Role of Hsp70 in Post-Translational Protein Targeting: Tail-Anchored Membrane Proteins and Beyond
Abstract
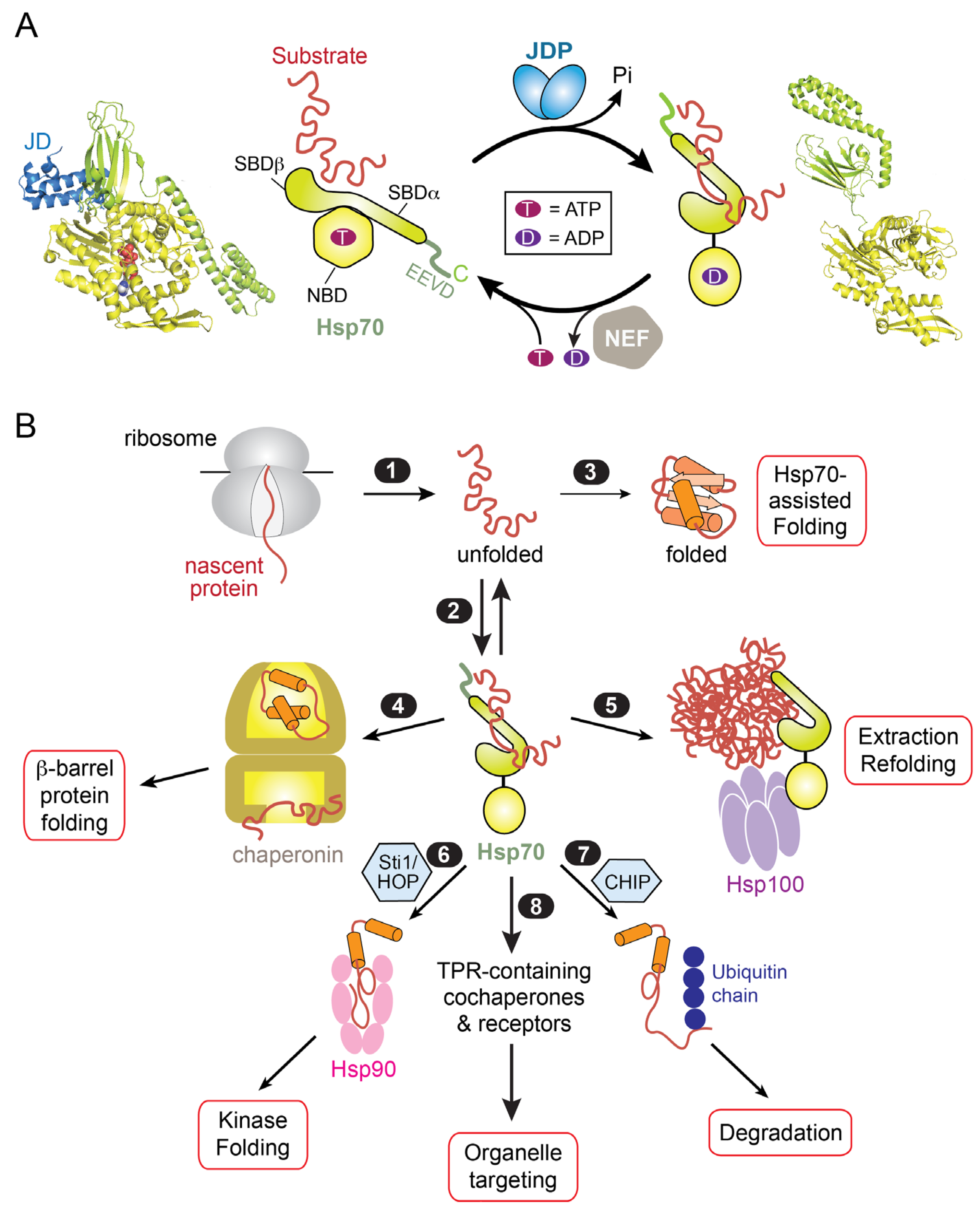
1. Hsp70: An Allosteric Machine with Diverse Functions in Protein Biogenesis
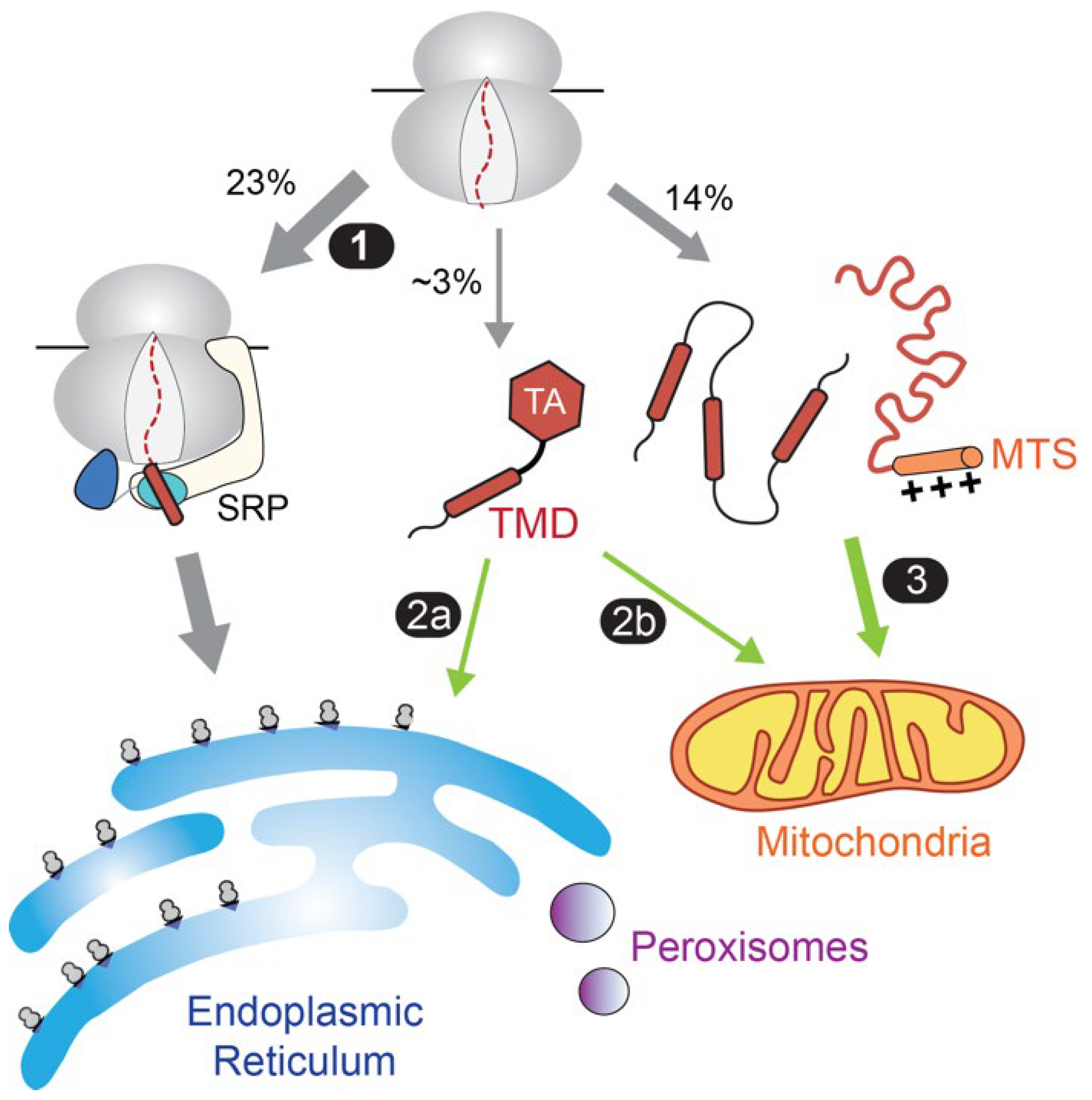
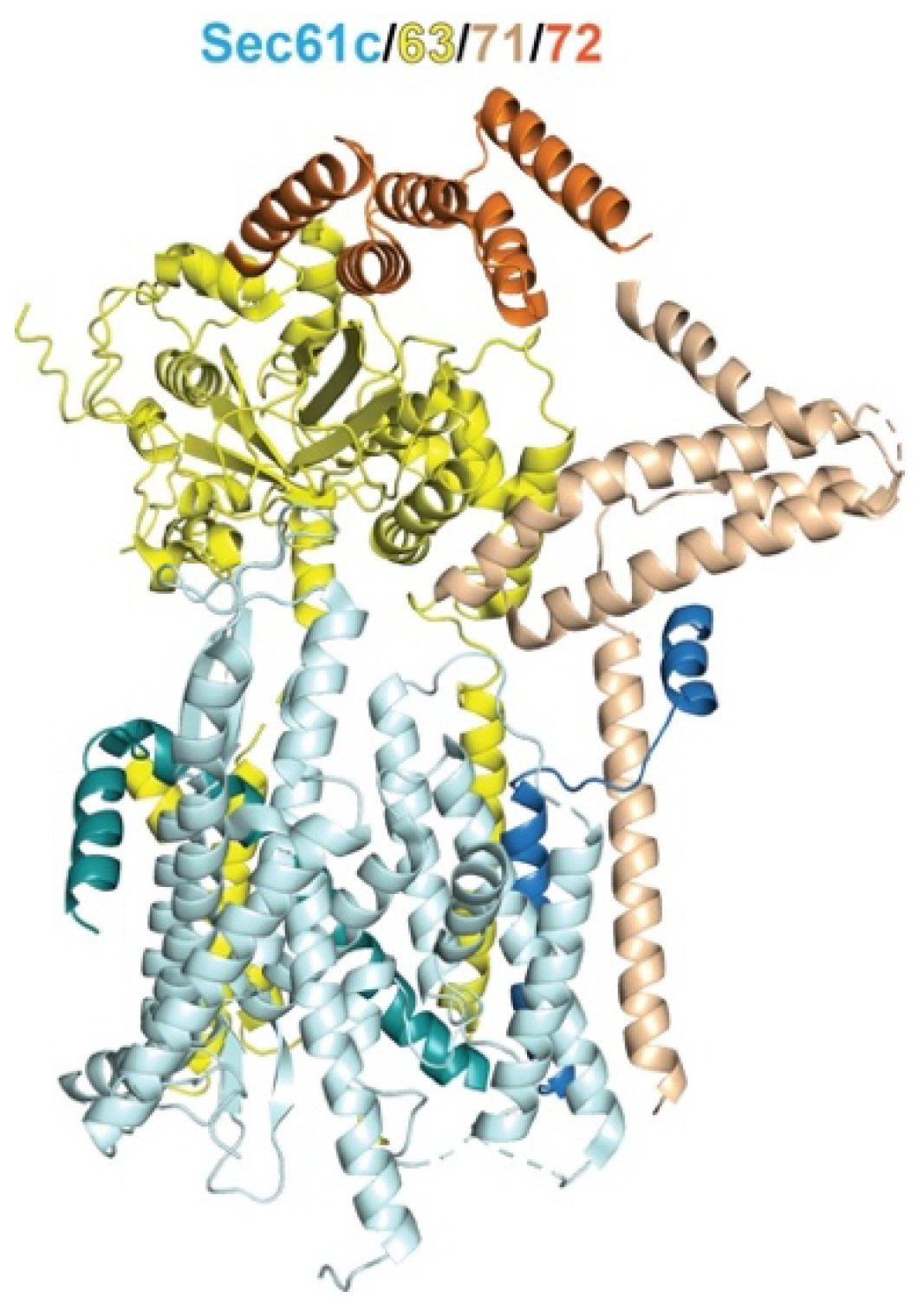
2. Overview of Protein Targeting and the Role of Hsp70
3. An Hsp70-Cochaperone Cascade Guides TAs to the ER
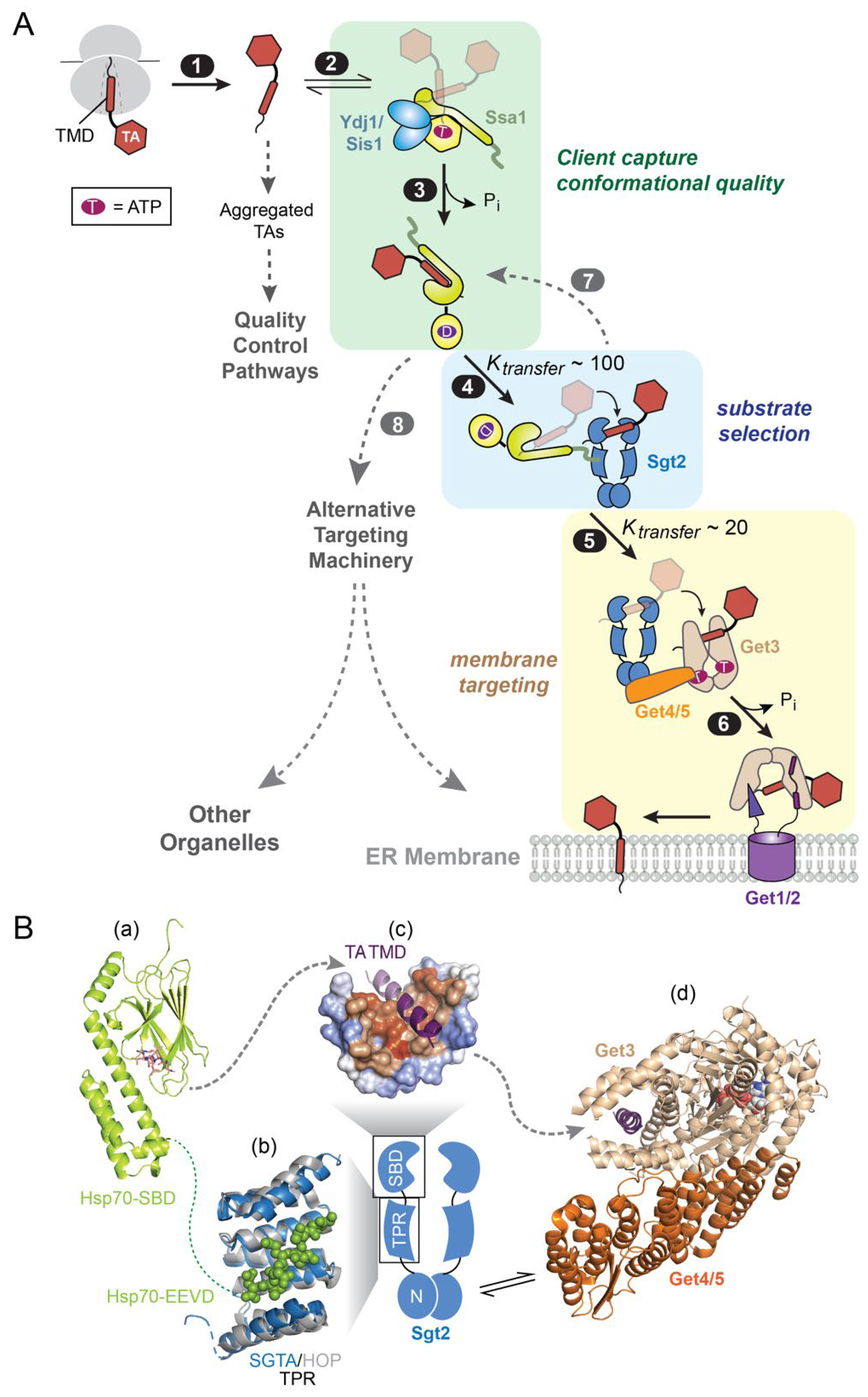
3.1. Client Conformational Quality
3.2. Privileged Substrate Relays
3.3. Driving Force and Organizational Principles
4. Role of Hsp70 in Protein Targeting to Mitochondria
5. Additional Hsp70 Involvements in Protein Targeting
6. Conclusions
7. Additional Outstanding Questions
- What provides the specificity of protein targeting when the same Hsp70s engage with proteins destined to different organelles? This question is especially intriguing given the observation that the major housekeeping Hsp70 and JDPs in yeast, Ssa1 and Ydj1/Sis1, respectively, are involved in the targeting of TAs to both the ER and mitochondria. It is possible that specificity is conferred by the ability of the downstream cochaperone/receptor to receive client proteins from Hsp70, but this hypothesis remains to be tested.
- What are the NEFs associated with each of the distinct Hsp70-mediated targeting pathways? As described in Section 1, NEFs are an essential component of the Hsp70 ATPase cycle during protein folding and remodelling. Although the roles of Hsp70 and JDPs in protein targeting has been extensively documented, little is known about whether NEFs participate in Hsp70-dependent protein targeting pathways and if so, what their precise roles are. The observations that a model substrate protein could be rapidly transferred from Ssa1 to a downstream cochaperone (Sgt2) or receptor (Tom70) in purified systems raises the possibility that NEFs may not be required to drive substrate release from Hsp70 during protein targeting. However, NEFs may still play critical roles in returning Hsp70 to the ATP-state for additional rounds of protein targeting. Both of these possibilities remain to be tested.
- Are Hsp70s involved in Hsp40-dependent protein targeting pathways? As described in Section 4, in multiple cases the involvement of Hsp40 in mitochondrial protein targeting has been described, but the involvement of Hsp70 in these processes was unclear. In many of these cases, testing the effect of mutations in the signature HPQ motif of the J-domain, which is essential for inducing ATPase activation in Hsp70, would provide key answers to this question and may uncover additional Hsp70-JDP pairs that participate in protein targeting.
- What happens to protein targeting under different environmental conditions? Besides the housekeeping Hsp70s, many Hsp70 isoforms and paralogs are induced by proteostatic stress, such as heat shock. In addition, the aggregation propensity of MPs and organellar proteins in the cytosol is likely exacerbated under stress conditions. This raises the possibility that Hsp70-dependent protein targeting pathways could be regulated under different environmental conditions. The fate of nascent post-translationally targeted proteins and how the Hsp70 network is rewired to handle them during proteostatic stress remain outstanding questions.
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular Chaperone Functions in Protein Folding and Proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U. Protein Misfolding Diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Mashaghi, A.; Bezrukavnikov, S.; Minde, D.P.; Wentink, A.S.; Kityk, R.; Zachmann-Brand, B.; Mayer, M.P.; Kramer, G.; Bukau, B.; Tans, S.J. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 2016, 539, 448–451. [Google Scholar] [CrossRef]
- Rapoport, T.A.; Li, L.; Park, E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef]
- Backes, S.; Herrmann, J.M. Protein Translocation into the Intermembrane Space and Matrix of Mitochondria: Mechanisms and Driving Forces. Front. Mol. Biosci. 2017, 4, 83. [Google Scholar] [CrossRef]
- Clerico, E.M.; Tilitsky, J.M.; Meng, W.; Gierasch, L.M. How Hsp70 Molecular Machines Interact with Their Substrates to Mediate Diverse Physiological Functions. J. Mol. Biol. 2015, 427, 1575–1588. [Google Scholar] [CrossRef]
- Clerico, E.M.; Pozhidaeva, A.K.; Jansen, R.M.; Özden, C.; Tilitsky, J.M.; Gierasch, L.M. Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2016962118. [Google Scholar] [CrossRef]
- Nordquist, E.B.; Clerico, E.M.; Chen, J.; Gierasch, L.M. Computationally-Aided Modeling of Hsp70-Client Interactions: Past, Present, and Future. J. Phys. Chem. B 2022, 126, 6780–6791. [Google Scholar] [CrossRef]
- Bascos, N.A.D.; Mayer, M.P.; Bukau, B.; Landry, S.J. The Hsp40 J-domain modulates Hsp70 conformation and ATPase activity with a semi-elliptical spring. Protein Sci. 2017, 26, 1838–1851. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592, Erratum in Nat. Rev. Mol. Cell Biol. 2010, 11, 750. [Google Scholar] [CrossRef] [PubMed]
- Abrams, J.L.; Verghese, J.; Gibney, P.A.; Morano, K.A. Hierarchical Functional Specificity of Cytosolic Heat Shock Protein 70 (Hsp70) Nucleotide Exchange Factors in Yeast. J. Biol. Chem. 2014, 289, 13155–13167. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, C.; Fiaux, J.; Rampelt, H.; Mayer, M.; Bukau, B. Hsp110 Is a Nucleotide-activated Exchange Factor for Hsp70. J. Biol. Chem. 2008, 283, 8877–8884. [Google Scholar] [CrossRef]
- Rampelt, H.; Mayer, M.P.; Bukau, B. Nucleotide Exchange Factors for Hsp70 Chaperones. In Chaperones; Humana Press: New York, NY, USA, 2017; Volume 1709, pp. 179–188. [Google Scholar] [CrossRef]
- Polier, S.; Dragovic, Z.; Hartl, F.U.; Bracher, A. Structural Basis for the Cooperation of Hsp70 and Hsp110 Chaperones in Protein Folding. Cell 2008, 133, 1068–1079. [Google Scholar] [CrossRef]
- Sharma, S.K.; De Los Rios, P.; Christen, P.; Lustig, A.; Goloubinoff, P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat. Chem. Biol. 2010, 6, 914–920. [Google Scholar] [CrossRef]
- Imamoglu, R.; Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Bacterial Hsp70 resolves misfolded states and accelerates productive folding of a multi-domain protein. Nat. Commun. 2020, 11, 365. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Andreasson, C.; Barducci, A.; Cheetham, M.E.; Cyr, D.; Emanuelsson, C.; Genevaux, P.; Gestwicki, J.E.; Goloubinoff, P.; Huerta-Cepas, J.; et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperon 2018, 24, 7–15. [Google Scholar] [CrossRef]
- Zhang, R.; Malinverni, D.; Cyr, D.M.; Rios, P.D.L.; Nillegoda, N.B. J-domain protein chaperone circuits in proteostasis and disease. Trends Cell Biol. 2022, 33, 30–47. [Google Scholar] [CrossRef]
- Hayer-Hartl, M.; Bracher, A.; Hartl, F.U. The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem. Sci. 2016, 41, 62–76. [Google Scholar] [CrossRef]
- Horwich, A.L.; Farr, G.W.; Fenton, W.A. GroEL-GroES-mediated protein folding. Chem. Rev. 2006, 106, 1917–1930. [Google Scholar] [CrossRef] [PubMed]
- Yam, A.Y.; Xia, Y.; Lin, H.-T.J.; Burlingame, A.; Gerstein, M.; Frydman, J. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat. Struct. Mol. Biol. 2008, 15, 1255–1262. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Moradi, S.; Zarrine-Afsar, A.; Glover, J.R.; Kay, L.E. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 2013, 339, 1080–1083. [Google Scholar] [CrossRef] [PubMed]
- Seyffer, F.; Kummer, E.; Oguchi, Y.; Winkler, J.; Kumar, M.; Zahn, R.; Sourjik, V.; Bukau, B.; Mogk, A. Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat. Struct. Mol. Biol. 2012, 19, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Glover, J.R.; Lindquist, S. Hsp104, Hsp70, and Hsp40: A Novel Chaperone System that Rescues Previously Aggregated Proteins. Cell 1998, 94, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Nillegoda, N.B.; Kirstein, J.; Szlachcic, A.; Berynskyy, M.; Stank, A.; Stengel, F.; Arnsburg, K.; Gao, X.; Scior, A.; Aebersold, R.; et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 2015, 524, 247–251. [Google Scholar] [CrossRef]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.; Moarefi, I. Structure of TPR Domain–Peptide Complexes: Critical Elements in the Assembly of the Hsp70–Hsp90 Multichaperone Machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Zhang, M.; Windheim, M.; Roe, S.M.; Peggie, M.; Cohen, P.; Prodromou, C.; Pearl, L.H. Chaperoned Ubiquitylation—Crystal Structures of the CHIP U Box E3 Ubiquitin Ligase and a CHIP-Ubc13-Uev1a Complex. Mol. Cell 2005, 20, 525–538. [Google Scholar] [CrossRef]
- Alvira, S.; Cuéllar, J.; Röhl, A.; Yamamoto, S.; Itoh, H.; Alfonso, C.; Rivas, G.; Buchner, J.; Valpuesta, J.M. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nat. Commun. 2014, 5, 5484. [Google Scholar] [CrossRef] [PubMed]
- Biebl, M.M.; Buchner, J. Structure, Function, and Regulation of the Hsp90 Machinery. Cold Spring Harb. Perspect. Biol. 2019, 11, a034017. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.R.; Noddings, C.M.; Kirschke, E.; Myasnikov, A.G.; Johnson, J.L.; Agard, D.A. Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism. Nature 2022, 601, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Wegele, H.; Wandinger, S.K.; Schmid, A.B.; Reinstein, J.; Buchner, J. Substrate Transfer from the Chaperone Hsp70 to Hsp90. J. Mol. Biol. 2006, 356, 802–811. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Becker, T. Fidelity of organellar protein targeting. Curr. Opin. Cell Biol. 2022, 75, 102071. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Shan, S. Substrate relay in an Hsp70-cochaperone cascade safeguards tail-anchored membrane protein targeting. EMBO J. 2018, 37, e99264. [Google Scholar] [CrossRef]
- Jaru-Ampornpan, P.; Shen, K.; Lam, V.Q.; Ali, M.; Doniach, S.; Jia, T.Z.; Shan, S.-O. ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat. Struct. Mol. Biol. 2010, 17, 696–702. [Google Scholar] [CrossRef]
- Costa, E.A.; Subramanian, K.; Nunnari, J.; Weissman, J.S. Defining the physiological role of SRP in protein-targeting efficiency and specificity. Science 2018, 359, 689–692. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. [53] Signal recognition particle: A ribonucleoprotein required for cotranslational translocation of proteins, isolation and properties. Methods Enzymol. 1983, 96, 682–691. [Google Scholar] [CrossRef]
- Walter, P.; Gilmore, R.; Blobel, G. Protein translocation across the endoplasmic reticulum. Cell 1984, 38, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Johnson, A.E. Signal Sequence Recognition and Protein Targeting to the Endoplasmic Reticulum Membrane. Annu. Rev. Cell Biol. 1994, 10, 87–119. [Google Scholar] [CrossRef] [PubMed]
- Akopian, D.; Shen, K.; Zhang, X.; Shan, S.-O. Signal Recognition Particle: An Essential Protein-Targeting Machine. Annu. Rev. Biochem. 2013, 82, 693–721. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-H.; Shan, S.-O. Fidelity of Cotranslational Protein Targeting to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2021, 23, 281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shan, S.-O. Fidelity of Cotranslational Protein Targeting by the Signal Recognition Particle. Annu. Rev. Biophys. 2014, 43, 381–408. [Google Scholar] [CrossRef] [PubMed]
- Chartron, J.; Clemons, W.M.; Suloway, C. The complex process of GETting tail-anchored membrane proteins to the ER. Curr. Opin. Struct. Biol. 2012, 22, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Chio, U.S.; Cho, H.; Shan, S.-O. Mechanisms of Tail-Anchored Membrane Protein Targeting and Insertion. Annu. Rev. Cell Dev. Biol. 2017, 33, 417–438. [Google Scholar] [CrossRef]
- Hegde, R.S.; Keenan, R.J. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2011, 12, 787–798. [Google Scholar] [CrossRef]
- Kutay, U.; Hartmann, E.; A Rapoport, T. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993, 3, 72–75. [Google Scholar] [CrossRef]
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138. [Google Scholar] [CrossRef]
- Guna, A.; Volkmar, N.; Christianson, J.C.; Hegde, R.S. The ER membrane protein complex is a transmembrane domain insertase. Science 2018, 359, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Caplan, A.J.; Cyr, D.M.; Douglas, M.G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 1992, 71, 1143–1155. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Koch, B.D.; Werner-Washburne, M.; Craig, E.A.; Schekman, R. 70 kD stress protein homologues facilitate translocation of secretory and mitochondrial precursor polypeptides. Nature 1988, 332, 800–805. [Google Scholar] [CrossRef]
- Kim, S.; Schilke, B.; Craig, E.A.; Horwich, A.L. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 12860–12865. [Google Scholar] [CrossRef] [PubMed]
- McClellan, A.J.; Scott, M.D.; Frydman, J. Folding and Quality Control of the VHL Tumor Suppressor Proceed through Distinct Chaperone Pathways. Cell 2005, 121, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; Walter, W.; Yan, W.; Craig, E.A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol. 1996, 16, 4378–4386. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Sanders, S.L.; Feldheim, D.A.; Schekman, R. Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 1991, 349, 806–808. [Google Scholar] [CrossRef]
- Feldheim, D.; Schekman, R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J. Cell Biol. 1994, 126, 935–943. [Google Scholar] [CrossRef]
- Panzner, S.; Dreier, L.; Hartmann, E.; Kostka, S.; Rapoport, T.A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 1995, 81, 561–570. [Google Scholar] [CrossRef]
- Chirico, W.J.; Waters, M.G.; Blobel, G. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature 1988, 332, 805–810. [Google Scholar] [CrossRef]
- Plath, K.; Rapoport, T.A. Spontaneous Release of Cytosolic Proteins from Posttranslational Substrates before Their Transport into the Endoplasmic Reticulum. J. Cell Biol. 2000, 151, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, A.; Mandon, E.C.; Gilmore, R.; Rapoport, T.A. Two alternative binding mechanisms connect the protein translocation Sec71-Sec72 complex with heat shock proteins. J. Biol. Chem. 2017, 292, 8007–8018. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cabanos, C.; Rapoport, T.A. Structure of the post-translational protein translocation machinery of the ER membrane. Nature 2018, 566, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Jonikas, M.C.; Collins, S.R.; Denic, V.; Oh, E.; Quan, E.M.; Schmid, V.; Weibezahn, J.; Schwappach, B.; Walter, P.; Weissman, J.S.; et al. Comprehensive Characterization of Genes Required for Protein Folding in the Endoplasmic Reticulum. Science 2009, 323, 1693–1697. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epi-static Miniarray Profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET Complex Mediates Insertion of Tail-Anchored Proteins into the ER Membrane. Cell 2008, 134, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, S.; Hegde, R.S. Identification of a targeting factor for post-translational membrane protein insertion into the ER. Cell 2007, 128, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Mariappan, M.; Li, X.; Stefanovic, S.; Sharma, A.; Mateja, A.; Keenan, R.J.; Hegde, R.S. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 2010, 466, 1120–1124. [Google Scholar] [CrossRef]
- Wang, F.; Brown, E.C.; Mak, G.; Zhuang, J.; Denic, V. A Chaperone Cascade Sorts Proteins for Posttranslational Membrane Insertion into the Endoplasmic Reticulum. Mol. Cell 2010, 40, 159–171. [Google Scholar] [CrossRef]
- Mock, J.-Y.; Chartron, J.W.; Zaslaver, M.; Xu, Y.; Ye, Y.; Clemons, W.M. Bag6 complex contains a minimal tail-anchor–targeting module and a mock BAG domain. Proc. Natl. Acad. Sci. USA 2014, 112, 106–111. [Google Scholar] [CrossRef]
- Shao, S.; Rodrigo-Brenni, M.C.; Kivlen, M.H.; Hegde, R.S. Mechanistic basis for a molecular triage reaction. Science 2017, 355, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.-F.; Fry, M.Y.; Saladi, S.M.; Clemons, W.M. Molecular basis of tail-anchored integral membrane protein recognition by the cochaperone Sgt2. J. Biol. Chem. 2021, 296, 100441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; De Laurentiis, E.; Bohnsack, K.E.; Wahlig, M.; Ranjan, N.; Gruseck, S.; Hackert, P.; Wölfle, T.; Rodnina, M.V.; Schwappach, B.; et al. Ribosome-bound Get4/5 facilitates the capture of tail-anchored proteins by Sgt2 in yeast. Nat. Commun. 2021, 12, 782. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Okreglak, V.; Chio, U.S.; Cho, H.; Walter, P.; Shan, S.-o. Multiple selection filters ensure accurate tail-anchored membrane protein targeting. Elife 2016, 5, e21301. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Shim, W.J.; Liu, Y.; Shan, S.-O. J-domain proteins promote client relay from Hsp70 during tail-anchored membrane protein targeting. J. Biol. Chem. 2021, 296, 100546. [Google Scholar] [CrossRef]
- Abe, Y.; Shodai, T.; Muto, T.; Mihara, K.; Torii, H.; Nishikawa, S.-I.; Endo, T.; Kohda, D. Structural Basis of Presequence Recognition by the Mitochondrial Protein Import Receptor Tom20. Cell 2000, 100, 551–560. [Google Scholar] [CrossRef]
- Chartron, J.W.; Gonzalez, G.M.; Clemons, W.M., Jr. A structural model of the Sgt2 protein and its interactions with chaperones and the Get4/Get5 complex. J. Biol. Chem. 2011, 286, 34325–34334. [Google Scholar] [CrossRef]
- Chio, U.S.; Chung, S.; Weiss, S.; Shan, S.-O. A Chaperone Lid Ensures Efficient and Privileged Client Transfer during Tail-Anchored Protein Targeting. Cell Rep. 2019, 26, 37–44.e7. [Google Scholar] [CrossRef]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 2020, 594, 2770–2781. [Google Scholar] [CrossRef]
- Costello, S.M.; Plummer, A.M.; Fleming, P.J.; Fleming, K.G. Dynamic periplasmic chaperone reservoir facilitates biogenesis of outer membrane proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E4794–E4800. [Google Scholar] [CrossRef]
- Chang, Y.-W.; Chuang, Y.-C.; Ho, Y.-C.; Cheng, M.-Y.; Sun, Y.-J.; Hsiao, C.-D.; Wang, C. Crystal Structure of Get4-Get5 Complex and Its Interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 2010, 285, 9962–9970. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.C.; Simpson, P.J.; Goldstone, R.M.; Krysztofinska, E.M.; Murray, J.W.; High, S.; Isaacson, R.L. Structure of the Sgt2/Get5 complex provides insights into GET-mediated targeting of tail-anchored membrane proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 1327–1332. [Google Scholar] [CrossRef]
- Chartron, J.W.; VanderVeide, D.G.; Rao, M.; Clemons, W.M., Jr. Get5 carboxyl-terminal domain is a novel dimerization motif that tethers an extended Get4/Get5 complex. J. Biol. Chem. 2012, 287, 8310–8317. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, G.; Stjepanovic, G.; Vilardi, F.; Amlacher, S.; Wild, K.; Bange, G.; Favaloro, V.; Rippe, K.; Hurt, E.; Dobberstein, B.; et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc. Natl. Acad. Sci. USA 2009, 106, 21131–21136. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Li, J.; Qian, X.; Denic, V.; Sha, B. The Crystal Structures of Yeast Get3 Suggest a Mechanism for Tail-Anchored Protein Membrane Insertion. PLoS ONE 2009, 4, e8061. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Szlachcic, A.; Downing, M.E.; Dobosz, M.; Mariappan, M.; Hegde, R.S.; Keenan, R.J. The structural basis of tail-anchored membrane protein recognition by Get3. Nature 2009, 461, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Suloway, C.J.M.; Chartron, J.W.; Zaslaver, M.; Clemons, W.M. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc. Natl. Acad. Sci. USA 2009, 106, 14849–14854. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Paduch, M.; Chang, H.-Y.; Szydlowska, A.; Kossiakoff, A.A.; Hegde, R.S.; Keenan, R.J. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science 2015, 347, 1152–1155. [Google Scholar] [CrossRef]
- Gristick, H.B.; Rao, M.; Chartron, J.W.; Rome, M.E.; Shan, S.-O.; Clemons, W.M., Jr. Crystal structure of ATP-bound Get3–Get4–Get5 complex reveals regulation of Get3 by Get4. Nat. Struct. Mol. Biol. 2014, 21, 437–442. [Google Scholar] [CrossRef]
- Gristick, H.B.; Rome, M.E.; Chartron, J.W.; Rao, M.; Hess, S.; Shan, S.-O.; Clemons, W.M. Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting. J. Biol. Chem. 2015, 290, 30006–30017. [Google Scholar] [CrossRef]
- Rome, M.E.; Rao, M.; Clemons, W.M.; Shan, S.-O. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 7666–7671. [Google Scholar] [CrossRef] [PubMed]
- Rome, M.E.; Chio, U.S.; Rao, M.; Gristick, H.; Shan, S.-O. Differential gradients of interaction affinities drive efficient targeting and recycling in the GET pathway. Proc. Natl. Acad. Sci. USA 2014, 111, E4929–E4935. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Gierasch, L.M. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J. Biol. Chem. 2019, 294, 2085–2097, Correction in J. Biol. Chem. 2020, 295, 288. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, R.; Sekhar, A.; Nagesh, J.; E Kay, L. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. Elife 2017, 6. [Google Scholar] [CrossRef]
- Chio, U.S.; Chung, S.; Weiss, S.; Shan, S.O. A protean clamp guides membrane targeting of tail-anchored proteins. Proc. Natl. Acad. Sci. USA 2017, 114, E8585–E8594. [Google Scholar] [CrossRef]
- Fry, M.Y.; Najdrová, V.; Maggiolo, A.O.; Saladi, S.M.; Doležal, P.; Clemons, W.M. Structurally derived universal mechanism for the catalytic cycle of the tail-anchored targeting factor Get3. Nat. Struct. Mol. Biol. 2022, 29, 820–830. [Google Scholar] [CrossRef]
- Beilharz, T.; Egan, B.; Silver, P.A.; Hofmann, K.; Lithgow, T. Bipartite Signals Mediate Subcellular Targeting of Tail-anchored Membrane Proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003, 278, 8219–8223. [Google Scholar] [CrossRef]
- Pedrazzini, E.; Villa, A.; Borgese, N. A mutant cytochrome b5 with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc. Natl. Acad. Sci. USA 1996, 93, 4207–4212. [Google Scholar] [CrossRef]
- Fry, M.Y.; Saladi, S.M.; Cunha, A.; Clemons, W.M. Sequence-based features that are determinant for tail-anchored membrane protein sorting in eukaryotes. Traffic 2021, 22, 306–318. [Google Scholar] [CrossRef]
- Morgenstern, M.; Peikert, C.D.; Lübbert, P.; Suppanz, I.; Klemm, C.; Alka, O.; Steiert, C.; Naumenko, N.; Schendzielorz, A.; Melchionda, L.; et al. Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab. 2021, 33, 2464–2483.e18. [Google Scholar] [CrossRef]
- Morgenstern, M.; Stiller, S.B.; Lübbert, P.; Peikert, C.D.; Dannenmaier, S.; Drepper, F.; Weill, U.; Höß, P.; Feuerstein, R.; Gebert, M.; et al. Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale. Cell Rep. 2017, 19, 2836–2852. [Google Scholar] [CrossRef] [PubMed]
- Dimogkioka, A.-R.; Lees, J.; Lacko, E.; Tokatlidis, K. Protein import in mitochondria biogenesis: Guided by targeting signals and sustained by dedicated chaperones. RSC Adv. 2021, 11, 32476–32493. [Google Scholar] [CrossRef] [PubMed]
- Becker, T.; Song, J.; Pfanner, N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends Cell Biol. 2019, 29, 534–548. [Google Scholar] [CrossRef] [PubMed]
- Araiso, Y.; Tsutsumi, A.; Qiu, J.; Imai, K.; Shiota, T.; Song, J.; Lindau, C.; Wenz, L.-S.; Sakaue, H.; Yunoki, K.; et al. Structure of the mitochondrial import gate reveals distinct preprotein paths. Nature 2019, 575, 395–401. [Google Scholar] [CrossRef]
- Su, J.; Liu, D.; Yang, F.; Zuo, M.-Q.; Li, C.; Dong, M.-Q.; Sun, S.; Sui, S.-F. Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors. Proc. Natl. Acad. Sci. USA 2022, 119, e2200158119. [Google Scholar] [CrossRef]
- Tucker, K.; Park, E. Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Nat. Struct. Mol. Biol. 2019, 26, 1158–1166. [Google Scholar] [CrossRef]
- Bausewein, T.; Mills, D.J.; Langer, J.D.; Nitschke, B.; Nussberger, S.; Kühlbrandt, W. Cryo-EM Structure of the TOM Core Complex from Neurospora crassa. Cell 2017, 170, 693–700.e7. [Google Scholar] [CrossRef]
- Wang, W.; Chen, X.; Zhang, L.; Yi, J.; Ma, Q.; Yin, J.; Zhuo, W.; Gu, J.; Yang, M. Atomic structure of human TOM core complex. Cell Discov. 2020, 6, 67. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Drwesh, L.; Heim, B.; Graf, M.; Kehr, L.; Hansen-Palmus, L.; Franz-Wachtel, M.; Macek, B.; Kalbacher, H.; Buchner, J.; Rapaport, D. A network of cytosolic (co)chaperones promotes the biogenesis of mitochondrial signal-anchored outer membrane proteins. Elife 2022, 11. [Google Scholar] [CrossRef]
- Hoseini, H.; Pandey, S.; Jores, T.; Schmitt, A.; Franz-Wachtel, M.; Macek, B.; Buchner, J.; Dimmer, K.S.; Rapaport, D. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 2016, 283, 3338–3352. [Google Scholar] [CrossRef]
- Jores, T.; Lawatscheck, J.; Beke, V.; Franz-Wachtel, M.; Yunoki, K.; Fitzgerald, J.C.; Macek, B.; Endo, T.; Kalbacher, H.; Buchner, J.; et al. Cytosolic Hsp70 and Hsp40 chaperones enable the biogenesis of mitochondrial beta-barrel proteins. J. Cell Biol. 2018, 217, 3091–3108. [Google Scholar] [CrossRef]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F. Molecular Chaperones Hsp90 and Hsp70 Deliver Preproteins to the Mitochondrial Import Receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Bhangoo, M.K.; Tzankov, S.; Fan, A.C.Y.; Dejgaard, K.; Thomas, D.; Young, J.C. Multiple 40-kDa Heat-Shock Protein Chaperones Function in Tom70-dependent Mitochondrial Import. Mol. Biol. Cell 2007, 18, 3414–3428. [Google Scholar] [CrossRef]
- Li, J.; Qian, X.; Hu, J.; Sha, B. Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 2009, 284, 23852–23859. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sha, B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006, 13, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.D.; Trewhella, J.; Qiu, T.W.; Welte, T.; Ryan, T.M.; Hanley, T.; Knott, R.B.; Lithgow, T.; Mulhern, T.D. Domain Organization of the Monomeric Form of the Tom70 Mitochondrial Import Receptor. J. Mol. Biol. 2009, 388, 1043–1058. [Google Scholar] [CrossRef] [PubMed]
- Backes, S.; Bykov, Y.S.; Flohr, T.; Räschle, M.; Zhou, J.; Lenhard, S.; Krämer, L.; Mühlhaus, T.; Bibi, C.; Jann, C.; et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 2021, 35, 108936. [Google Scholar] [CrossRef] [PubMed]
- Melin, J.; Kilisch, M.; Neumann, P.; Lytovchenko, O.; Gomkale, R.; Schendzielorz, A.; Schmidt, B.; Liepold, T.; Ficner, R.; Jahn, O.; et al. A presequence-binding groove in Tom70 supports import of Mdl1 into mitochondria. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 1850–1859. [Google Scholar] [CrossRef]
- Brix, J.; Dietmeier, K.; Pfanner, N. Differential Recognition of Preproteins by the Purified Cytosolic Domains of the Mitochondrial Import Receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997, 272, 20730–20735. [Google Scholar] [CrossRef]
- Backes, S.; Hess, S.; Boos, F.; Woellhaf, M.W.; Gödel, S.; Jung, M.; Mühlhaus, T.; Herrmann, J.M. Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences. J. Cell Biol. 2018, 217, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Fukui, K.; Takahashi, H.; Kitamura, S.; Shiota, T.; Terao, K.; Uchida, M.; Esaki, M.; Nishikawa, S.-I.; Yoshihisa, T.; et al. Roles of Tom70 in Import of Presequence-containing Mitochondrial Proteins. J. Biol. Chem. 2009, 284, 31635–31646. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Hiatt, J.; Bouhaddou, M.; Rezelj, V.V.; Ulferts, S.; Braberg, H.; Jureka, A.S.; Obernier, K.; Guo, J.Z.; Batra, J.; et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020, 370, eabe9403. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Jiang, H.W.; Zhang, H.N.; Meng, Q.F.; Xie, J.; Li, Y.; Chen, H.; Zheng, Y.X.; Wang, X.N.; Qi, H.; Zhang, J.; et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol. Immunol. 2020, 17, 998–1000. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, K.; Qin, B.; Olieric, V.; Wang, M.; Cui, S. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat. Commun. 2021, 12, 2843. [Google Scholar] [CrossRef]
- Meier, C.; Aricescu, A.R.; Assenberg, R.; Aplin, R.T.; Gilbert, R.J.; Grimes, J.M.; Stuart, D.I. The Crystal Structure of ORF-9b, a Lipid Binding Protein from the SARS Coronavirus. Structure 2006, 14, 1157–1165. [Google Scholar] [CrossRef]
- Opaliński, Ł.; Song, J.; Priesnitz, C.; Wenz, L.-S.; Oeljeklaus, S.; Warscheid, B.; Pfanner, N.; Becker, T. Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis. Cell Rep. 2018, 25, 2036–2043.e5. [Google Scholar] [CrossRef]
- Papić, D.; Elbaz-Alon, Y.; Koerdt, S.N.; Leopold, K.; Worm, D.; Jung, M.; Schuldiner, M.; Rapaport, D. The Role of Djp1 in Import of the Mitochondrial Protein Mim1 Demonstrates Specificity between a Cochaperone and Its Substrate Protein. Mol. Cell. Biol. 2013, 33, 4083–4094. [Google Scholar] [CrossRef]
- Hansen, K.G.; Aviram, N.; Laborenz, J.; Bibi, C.; Meyer, M.; Spang, A.; Schuldiner, M.; Herrmann, J.M. An ER surface retrieval pathway safeguards the import of mitochondrial membrane proteins in yeast. Science 2018, 361, 1118–1122. [Google Scholar] [CrossRef]
- Yamano, K.; Yatsukawa, Y.-I.; Esaki, M.; Hobbs, A.E.A.; Jensen, R.E.; Endo, T. Tom20 and Tom22 Share the Common Signal Recognition Pathway in Mitochondrial Protein Import. J. Biol. Chem. 2008, 283, 3799–3807. [Google Scholar] [CrossRef] [PubMed]
- Muto, T.; Obita, T.; Abe, Y.; Shodai, T.; Endo, T.; Kohda, D. NMR identification of the Tom20 binding segment in mitochondrial presequences. J. Mol. Biol. 2001, 306, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Obita, T.; Muto, T.; Endo, T.; Kohda, D. Peptide Library Approach with a Disulfide Tether to Refine the Tom20 Recognition Motif in Mitochondrial Presequences. J. Mol. Biol. 2003, 328, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Harano, T.; Nose, S.; Uezu, R.; Shimizu, N.; Fujiki, Y. Hsp70 regulates the interaction between the peroxisome targeting signal type 1 (PTS1)-receptor Pex5p and PTS1. Biochem. J. 2001, 357 Pt 1, 157–165. [Google Scholar] [CrossRef]
- Harper, C.C.; Berg, J.M.; Gould, S.J. PEX5 Binds the PTS1 Independently of Hsp70 and the Peroxin PEX12. J. Biol. Chem. 2003, 278, 7897–7901. [Google Scholar] [CrossRef]
- Legakis, J.E.; Terlecky, S.R. PTS2 protein import into mammalian peroxisomes. Traffic 2001, 2, 252–260. [Google Scholar] [CrossRef]
- Ast, T.; Cohen, G.; Schuldiner, M. A Network of Cytosolic Factors Targets SRP-Independent Proteins to the Endoplasmic Reticulum. Cell 2013, 152, 1134–1145. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, S.-o. Role of Hsp70 in Post-Translational Protein Targeting: Tail-Anchored Membrane Proteins and Beyond. Int. J. Mol. Sci. 2023, 24, 1170. https://doi.org/10.3390/ijms24021170
Shan S-o. Role of Hsp70 in Post-Translational Protein Targeting: Tail-Anchored Membrane Proteins and Beyond. International Journal of Molecular Sciences. 2023; 24(2):1170. https://doi.org/10.3390/ijms24021170
Chicago/Turabian StyleShan, Shu-ou. 2023. "Role of Hsp70 in Post-Translational Protein Targeting: Tail-Anchored Membrane Proteins and Beyond" International Journal of Molecular Sciences 24, no. 2: 1170. https://doi.org/10.3390/ijms24021170
APA StyleShan, S.-o. (2023). Role of Hsp70 in Post-Translational Protein Targeting: Tail-Anchored Membrane Proteins and Beyond. International Journal of Molecular Sciences, 24(2), 1170. https://doi.org/10.3390/ijms24021170





