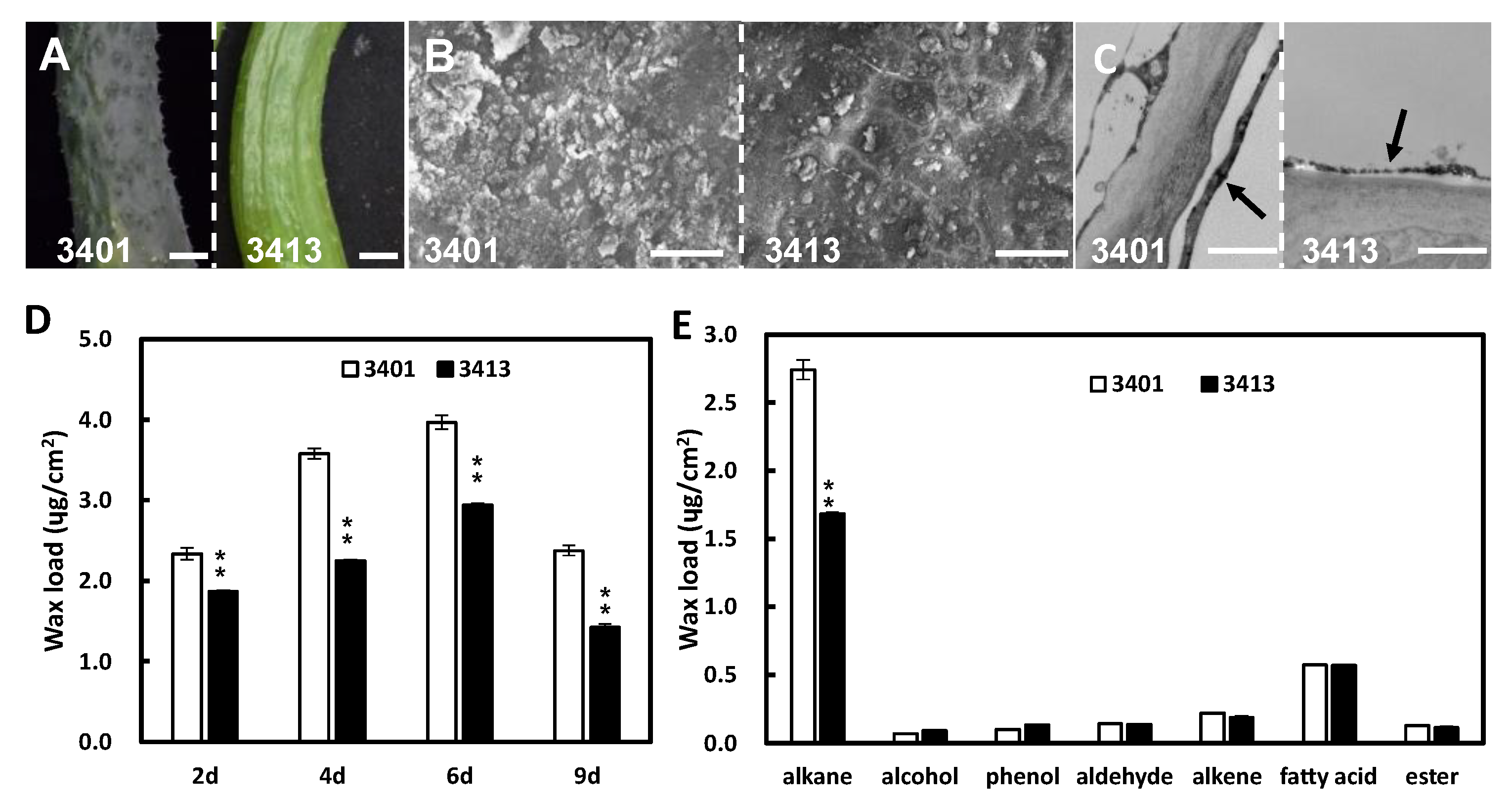
2.1. Fruit Cuticular Waxes Influence Fruit Brightness in Cucumber
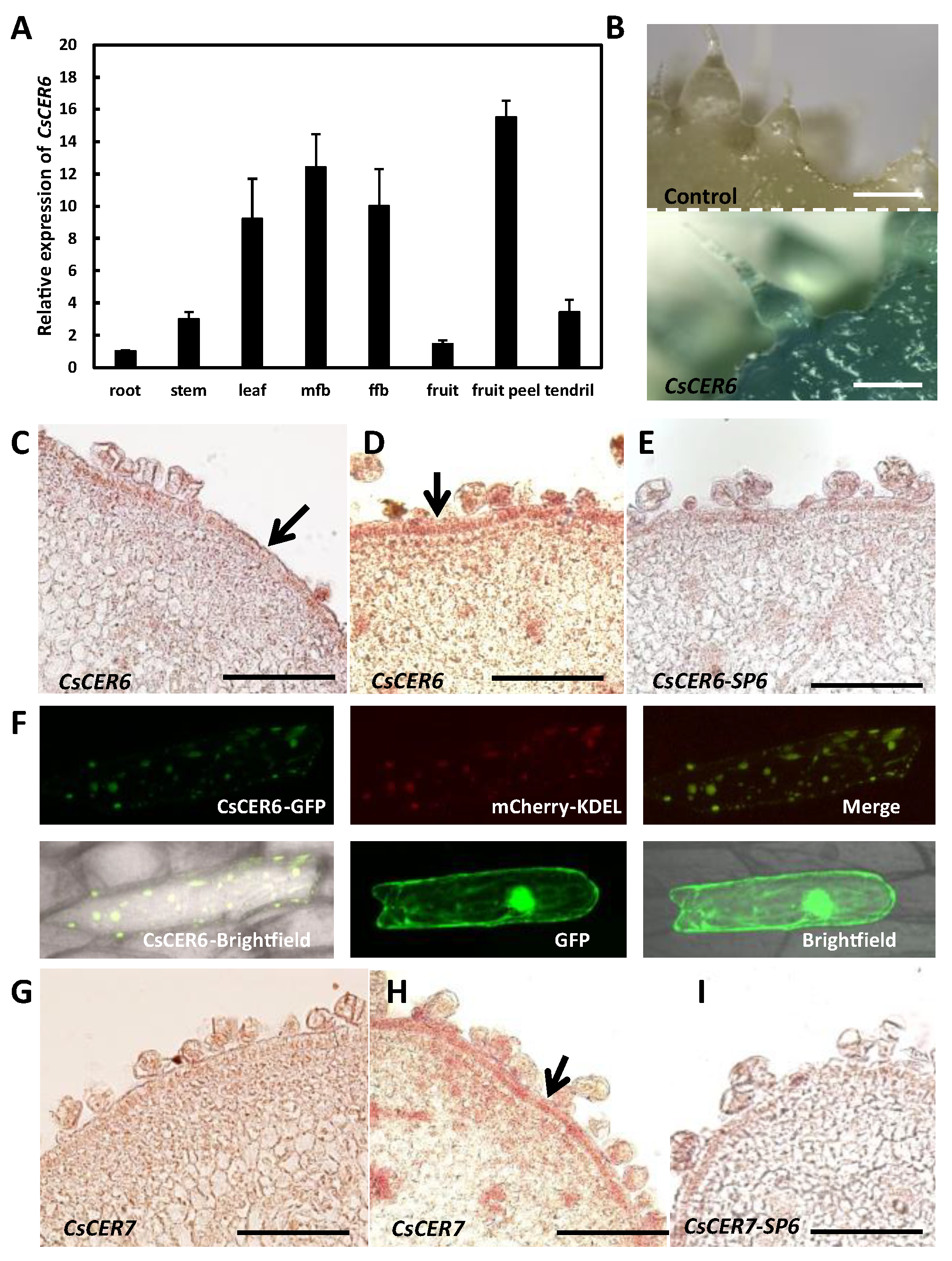
2.2. Wax-Related Genes Are Differently Expressed between 3401 and 3413
2.3. ABA Can Induce Fruit Wax Deposition and the Expression of Wax Biosynthesis-Related Genes
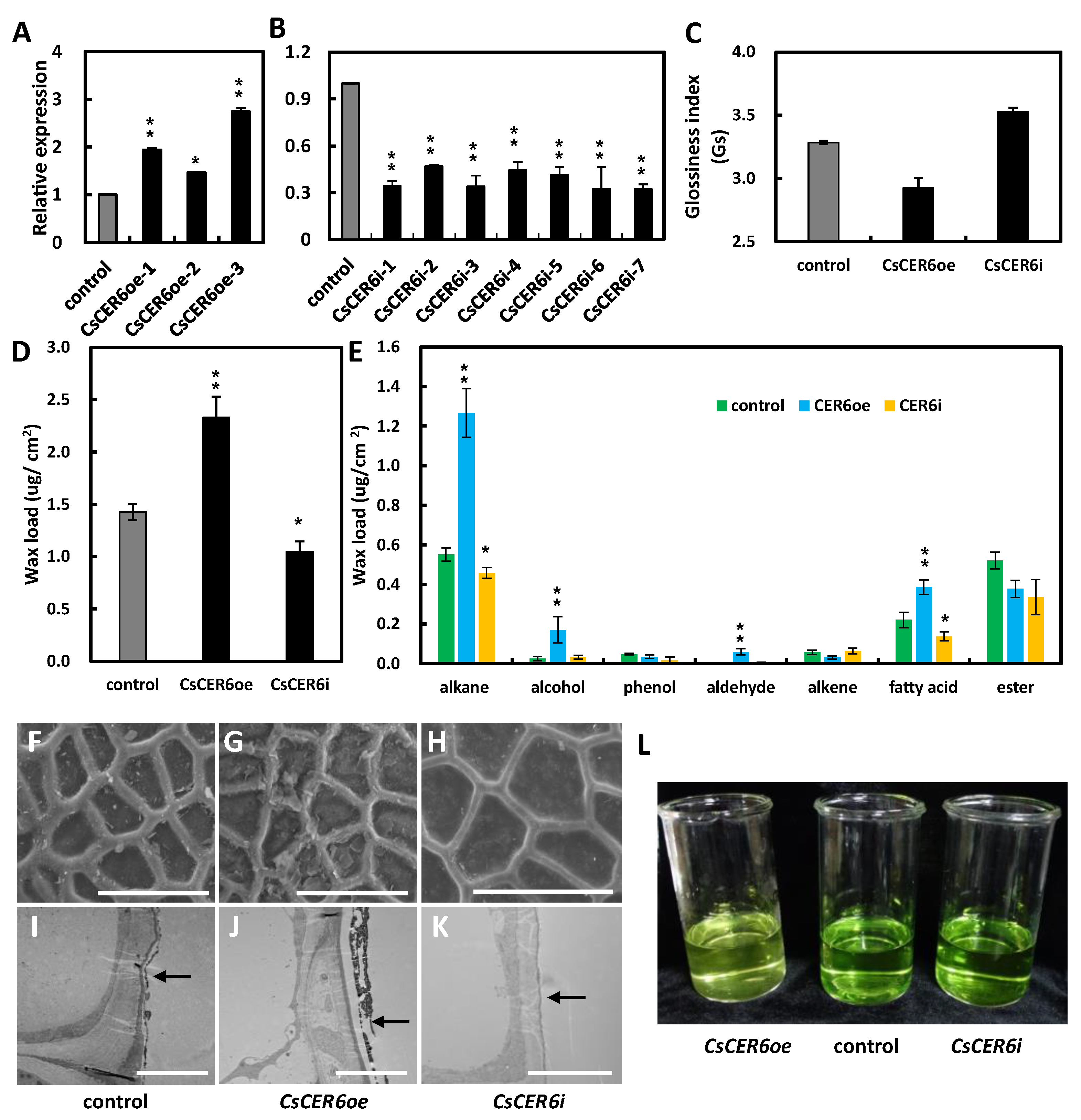
2.4. CsCER6 Positively Regulates Fruit Cuticular Wax Biosynthesis in Cucumber
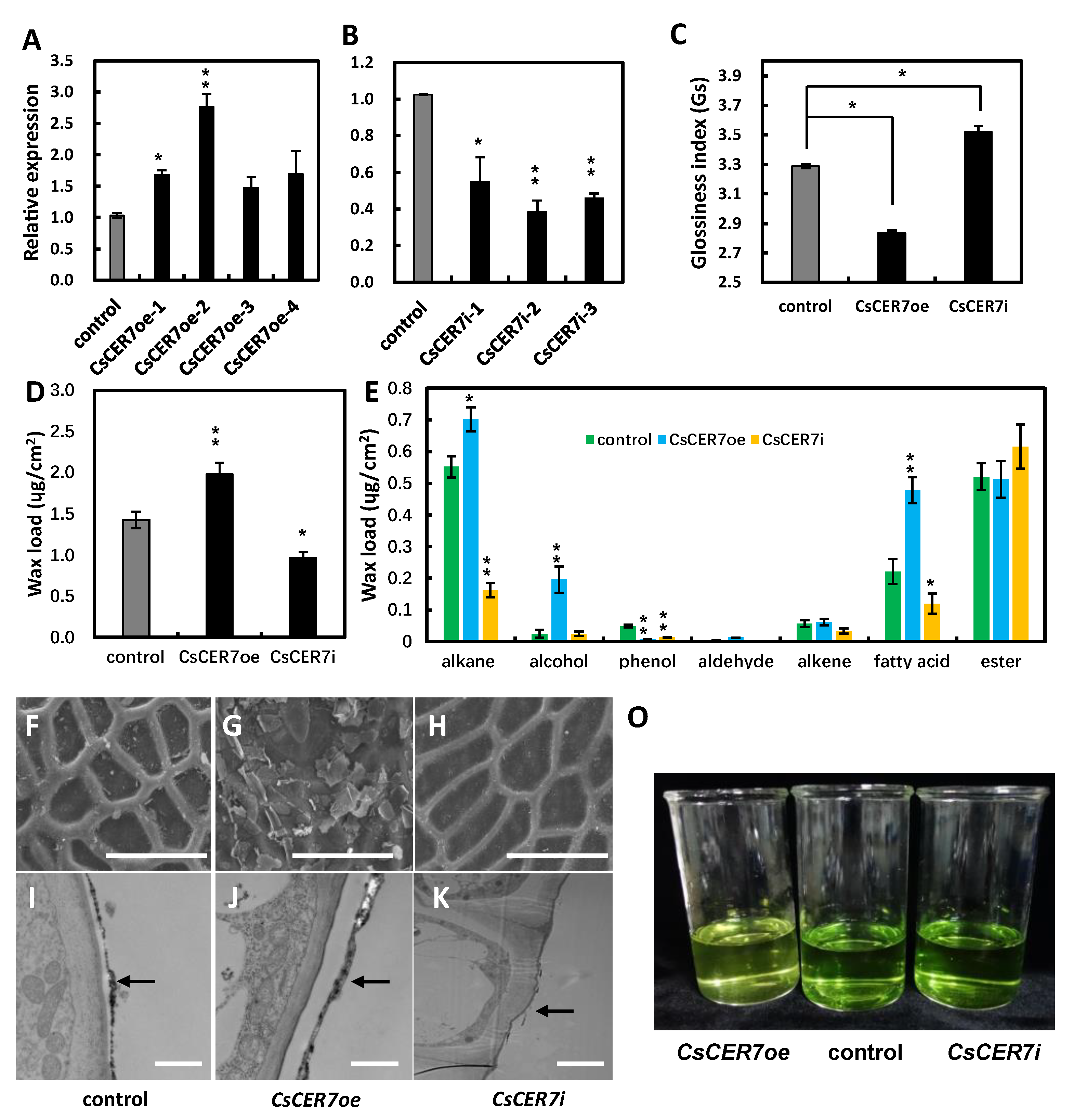
2.5. CsCER7 Positively Regulates Fruit Cuticular Wax Biosynthesis in Cucumber
3. Discussion
3.1. Fruit Cuticular Wax Loads Are Negatively Correlated with Fruit Glossiness in Cucumber
3.2. ER-Localized CsCER6 Plays an Important Role in Wax Biosynthesis in Cucumber Fruits
3.3. CsCER7 Is Involved in Regulating Fruit Cuticular Wax Contents in Cucumber
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Scanning Electron Microscopy (SEM) and Transmission Electron Microscopy (TEM) Analysis
4.3. Wax Content Test
4.4. Transcript Abundance Analysis
4.5. Subcellular Localization
4.6. Cucumber Transformation
4.7. GUS Staining
4.8. In situ Hybridization
4.9. Chlorophyll Leaching Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weng, Y. Cucumis sativus chromosome evolution, domestication, and genetic diversity–implications for cucumber breeding. Plant Breed Rev. 2021, 49, 77–111. [Google Scholar]
- Pollack, S.L. Consumer demand for fruit and vegetables: The US example. In Changing Structure of Global Food Consumption and Trade, Economic Research Service USDA; Regmi, A., Ed.; Diane: Collingdale, PA, USA, 2001; pp. 49–54. [Google Scholar]
- Mizrach, A.; Lu, R.; Rubino, M. Gloss Evaluation of Curved-surface Fruits and Vegetables. Food Bioprocess Technol. 2009, 2, 300–307. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, W.; Li, Y.; He, H.; Bie, B.; Ren, G.; Zhao, J.; Wang, Y.; Nie, J.; Pan, J. High-resolution mapping of the dull fruit skin gene D in cucumber (Cucumis sativus L.). Mol. Breed 2014, 33, 15–22. [Google Scholar] [CrossRef]
- Althaus, B.; Blanke, M. Non-Destructive, Opto-Electronic Determination of the Freshness and Shrivel of Bell Pepper Fruits. J. Imaging 2020, 6, 122. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Clemens, S.; Zachgo, S.; Giblin, E.M.; Taylor, D.C.; Kunst, L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 1999, 11, 825–838. [Google Scholar] [CrossRef]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 1999, 16, 2463–2480. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, P.; Lu, S.; Ou-yang, Q.; Zhu-ge, Y.; Tian, R.; Jia, H.; Fang, J. Comparative Analysis of Cuticular Wax in Various Grape Cultivars During Berry Development and After Storage. Front Nutr. 2021, 28, 817796. [Google Scholar] [CrossRef]
- Mukhtar, A.; Damerow, L.; Blanke, M. Non-invasive assessment of glossiness and polishing of the wax bloom of European plum. Postharvest Biol Tec. 2014, 87, 144–151. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, A. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Blanke, M.M.; Bacher, W.; Pring, R.J.; Baker, E.A. Ammonium Nutrition Enhances Chlorophyll and Glaucousness in Kohlrabi. Ann Bot. 1996, 78, 599–604. [Google Scholar] [CrossRef]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Jetter, R.; Klinger, A.; Schäffer, S. Very long-chain phenylpropyl and phenylbutyl esters from Taxus baccata needle cuticular waxes. Phytochemistry 2002, 61, 579–587. [Google Scholar] [CrossRef]
- Ji, X.; Jetter, R. Very long chain alkylresorcinols accumulate in the intracuticular wax of rye (Secale cereale L.) leaves near the tissue surface. Phytochemistry 2008, 69, 1197–1207. [Google Scholar] [CrossRef]
- Jenks, M.A.; Tuttle, H.A.; Eigenbrode, S.D.; Feldmann, K.A. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 1995, 108, 369–377. [Google Scholar] [CrossRef]
- Bourdenx, B.; Bernard, A.; Domergue, F.; Pascal, S.; Léger, A.; Roby, D.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011, 156, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Hooker, T.S.; Millar, A.A.; Kunst, L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 2002, 129, 1568–1580. [Google Scholar] [CrossRef]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef]
- Greer, S.; Wen, M.; Bird, D.; Wu, X.; Samuels, L.; Kunst, L.; Jetter, R. The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 2007, 145, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Pighin, J.A.; Zheng, H.; Balakshin, L.J.; Goodman, I.P.; Western, T.L.; Jetter, R.; Kunst, L.; Samuels, A.L. Plant cuticular lipid export requires an ABC transporter. Science 2004, 306, 702–704. [Google Scholar] [CrossRef]
- Panikashvili, D.; Aharoni, A. ABC-type transporters and cuticle assembly: Linking function to polarity in epidermis cells. Plant Signal Behav. 2008, 3, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Hooker, T.S.; Lam, P.; Zheng, H.; Kunst, H. A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 2007, 19, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Hofte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-Like Transcription Factors and WAX INDUCER1/SHINE1 Coordinately Regulate Cuticle Development in Arabidopsis and Torenia fournieri. Plant cell 2013, 25, 1609–1624. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol Biol. 2015, 87, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, X.; Gai, X.; Ren, J.; Liu, X.; Cai, Y.; Wang, Q.; Ren, H. Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 2015, 56, 1339–1354. [Google Scholar] [CrossRef]
- Zhai, X.; Wu, H.; Wang, Y.; Zhang, Z.; Shan, L.; Zhao, X.; Wang, R.; Liu, C.; Weng, Y.; Wang, Y.; et al. The fruit glossiness locus, dull fruit (D), encodes a C2H2-type zinc finger transcription factor, CsDULL, in cucumber (Cucumis sativus L.). Hortic. Res. 2022, 9, uhac146. [Google Scholar] [CrossRef]
- Verslues, P.E. ABA and cytokinins: Challenge and opportunity for plant stress research. Plant Mol. Biol. 2016, 91, 629–640. [Google Scholar] [CrossRef]
- Xu, X.; Feng, J.; Lü, S.; Lohrey, G.T.; An, H.; Zhou, Y.; Jenks, M.A. Leaf cuticular lipids on the Shandong and Yukon ecotypes of saltwater cress, Eutrema salsugineum, and their response to water deficiency and impact on cuticle permeability. Physiol. Plant 2014, 151, 446–458. [Google Scholar] [CrossRef]
- Islam, M.A.; Du, H.; Ning, J.; Ye, H.; Xiong, L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009, 70, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; You, C.; Li, Y.; Hao, Y. Advances in Biosynthesis, Regulation, and Function of Apple Cuticular Wax. Front. Plant Sci. 2020, 11, 1165. [Google Scholar] [CrossRef]
- Liu, D.; Zeng, Q.; Ji, Q.; Liu, C.; Liu, S.; Liu, Y. A comparison of the ultrastructure and composition of fruits’ cuticular wax from the wild-type ’Newhall’ navel orange (Citrus sinensis [L.] Osbeck cv. Newhall) and its glossy mutant. Plant Cell Rep. 2012, 31, 2239–2246. [Google Scholar] [CrossRef] [PubMed]
- Haslam, T.M.; Mañas-Fernández, A.; Zhao, L.; Kunst, L. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol. 2012, 160, 1164–1174. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Cheng, Z.; Lei, C.; Xu, F.; Gao, S.; Ren, Y.; Wang, J.; Zhang, X.; Wang, J.; Wu, F.; et al. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 2012, 235, 39–52. [Google Scholar] [CrossRef]
- Laila, R.; Robin, A.H.K.; Yang, K.; Park, J.; Suh, M.C.; Kim, J.; Nou, I. Developmental and Genotypic Variation in Leaf Wax Content and Composition, and in Expression of Wax Biosynthetic Genes in Brassica oleracea var. capitata. Front Plant Sci. 2017, 7, 1972. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Xie, Q.; Yang, Q.; Sun, P.; Gao, S.; Li, H.; Zhang, J.; Wang, T.; Ye, Z.; Yang, C. WOOLLY, interacting with MYB transcription factor MYB31, regulates cuticular wax biosynthesis by modulating CER6 expression in tomato. Plant J. 2020, 103, 323–337. [Google Scholar] [CrossRef]
- Lassner, M.W.; Lardizabal, K.; Metz, J.G. A jojoba-ketoacyl-CoA synthase cDNA complements the canola fatty acid elongation mutation in transgenic plants. Plant Cell 1996, 8, 281–292. [Google Scholar]
- Millar, A.A.; Kunst, L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997, 12, 121–131. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA synthase is involved in rice leaf cuticular wax synthesis and requires a CER2-LIKE protein as a cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef]
- Zheng, H.; Rowland, O.; Kunst, J. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.; Zhao, L.; McFarlane, H.E.; Aiga, M.; Lam, V.; Hooker, T.S.; Kunst, L. RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol. 2012, 159, 1385–1395. [Google Scholar] [CrossRef]
- Lam, P.; Zhao, L.; Eveleigh, N.; Yu, Y.; Chen, X.; Kunst, L. The exosome and trans-acting small interfering RNAs regulate cuticular wax biosynthesis during Arabidopsis inflorescence stem development. Plant Physiol. 2015, 167, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Kunst, J. SUPERKILLER complex components are required for the RNA Exosome-Mediated control of cuticular wax biosynthesis in Arabidopsis inflorescence stems. Plant Physiol. 2016, 171, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Matschi, S.; Vasquez, M.; Chamness, J.; Kaczmar, N.; Baseggio, M.; Miller, M.; Stewart, E.L.; Qiao, P.; Scanlon, M.J.; et al. Genome-Wide association study for maize leaf cuticular conductance identifies candidate genes involved in the regulation of cuticle development. G3 2020, 10, 1671–1683. [Google Scholar] [CrossRef]
- Lee, S.B.; Jung, S.J.; Go, Y.S.; Kim, H.U.; Kim, J.K.; Cho, H.J.; Park, O.K.; Suh, M.C. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009, 60, 462–475. [Google Scholar] [CrossRef]
- Smirnova, A.; Leide, J.; Riederer, M. Deficiency in a very-long-chain fatty acid beta-ketoacyl-coenzyme a synthase of tomato impairs microgametogenesis and causes floral organ fusion. Plant Physiol. 2013, 161, 196–209. [Google Scholar] [CrossRef]
- Varagona, M.J.; Schmidt, R.J.; Raikhel, N.V. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 1992, 4, 1213–1227. [Google Scholar]
- Wang, H.; Sui, X.; Guo, J.; Wang, Z.; Cheng, J.; Ma, S.; Li, X.; Zhang, Z. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant Cell Environ. 2014, 7, 795–810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, Y.; Ding, L.; Wu, Z.; Liu, R.; Meyerowitz, E.M. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 2013, 25, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Lolle, S.J.; Berlyn, G.P.; Engstrom, E.M.; Krolikowski, K.A.; Reiter, W.D.; Pruitt, R.E. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Dev Biol. 1997, 189, 311–321. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Ge, X.; An, J.; Liu, X.; Ren, H. CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber. Int. J. Mol. Sci. 2023, 24, 1135. https://doi.org/10.3390/ijms24021135
Liu X, Ge X, An J, Liu X, Ren H. CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber. International Journal of Molecular Sciences. 2023; 24(2):1135. https://doi.org/10.3390/ijms24021135
Chicago/Turabian StyleLiu, Xiaofeng, Xinshuang Ge, Jingbo An, Xingwang Liu, and Huazhong Ren. 2023. "CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber" International Journal of Molecular Sciences 24, no. 2: 1135. https://doi.org/10.3390/ijms24021135
APA StyleLiu, X., Ge, X., An, J., Liu, X., & Ren, H. (2023). CsCER6 and CsCER7 Influence Fruit Glossiness by Regulating Fruit Cuticular Wax Accumulation in Cucumber. International Journal of Molecular Sciences, 24(2), 1135. https://doi.org/10.3390/ijms24021135






