PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning
Abstract
:1. Introduction
2. Results
2.1. Evaluation Properties of the Generated Molecules
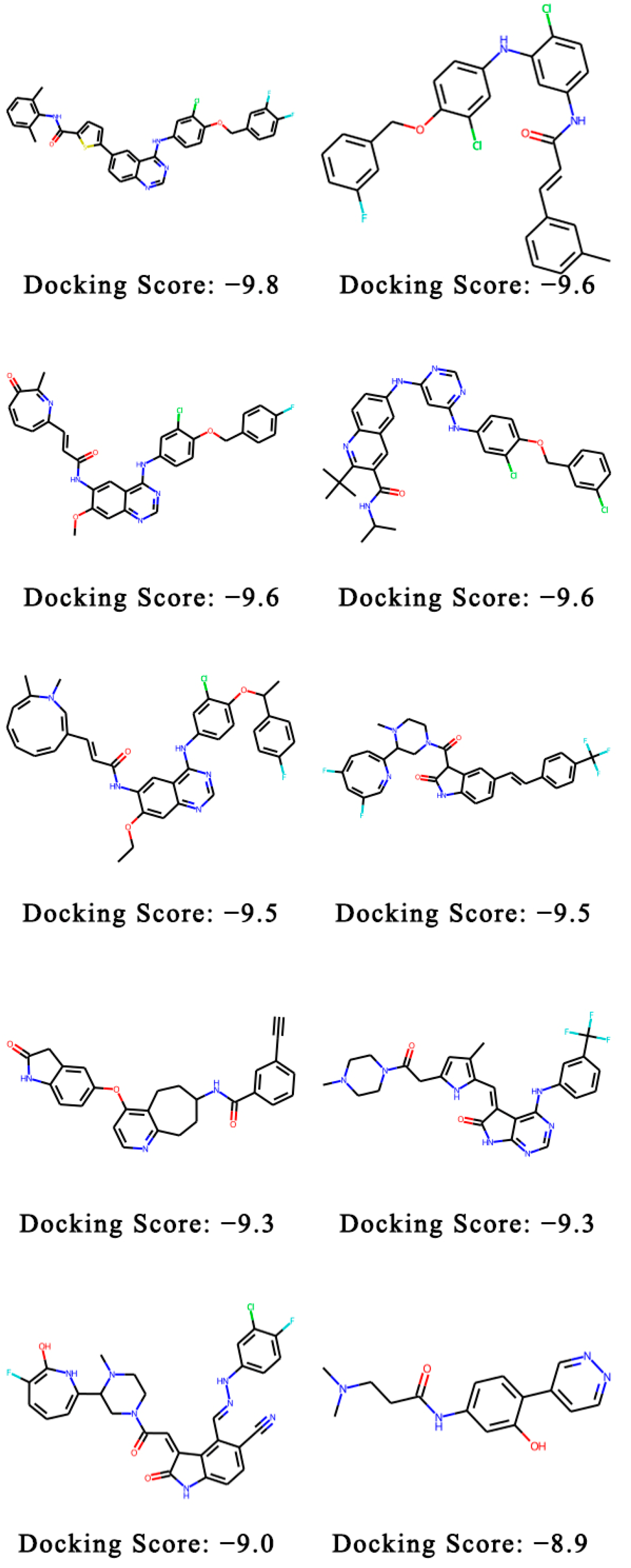
2.2. QuickVina-W Scores of the Generated Molecules
2.3. The Comparison of Properties
2.4. Experimental Results with the Dictionary of Protein Secondary Structure (DSSP)
2.5. Experimental Results with Control of the Docking Score
2.6. The Distribution of the Docking Scores
2.7. Shifting Distributions of Properties during Transfer Learning
2.8. Molecular Similarities between the Generated Molecules and the Known Active Compounds
3. Discussion
4. Materials and Methods
4.1. Datasets
4.1.1. The Drug Dataset (Pretrain)
4.1.2. Target Protein and Active Compounds (Transfer Learning)
4.2. Model Architecture
4.2.1. The Pretrained Model
4.2.2. Protein Preparation and Encoding
4.2.3. Experimental Setup
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A







References
- Cheng, T.; Li, Q.; Zhou, Z.; Wang, Y.; Bryant, S.H. Structure-Based Virtual Screening for Drug Discovery: A Problem-Centric Review. AAPS J. 2012, 14, 133–141. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, Z.; Zhang, C.; Meng, X.; Shi, X.; Qu, P. TransPhos: A Deep-Learning Model for General Phosphorylation Site Prediction Based on Transformer-Encoder Architecture. Int. J. Mol. Sci. 2022, 23, 4263. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, C.; Zhang, Y.; Meng, X.; Zhang, Z.; Shi, X.; Song, T. IMGG: Integrating Multiple Single-Cell Datasets through Connected Graphs and Generative Adversarial Networks. Int. J. Mol. Sci. 2022, 23, 2082. [Google Scholar] [CrossRef]
- Song, C.M.; Lim, S.J.; Tong, J.C. Recent Advances in Computer-Aided Drug Design. Brief. Bioinform. 2009, 10, 579–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavecchia, A.; di Giovanni, C. Virtual Screening Strategies in Drug Discovery: A Critical Review. Curr. Med. Chem. 2013, 20, 2839–2860. [Google Scholar] [CrossRef] [PubMed]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef] [Green Version]
- Caulfield, T.R.; Hayes, K.E.; Qiu, Y.; Coban, M.; Seok Oh, J.; Lane, A.L.; Yoshimitsu, T.; Hazlehurst, L.; Copland, J.A.; Tun, H.W. A Virtual Screening Platform Identifies Chloroethylagelastatin A as a Potential Ribosomal Inhibitor. Biomolecules 2020, 10, 1407. [Google Scholar] [CrossRef]
- von Roemeling, C.A.; Caulfield, T.R.; Marlow, L.; Bok, I.; Wen, J.; Miller, J.L.; Hughes, R.; Hazlehurst, L.; Pinkerton, A.B.; Radisky, D.C.; et al. Accelerated Bottom-up Drug Design Platform Enables the Discovery of Novel Stearoyl-CoA Desaturase 1 Inhibitors for Cancer Therapy. Oncotarget 2018, 9, 3–20. [Google Scholar] [CrossRef] [Green Version]
- Caulfield, T.; Coban, M.; Tek, A.; Flores, S.C. Molecular Dynamics Simulations Suggest a Non-Doublet Decoding Model of –1 Frameshifting by TRNASer3. Biomolecules 2019, 9, 745. [Google Scholar] [CrossRef] [Green Version]
- Coban, M.A.; Fraga, S.; Caulfield, T.R. Structural and Computational Perspectives of Selectively Targeting Mutant Proteins. Curr. Drug Discov. Technol. 2021, 18, 365–378. [Google Scholar] [CrossRef]
- Coban, M.A.; Morrison, J.; Maharjan, S.; Hernandez Medina, D.H.; Li, W.; Zhang, Y.S.; Freeman, W.D.; Radisky, E.S.; le Roch, K.G.; Weisend, C.M.; et al. Attacking COVID-19 Progression Using Multi-Drug Therapy for Synergetic Target Engagement. Biomolecules 2021, 11, 787. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, Y.; Mao, J.; Jeon, H.-N.; Jin, H.; Zeb, A.; Jang, Y.; Cho, K.-H.; Song, T.; No, K.T. De Novo Molecular Design with Deep Molecular Generative Models for PPI Inhibitors. Brief. Bioinform. 2022, 23, bbac285. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Pang, Y.; Xia, Y.; Chen, Z.; Luo, J. TuiGAN: Learning Versatile Image-to-Image Translation with Two Unpaired Images. In Proceedings of the 16th European Conference, Glasgow, UK, 23–28 August 2020; pp. 18–35. [Google Scholar]
- Chen, X.; Duan, Y.; Houthooft, R.; Schulman, J.; Sutskever, I.; Abbeel, P. InfoGAN: Interpretable Representation Learning by Information Maximizing Generative Adversarial Nets. In Proceedings of the Advances in Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; Lee, D., Sugiyama, M., Luxburg, U., Guyon, I., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2016; Volume 29. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Guyon, I., von Luxburg, U., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Hsu, S.T.; Moon, C.; Jones, P.; Samatova, N. An Interpretable Generative Adversarial Approach to Classification of Latent Entity Relations in Unstructured Sentences. In Proceedings of the AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; Volume 32. [Google Scholar]
- Cheng, Y.; Gong, Y.; Liu, Y.; Song, B.; Zou, Q. Molecular Design in Drug Discovery: A Comprehensive Review of Deep Generative Models. Brief. Bioinform. 2021, 22, bbab344. [Google Scholar] [CrossRef]
- Krishnan, S.R.; Bung, N.; Bulusu, G.; Roy, A. Accelerating De Novo Drug Design against Novel Proteins Using Deep Learning. J. Chem. Inf. Model. 2021, 61, 621–630. [Google Scholar] [CrossRef]
- Xue, D.; Gong, Y.; Yang, Z.; Chuai, G.; Qu, S.; Shen, A.; Yu, J.; Liu, Q. Advances and Challenges in Deep Generative Models for de Novo Molecule Generation. WIREs Comput. Mol. Sci. 2019, 9, e1395. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; Meng, X.; Wang, S.; Zhang, Y.; Rodriguez-Paton, A.; Wang, J.; Wang, X. Molormer: A Lightweight Self-Attention-Based Method Focused on Spatial Structure of Molecular Graph for Drug–Drug Interactions Prediction. Brief. Bioinform. 2022, 23, bbac296. [Google Scholar] [CrossRef]
- Song, T.; Zhang, X.; Ding, M.; Rodriguez-Paton, A.; Wang, S.; Wang, G. DeepFusion: A Deep Learning Based Multi-Scale Feature Fusion Method for Predicting Drug-Target Interactions. Methods 2022, 204, 269–277. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Ivanenkov, Y.A.; Aliper, A.; Veselov, M.S.; Aladinskiy, V.A.; Aladinskaya, A.V.; Terentiev, V.A.; Polykovskiy, D.A.; Kuznetsov, M.D.; Asadulaev, A.; et al. Deep Learning Enables Rapid Identification of Potent DDR1 Kinase Inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Bombarelli, R.; Wei, J.N.; Duvenaud, D.; Hernández-Lobato, J.M.; Sánchez-Lengeling, B.; Sheberla, D.; Aguilera-Iparraguirre, J.; Hirzel, T.D.; Adams, R.P.; Aspuru-Guzik, A. Automatic Chemical Design Using a Data-Driven Continuous Representation of Molecules. ACS Cent. Sci. 2018, 4, 268–276. [Google Scholar] [CrossRef]
- Segler, M.H.S.; Kogej, T.; Tyrchan, C.; Waller, M.P. Generating Focused Molecule Libraries for Drug Discovery with Recurrent Neural Networks. ACS Cent. Sci. 2018, 4, 120–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weininger, D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Elton, D.C.; Boukouvalas, Z.; Fuge, M.D.; Chung, P.W. Deep Learning for Molecular Design—A Review of the State of the Art. Mol. Syst. Des. Eng. 2019, 4, 828–849. [Google Scholar] [CrossRef] [Green Version]
- Pang, S.; Zhang, Y.; Song, T.; Zhang, X.; Wang, X.; Rodriguez-Patón, A. AMDE: A Novel Attention-Mechanism-Based Multidimensional Feature Encoder for Drug–Drug Interaction Prediction. Brief. Bioinform. 2022, 23, bbab545. [Google Scholar] [CrossRef]
- Grechishnikova, D. Transformer Neural Network for Protein-Specific de Novo Drug Generation as a Machine Translation Problem. Sci. Rep. 2021, 11, 321. [Google Scholar] [CrossRef]
- Xu, Z.; Wauchope, O.R.; Frank, A.T. Navigating Chemical Space by Interfacing Generative Artificial Intelligence and Molecular Docking. J. Chem. Inf. Model. 2021, 61, 5589–5600. [Google Scholar] [CrossRef]
- Olayioye, M.A. New EMBO Members’ Review: The ErbB Signaling Network: Receptor Heterodimerization in Development and Cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Abad, M.C.; Connolly, P.J.; Neeper, M.P.; Struble, G.T.; Springer, B.A.; Emanuel, S.L.; Pandey, N.; Gruninger, R.H.; Adams, M.; et al. 4-Amino-6-Arylamino-Pyrimidine-5-Carbaldehyde Hydrazones as Potent ErbB-2/EGFR Dual Kinase Inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 4615–4619. [Google Scholar] [CrossRef]
- Meng, X.; Li, X.; Wang, X. A Computationally Virtual Histological Staining Method to Ovarian Cancer Tissue by Deep Generative Adversarial Networks. Comput. Math. Methods Med. 2021, 2021, 4244157. [Google Scholar] [CrossRef]
- Yu, L.; He, L.; Gan, B.; Ti, R.; Xiao, Q.; Yang, X.; Hu, H.; Zhu, L.; Wang, S.; Ren, R. Structural Insights into Sphingosine-1-Phosphate Receptor Activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2117716119. [Google Scholar] [CrossRef]
- Xu, P.; Huang, S.; Zhang, H.; Mao, C.; Zhou, X.E.; Cheng, X.; Simon, I.A.; Shen, D.-D.; Yen, H.-Y.; Robinson, C.V.; et al. Structural Insights into the Lipid and Ligand Regulation of Serotonin Receptors. Nature 2021, 592, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gümüş, M.; Babacan, Ş.N.; Demir, Y.; Sert, Y.; Koca, İ.; Gülçin, İ. Discovery of Sulfadrug–Pyrrole Conjugates as Carbonic Anhydrase and Acetylcholinesterase Inhibitors. Arch. Pharm. 2022, 355, 2100242. [Google Scholar] [CrossRef] [PubMed]
- Dege, N.; Gökce, H.; Doğan, O.E.; Alpaslan, G.; Ağar, T.; Muthu, S.; Sert, Y. Quantum Computational, Spectroscopic Investigations on N-(2-((2-Chloro-4,5-Dicyanophenyl)Amino)Ethyl)-4-Methylbenzenesulfonamide by DFT/TD-DFT with Different Solvents, Molecular Docking and Drug-Likeness Researches. Colloids Surf. A Physicochem. Eng. Asp. 2022, 638, 128311. [Google Scholar] [CrossRef]
- Ma, B.; Terayama, K.; Matsumoto, S.; Isaka, Y.; Sasakura, Y.; Iwata, H.; Araki, M.; Okuno, Y. Structure-Based de Novo Molecular Generator Combined with Artificial Intelligence and Docking Simulations. J. Chem. Inf. Model. 2021, 61, 3304–3313. [Google Scholar] [CrossRef]
- Hassan, N.M.; Alhossary, A.A.; Mu, Y.; Kwoh, C.-K. Protein-Ligand Blind Docking Using QuickVina-W with Inter-Process Spatio-Temporal Integration. Sci. Rep. 2017, 7, 15451. [Google Scholar] [CrossRef] [Green Version]
- PyMOL by Schrödinger. Available online: https://pymol.org/ (accessed on 18 October 2022).
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved Protein–Ligand Blind Docking by Integrating Cavity Detection, Docking and Homologous Template Fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, Y.; Gan, J.; Xiao, Z.-X.; Cao, Y. FitDock: Protein–Ligand Docking by Template Fitting. Brief. Bioinform. 2022, 23, bbac087. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, Z.; Sun, H.; Wang, J.; Shen, C.; Weng, G.; Chai, X.; Li, H.; Cao, D.; Hou, T. Deep Learning Approaches for de Novo Drug Design: An Overview. Curr. Opin. Struct. Biol. 2022, 72, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhang, X.; Pan, Z.; Rodríguez Patón, A.; Wang, S.; Song, T.; Gu, Y. Multi-TransDTI: Transformer for Drug–Target Interaction Prediction Based on Simple Universal Dictionaries with Multi-View Strategy. Biomolecules 2022, 12, 644. [Google Scholar] [CrossRef] [PubMed]
- Polykovskiy, D.; Zhebrak, A.; Sanchez-Lengeling, B.; Golovanov, S.; Tatanov, O.; Belyaev, S.; Kurbanov, R.; Artamonov, A.; Aladinskiy, V.; Veselov, M.; et al. Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models. Front. Pharmacol. 2020, 11, 565644. [Google Scholar] [CrossRef]
- Teague, S.J.; Davis, A.M.; Leeson, P.D.; Oprea, T. The Design of Leadlike Combinatorial Libraries. Angew. Chem. Int. Ed. 1999, 38, 3743–3748. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Jeliazkova, N.; Chupakhin, V.; Golib-Dzib, J.-F.; Engkvist, O.; Carlsson, L.; Wegner, J.; Ceulemans, H.; Georgiev, I.; Jeliazkov, V.; et al. ExCAPE-DB: An Integrated Large Scale Dataset Facilitating Big Data Analysis in Chemogenomics. J. Cheminform. 2017, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Landrum, G. RDKit: Open-Source Cheminformatics Software. Available online: http://www.rdkit.org/ (accessed on 18 October 2022).
- Bemis, G.W.; Murcko, M.A. The Properties of Known Drugs. 1. Molecular Frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar] [CrossRef] [PubMed]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. Available online: https://cdn.openai.com/research-covers/language-unsupervised/language_understanding_paper.pdf (accessed on 18 October 2022).
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. arXiv 2014. [Google Scholar] [CrossRef]
- Bagal, V.; Aggarwal, R.; Vinod, P.K.; Priyakumar, U.D. MolGPT: Molecular Generation Using a Transformer-Decoder Model. J. Chem. Inf. Model. 2022, 62, 2064–2076. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational Protein–Ligand Docking and Virtual Drug Screening with the AutoDock Suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open Chemical Toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.-C. Prediction of Protein Cellular Attributes Using Pseudo-Amino Acid Composition. Proteins Struct. Funct. Genet. 2001, 43, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, Q.; Ma, Q.; Yu, B. LightGBM-PPI: Predicting Protein-Protein Interactions through LightGBM with Multi-Information Fusion. Chemom. Intell. Lab. Syst. 2019, 191, 54–64. [Google Scholar] [CrossRef]
- Li, X.; Han, P.; Wang, G.; Chen, W.; Wang, S.; Song, T. SDNN-PPI: Self-Attention with Deep Neural Networks Effect on Protein-Protein Interaction Prediction. Res. Sq. 2022, 23, 474. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Zhang, X.; Cui, S.; Zhang, Q. Adaptive Coding for DNA Storage with High Storage Density and Low Coverage. NPJ Syst. Biol. Appl. 2022, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, P.; Li, F.; Leier, A.; Marquez-Lago, T.T.; Wang, Y.; Webb, G.I.; Smith, A.I.; Daly, R.J.; Chou, K.-C.; et al. IFeature: A Python Package and Web Server for Features Extraction and Selection from Protein and Peptide Sequences. Bioinformatics 2018, 34, 2499–2502. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Zhang, J.; Luo, X.; Zhu, W.; Yu, K.; Chen, K.; Li, Y.; Jiang, H. Predicting Protein–Protein Interactions Based Only on Sequences Information. Proc. Natl. Acad. Sci. USA 2007, 104, 4337–4341. [Google Scholar] [CrossRef]








| Docking Score | QED | SA Score | LogP | |
|---|---|---|---|---|
| SBMolGen | −7.416 ± 0.36 | 0.509 ± 0.01 | 3.233 ± 0.11 | 4.582 ± 0.88 |
| Transfer Learning Dataset | −7.493 ± 0.46 | 0.415 ± 0.03 | 2.769 ± 0.22 | 4.749 ± 1.90 |
| DUD-E | −7.366 ± 0.42 | 0.443 ± 0.03 | 2.747 ± 0.21 | 4.608 ± 1.35 |
| PETrans (without transfer learning) | −7.689 ± 0.60 | 0.421 ± 0.03 | 2.920 ± 0.48 | 5.185 ± 2.82 |
| PETrans (Ours) | −7.969 ± 0.58 | 0.452 ± 0.05 | 2.736 ± 0.57 | 4.567 ± 4.17 |
| Docking Score | QED | SA Score | |
|---|---|---|---|
| PETrans (EGFR) | −7.969 ± 0.58 | 0.452 ± 0.05 | 2.736 ± 0.57 |
| PETrans (EGFR) * | −8.013 ± 0.60 | 0.422 ± 0.03 | 2.921 ± 0.49 |
| PETrans (HTR1A) | −8.589 ± 0.63 | 0.529 ± 0.02 | 2.971 ± 0.25 |
| PETrans (HTR1A) * | −8.599 ± 0.61 | 0.419 ± 0.01 | 2.267 ± 0.14 |
| PETrans (S1PR1) | −9.579 ± 0.64 | 0.459 ± 0.02 | 2.559 ± 0.13 |
| PETrans (S1PR1) * | −9.602 ± 0.60 | 0.458 ± 0.02 | 2.561 ± 0.11 |
| Docking Score | Valid Ratio | Novelty Ratio | Unique Ratio | |
|---|---|---|---|---|
| PETrans (EGFR) | −7.969 ± 0.58 | 0.998 | 1.0 | 0.953 |
| PETrans (EGFR) * | −8.153 ± 0.52 | 0.895 | 1.0 | 0.719 |
| PETrans (HTR1A) | −8.589 ± 0.63 | 0.982 | 1.0 | 0.979 |
| PETrans (HTR1A) * | −8.784 ± 0.57 | 0.905 | 1.0 | 0.624 |
| PETrans (S1PR1) | −9.579 ± 0.64 | 0.959 | 1.0 | 0.880 |
| PETrans (S1PR1) * | −9.403 ± 0.52 | 0.815 | 1.0 | 0.420 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Gao, C.; Han, P.; Li, X.; Chen, W.; Rodríguez Patón, A.; Wang, S.; Zheng, P. PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning. Int. J. Mol. Sci. 2023, 24, 1146. https://doi.org/10.3390/ijms24021146
Wang X, Gao C, Han P, Li X, Chen W, Rodríguez Patón A, Wang S, Zheng P. PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning. International Journal of Molecular Sciences. 2023; 24(2):1146. https://doi.org/10.3390/ijms24021146
Chicago/Turabian StyleWang, Xun, Changnan Gao, Peifu Han, Xue Li, Wenqi Chen, Alfonso Rodríguez Patón, Shuang Wang, and Pan Zheng. 2023. "PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning" International Journal of Molecular Sciences 24, no. 2: 1146. https://doi.org/10.3390/ijms24021146
APA StyleWang, X., Gao, C., Han, P., Li, X., Chen, W., Rodríguez Patón, A., Wang, S., & Zheng, P. (2023). PETrans: De Novo Drug Design with Protein-Specific Encoding Based on Transfer Learning. International Journal of Molecular Sciences, 24(2), 1146. https://doi.org/10.3390/ijms24021146





