Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins
Abstract
1. Introduction
2. Results and Discussion
2.1. Common Diclofenac Binding Sites in OSA-DIC, CSA-DIC, and LSA-DIC
2.2. Unique Diclofenac Binding Sites in OSA-DIC and CSA-DIC
2.3. Comparison of Diclofenac Binding Sites in All Known Structures of SA-DIC
3. Materials and Methods
3.1. Protein Purification, Crystallization, and Diffraction Measurement
3.2. Diffraction Data Processing and Crystal Structures Determination
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, M.; Jiang, M.; Gao, Y.; Zheng, Y.; Liu, Z.; Zhou, C. Systems Pharmacology in Small Molecular Drug Discovery. Int. J. Mol. Sci. 2016, 17, 246. [Google Scholar] [CrossRef]
- Stepensky, D. Prediction of Drug Disposition on the Basis of Its Chemical Structure. Clin. Pharmacokinet. 2013, 52, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Peters, T., Jr. All About Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: San Diego, CA, USA, 1996. [Google Scholar]
- Fanali, G.; di Masi, A.; Trezza, V.; Marino, M.; Fasano, M.; Ascenzi, P. Human Serum Albumin: From Bench to Bedside. Mol. Asp. Med. 2012, 33, 209–290. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Grüne, T.; Curry, S. Crystallographic Analysis Reveals Common Modes of Binding of Medium and Long-Chain Fatty Acids to Human Serum Albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Further Characterization of Specific Drug Binding Sites on Human Serum Albumin. Mol. Pharmacol. 1976, 12, 1052–1061. [Google Scholar] [PubMed]
- Sulkowska, A. Interaction of Drugs with Bovine and Human Serum Albumin. J. Mol. Struct. 2002, 614, 227–232. [Google Scholar] [CrossRef]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural Basis of the Drug-Binding Specificity of Human Serum Albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef]
- Curry, S. Lessons from the Crystallographic Analysis of Small Molecule Binding to Human Serum Albumin. Drug Metab. Pharmacokinet. 2009, 24, 342–357. [Google Scholar] [CrossRef]
- Setoguchi, N.; Takamura, N.; Fujita, K.-I.; Ogata, K.; Tokunaga, J.; Nishio, T.; Chosa, E.; Arimori, K.; Kawai, K.; Yamamoto, R. A Diclofenac Suppository–nabumetone Combination Therapy for Arthritic Pain Relief and a Monitoring Method for the Diclofenac Binding Capacity of HSA Site II in Rheumatoid Arthritis. Biopharm. Drug Dispos. 2013, 34, 125–136. [Google Scholar] [CrossRef]
- Osaki, T.; Ozaki, M.; Takamura, N.; Ogata, K.; Tokunaga, J.; Setoguchi, N.; Arimori, K. Albumin-Binding of Diclofenac and the Effect of a Site II Inhibitor in the Aqueous Humor of Cataract Patients with the Instillation of Diclofenac. Biopharm. Drug Dispos. 2014, 35, 218–227. [Google Scholar] [CrossRef]
- Khodaei, A.; Bolandnazar, S.; Valizadeh, H.; Hasani, L.; Zakeri-Milani, P. Interactions between Sirolimus and Anti-Inflammatory Drugs: Competitive Binding for Human Serum Albumin. Adv. Pharm. Bull. 2016, 6, 227–233. [Google Scholar] [CrossRef]
- Hossain, K.; Khatun, A.; Rahman, M.; Akter, N.; Chowdhury, S.A.; Alam, S.M. Characterization of the Effect of Drug-Drug Interaction on Protein Binding in Concurrent Administration of Sulfamethoxazol and Diclofenac Sodium Using Bovine Serum Albumin. Adv. Pharm. Bull. 2016, 6, 589–595. [Google Scholar] [CrossRef]
- Hirohata, M.; Ono, K.; Naiki, H.; Yamada, M. Non-Steroidal Anti-Inflammatory Drugs Have Anti-Amyloidogenic Effects for Alzheimer’s β-Amyloid Fibrils in Vitro. Neuropharmacology 2005, 49, 1088–1099. [Google Scholar] [CrossRef] [PubMed]
- McGeer, P.L.; McGeer, E.G. McGeer. NSAIDs and Alzheimer Disease: Epidemiological, Animal Model and Clinical Studies. Neurobiol. Aging 2007, 28, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.K.Y. Biochemical Pharmacology of Nonsteroidal Anti-Inflammatory Drugs. Biochem. Pharmacol. 1998, 55, 543–547. [Google Scholar] [CrossRef]
- Singh, N.; Jabeen, T.; Sharma, S.; Somvanshi, R.K.; Dey, S.; Srinivasan, A.; Singh, T.P. Specific Binding of Non-Steroidal Anti-Inflammatory Drugs (NSAIDs) to Phospholipase A2: Structure of the Complex Formed between Phospholipase A2 and Diclofenac at 2.7 Resolution. Acta Crystallogr. D Biol. Crystallogr. 2006, 62, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.; Lirk, P.; Tan, C.; Seymour, R. An Evidence-Based Update on Nonsteroidal Anti-Inflammatory Drugs. Clin. Med. Res. 2007, 5, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.; Knaus, E.E. Evolution of Nonsteroidal Anti-Inflammatory Cyclooxygenase (COX) Inhibition and Beyond Drugs (NSAIDs). J. Pharm. Pharmaceut. Sci. 2008, 11, 81–110. [Google Scholar] [CrossRef]
- Bou-Abdallah, F.; Sprague, S.E.; Smith, B.M.; Giffune, T.R. Binding thermodynamics of Diclofenac and Naproxen with human and bovine serum albumins: A calorimetric and spectroscopic study. J. Chem. Thermodyn. 2016, 103, 299–309. [Google Scholar] [CrossRef]
- Sharma, R.; Choudhary, S.; Kishore, N. Insights into the Binding of the Drugs Diclofenac Sodium and Cefotaxime Sodium to Serum Albumin: Calorimetry and Spectroscopy. Eur. J. Pharm. Sci. 2012, 46, 435–445. [Google Scholar] [CrossRef]
- Indurthi, V.; Leclerc, S.K.E.; Vetter, S.W. Calorimetric Investigation of Diclofenac Drug Binding to a Panel of Moderately Glycated Serum Albumins. Eur. J. Pharm. Sci. 2014, 59, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Dohare, N.; Khan, A.B.; Athar, F.; Thakur, S.C.; Patel, R. Urea-Induced Binding between Diclofenac Sodium and Bovine Serum Albumin: A Spectroscopic Insight. Luminescence 2016, 31, 945–951. [Google Scholar] [CrossRef]
- Yasseen, Z.; El-Ghossain, M.O. Studies on Binding of Widely used Drugs with Human Serum Albumin at Different Temperatures and PHs. J. Biomed. Sci. 2016, 5, 19. [Google Scholar] [CrossRef]
- Cui, Y.; Hao, E.; Hui, G.; Guo, W.; Cui, F. Investigations on the Interactions of Diclofenac Sodium with HSA and ctDNA Using Molecular Modeling and Multispectroscopic Methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 110, 92–99. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Liu, Y.-N.; Wang, J. Estimation of Binding Constants for Diclofenac Sodium and Bovine Serum Albumin by Affinity Capillary Electrophoresis and Fluorescence Spectroscopy. J. Liq. Chromatogr. Relat. Technol. 2008, 31, 2077–2088. [Google Scholar] [CrossRef]
- Sekula, B.; Bujacz, A. Structural Insights into the Competitive Binding of Diclofenac and Naproxen by Equine Serum Albumin. J. Med. Chem. 2016, 59, 82–89. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, P.; Liang, S.; Zhou, Z.; Wu, X.; Yang, F.; Liang, H. Structural Basis of Non-Steroidal Anti-Inflammatory Drug Diclofenac Binding to Human Serum Albumin. Chem. Biol. Drug Des. 2015, 86, 1178–1184. [Google Scholar] [CrossRef]
- Oaks, J.L.; Gilbert, M.; Virani, M.Z.; Watson, R.T.; Meteyer, C.U.; Rideout, B.A.; Shivaprasad, H.L.; Ahmed, S.; Chaudhry, M.J.I.; Arshad, M.; et al. Diclofenac Residues as the Cause of Vulture Population Decline in Pakistan. Nature 2004, 427, 630–663. [Google Scholar] [CrossRef]
- Swan, G.E.; Cuthbert, R.; Quevedo, M.; Green, E.R.; Pain, D.J.; Bartels, P.; Cunningham, A.A.; Duncan, N.; Meharg, A.A.; Oaks, J.L.; et al. Toxicity of Diclofenac to Gyps Vultures. Biol. Lett. 2006, 2, 279–282. [Google Scholar] [CrossRef]
- Cuthbert, R.J.; Taggart, M.A.; Prakash, V.; Chakraborty, S.S.; Deori, P.; Galligan, T.; Kulkarni, M.; Ranade, S.; Saini, M.; Sharma, A.K.; et al. Avian Scavengers and the Threat from Veterinary Pharmaceuticals. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130574. [Google Scholar] [CrossRef]
- Auffinger, P.; Hays, F.A.; Westhof, E.; Ho, P.S. Halogen Bonds in Biological Molecules. Proc. Natl. Acad. Sci. USA 2004, 101, 16789–16794. [Google Scholar] [CrossRef]
- Bissantz, C.; Kuhn, B.; Stahl, M. A Medicinal Chemist’s Guide to Molecular Interactions. J. Med. Chem. 2010, 53, 5061–5084. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of Bovine, Equine and Leporine Serum Albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.; Darowski, N.; Fuchs, M.; Förster, R.; Hellmig, M.; Paithankar, K.; Pühringer, S.; Steffien, M.; Zocher, G.; Weiss, M.S. Facilities for Macromolecular Crystallography at the Helmholtz-Zentrum Berlin. J. Synchrotron Rad. 2012, 19, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Mueller, U.; Förster, R.; Hellmig, M.; Huschmann, F.U.; Kastner, A.; Malecki, P.; Pühringer, S.; Röwer, M.; Sparta, K.; Steffien, M.; et al. The macromolecular crystallography beamlines at BESSY II of the Helmholtz-Zentrum Berlin: Current status and perspectives. Eur. Phys. J. Plus 2015, 130, 141–150. [Google Scholar] [CrossRef]
- Berejnov, V.; Husseini, N.S.; Alsaied, O.A.; Thorne, R.E. Effects of Cryoprotectant Concentration and Cooling Rate on Vitrification of Aqueous Solutions. J. Appl. Crystallogr. 2006, 39, 244–251. [Google Scholar] [CrossRef]
- Bujacz, G.; Wrzesniewska, B.; Bujacz, A. Cryoprotection Properties of Salts of Organic Acids: A Case Study for a Tetragonal Crystal of HEW Lysozyme. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 789–796. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Cryst. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- Kabsch, W. Integration, scaling, space-group assignment and post refinement. Acta Crystallogr. 2010, 66, 133–144. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr. D Biol Crystallogr. 1997, 53, 240–255. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser Crystallographic Software. J Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Bujacz, A.; Talaj, J.A.; Zielinski, K.; Pietrzyk-Brzezinska, A.J.; Neumann, P. Crystal Structures of Serum Albumins from Domesticated Ruminants and Their Complexes with 3,5-Diiodosalicylic Acid. Acta Crystallogr. D Struct. Biol. 2017, 73, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.W.; McCoy, A.; et al. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder Maps: Improving OMIT Maps by Excluding Bulk-Solvent. Acta Crystallogr. D Struct. Biol. 2017, 73, 148–157. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-Atom Structure Validation for Macromolecular Crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Zielinski, K.; Sekula, B.; Bujacz, A.; Szymczak, I. Structural investigations of stereoselective profen binding by equine and leporine serum albumins. Chirality 2020, 32, 334–344. [Google Scholar] [CrossRef]
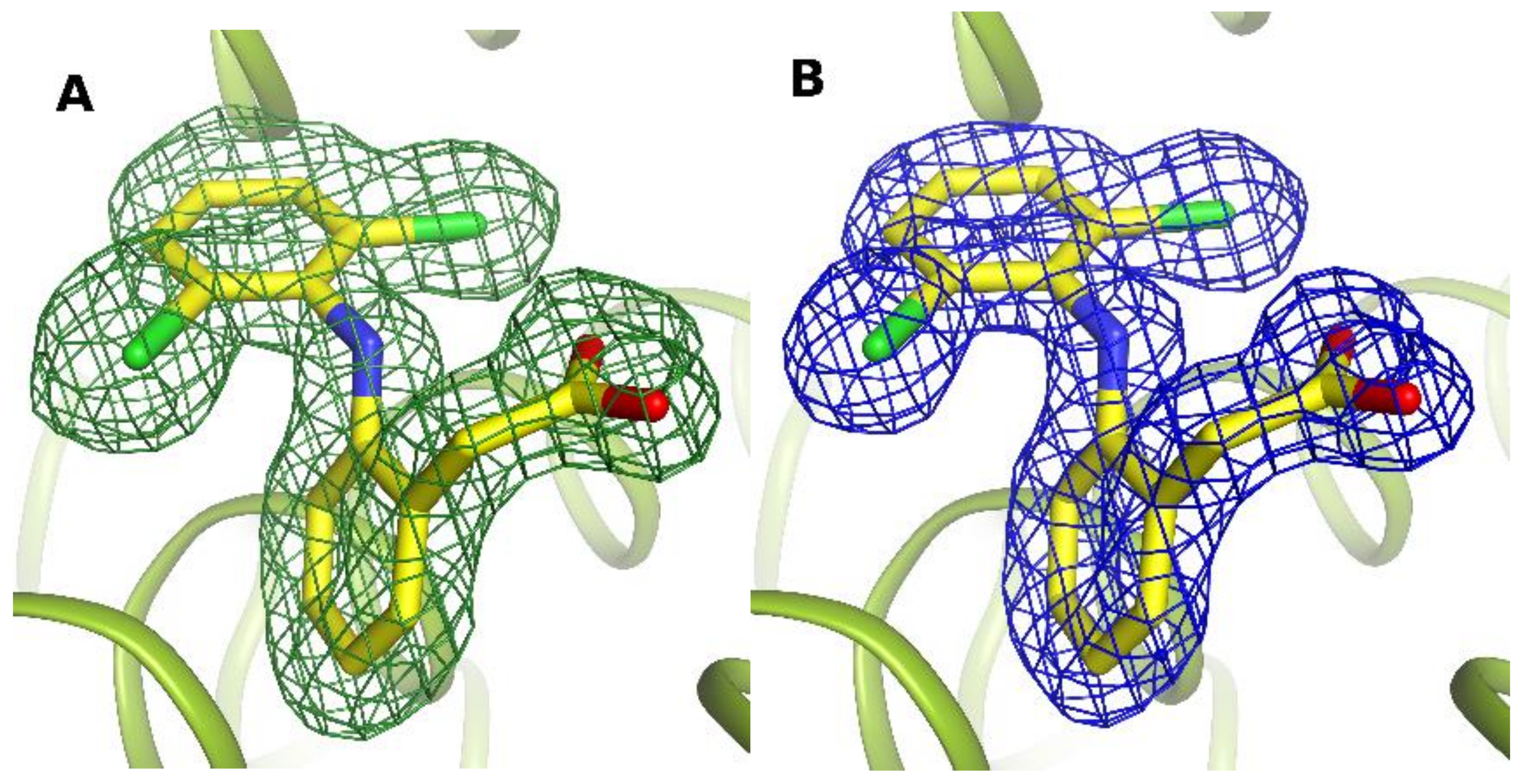
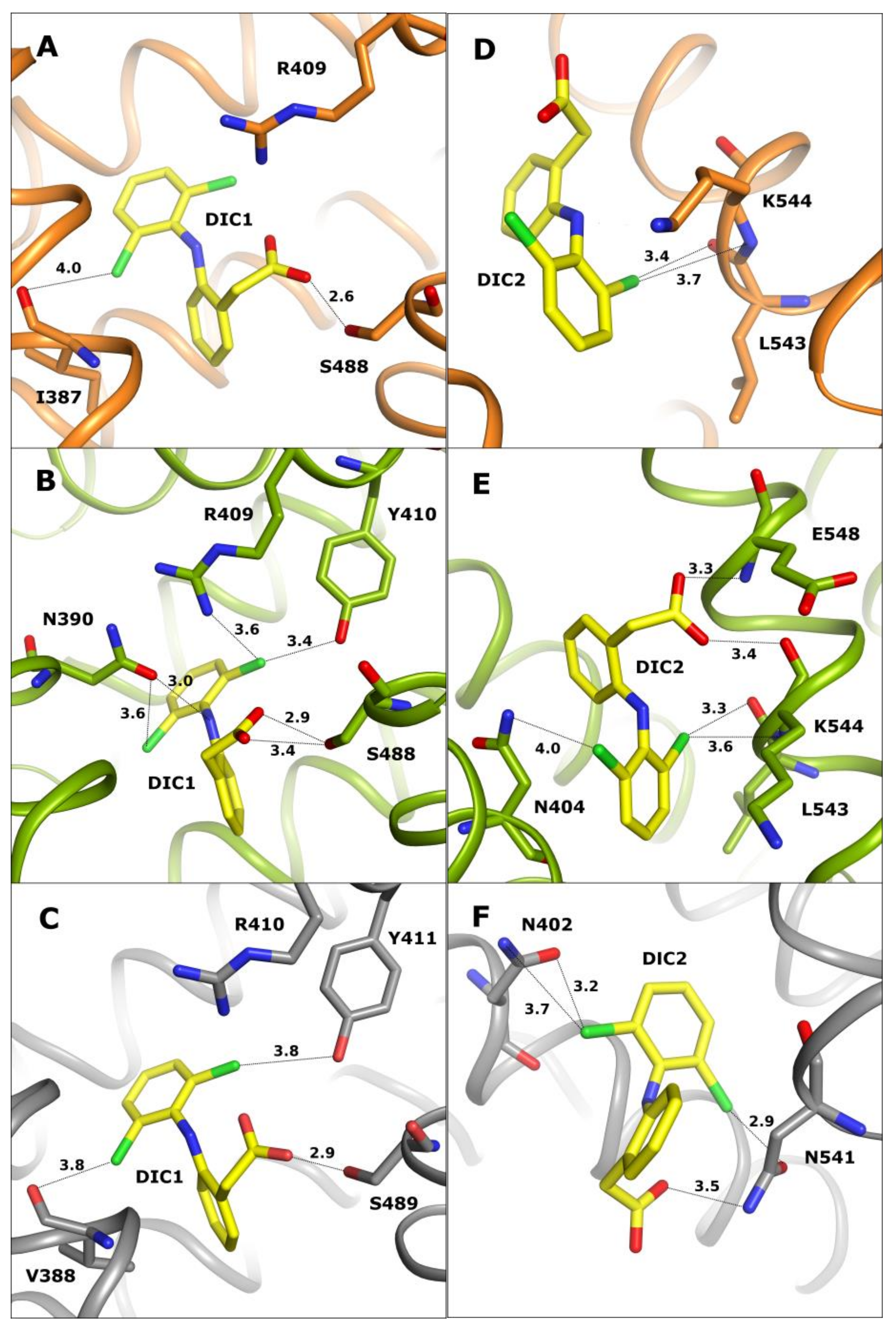

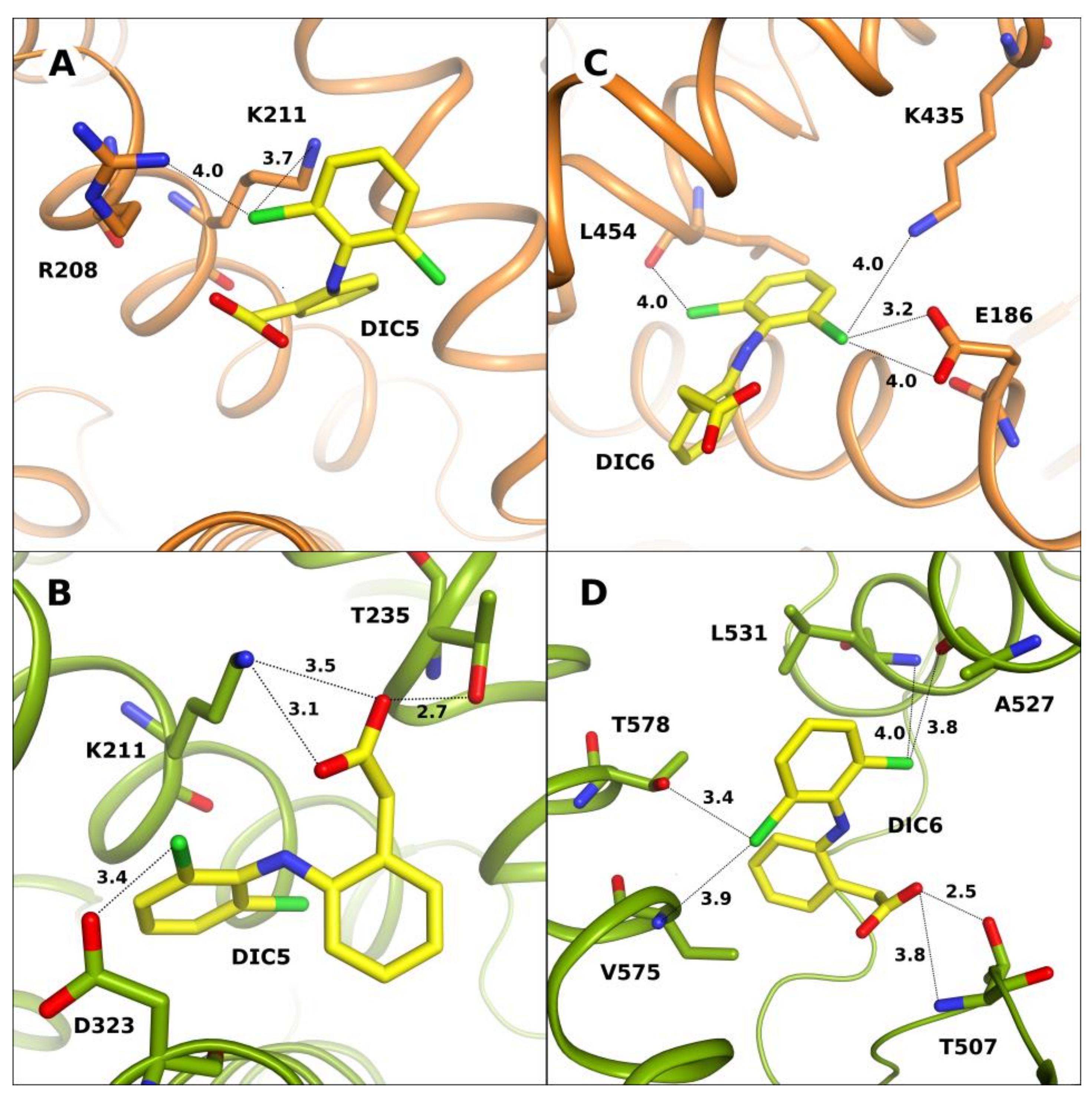
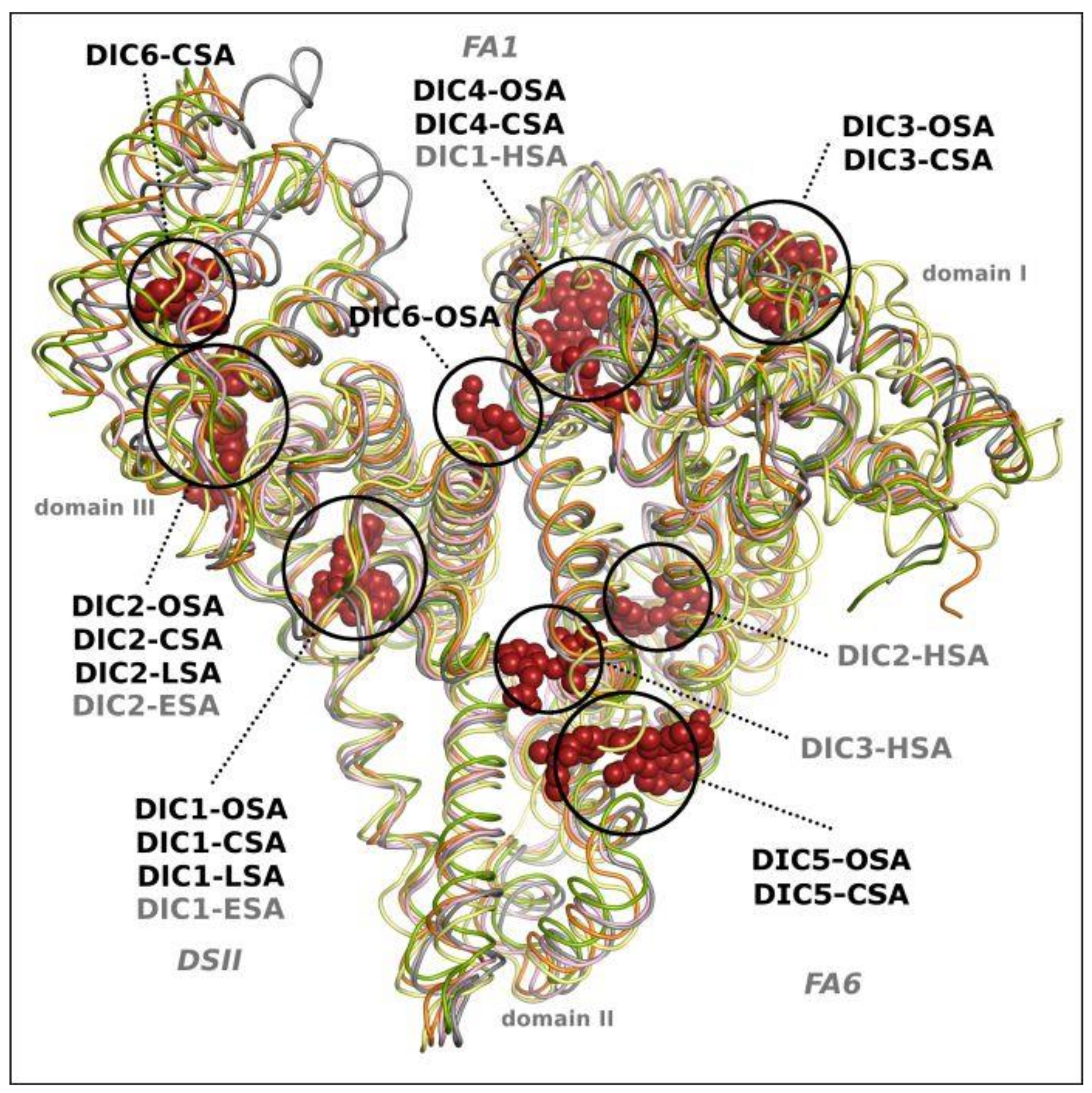
| PDB IDs of SA Complexes with DIC | Serum Albumin | Number of DIC | References |
|---|---|---|---|
| 6HN0 | Ovine (OSA-DIC) | 6 | This work |
| 6HN1 | Caprine (CSA-DIC) | 6 | This work |
| 8BSG | Leporine (LSA-DIC) | 2 | This work |
| 4ZBQ | Equine (ESA-DIC) | 2 | [27] |
| 4Z69 | Human (HSS-PA-DIC) | Chains: A/3, B/1 | [28] |
| Data Collection | OSA-DIC PDB ID: 6HN0 | CSA-DIC PDB ID: 6HN1 | LSA-DIC PDB ID: 8BSG |
|---|---|---|---|
| Space group | P3221 | P212121 | P212121 |
| Unit cell (Å, º) | a = 121.7, b = 121.7, c = 121.8 α = 90.0, β = 90.0, γ = 120.0 | a = 42.0, b = 66.3 c = 212.6 α = 90.0, β = 90.0, γ = 90.0 | a = 74.8, b = 80.1, c = 104.1 α = 90.0, β = 90.0, γ = 90.0 |
| VM [Å3/Da] | 3.9 | 2.2 | 2.3 |
| Radiation source | BX14 Petra | BESSY BL.14.2. | BESSY BL.14.2. |
| Resolution range (Å) | 50–2.12 (2.22–2.12) | 50–1.55 (1.65–1.55) | 50–1.89 (1.99–1.89) |
| Wavelength [Å] | 0.976 | 0.918 | 0.918 |
| Rmerge † (%) | 13.0 (142.7) | 4.9 (72.6) | 6.0 (132.7) |
| Mosaicity (º) | 0.064 | 0.328 | 0.430 |
| Completeness (%) | 98.9 (97.2) | 97.9 (91.9) | 99.7 (100) |
| Redundancy | 9.1 (8.7) | 5.9 (4.3) | 7.18 (6.95) |
| I/σI | 14.6 (2.1) | 22.1 (2.1) | 21.67 (2.09) |
| Rfree reflections | 2941 | 1111 | 2533 |
| Refinement | |||
| Molecules in asymmetric unit | 1 | 1 | 1 |
| R/Rfree ‡ (%) | 17.3/21.0 | 16.6/23.9 | 19.8/25.2 |
| Protein/ligand/water atoms | 4669/185/375 | 4714/115/501 | 4669/107/385 |
| R.m.s.d. bond lengths (Å) | 0.019 | 0.022 | 0.020 |
| R.m.s.d. bond angles (◦) | 1.822 | 1.981 | 2.040 |
| <B> protein (Å 2) overall | 45.1 | 28.0 | 49.95 |
| TLS bodies | 13 | - | 13 |
| Ramachandran plot | |||
| Most favored regions (%) | 97.7 | 97.2 | 95.65 |
| Allowed regions (%) | 2.3 | 2.8 | 4.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talaj, J.A.; Zielinski, K.; Bujacz, A. Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins. Int. J. Mol. Sci. 2023, 24, 1534. https://doi.org/10.3390/ijms24021534
Talaj JA, Zielinski K, Bujacz A. Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins. International Journal of Molecular Sciences. 2023; 24(2):1534. https://doi.org/10.3390/ijms24021534
Chicago/Turabian StyleTalaj, Julita A., Kamil Zielinski, and Anna Bujacz. 2023. "Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins" International Journal of Molecular Sciences 24, no. 2: 1534. https://doi.org/10.3390/ijms24021534
APA StyleTalaj, J. A., Zielinski, K., & Bujacz, A. (2023). Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins. International Journal of Molecular Sciences, 24(2), 1534. https://doi.org/10.3390/ijms24021534








