The Genome of Fusarium oxysporum f. sp. phaseoli Provides Insight into the Evolution of Genomes and Effectors of Fusarium oxysporum Species
Abstract
:1. Introduction
2. Results
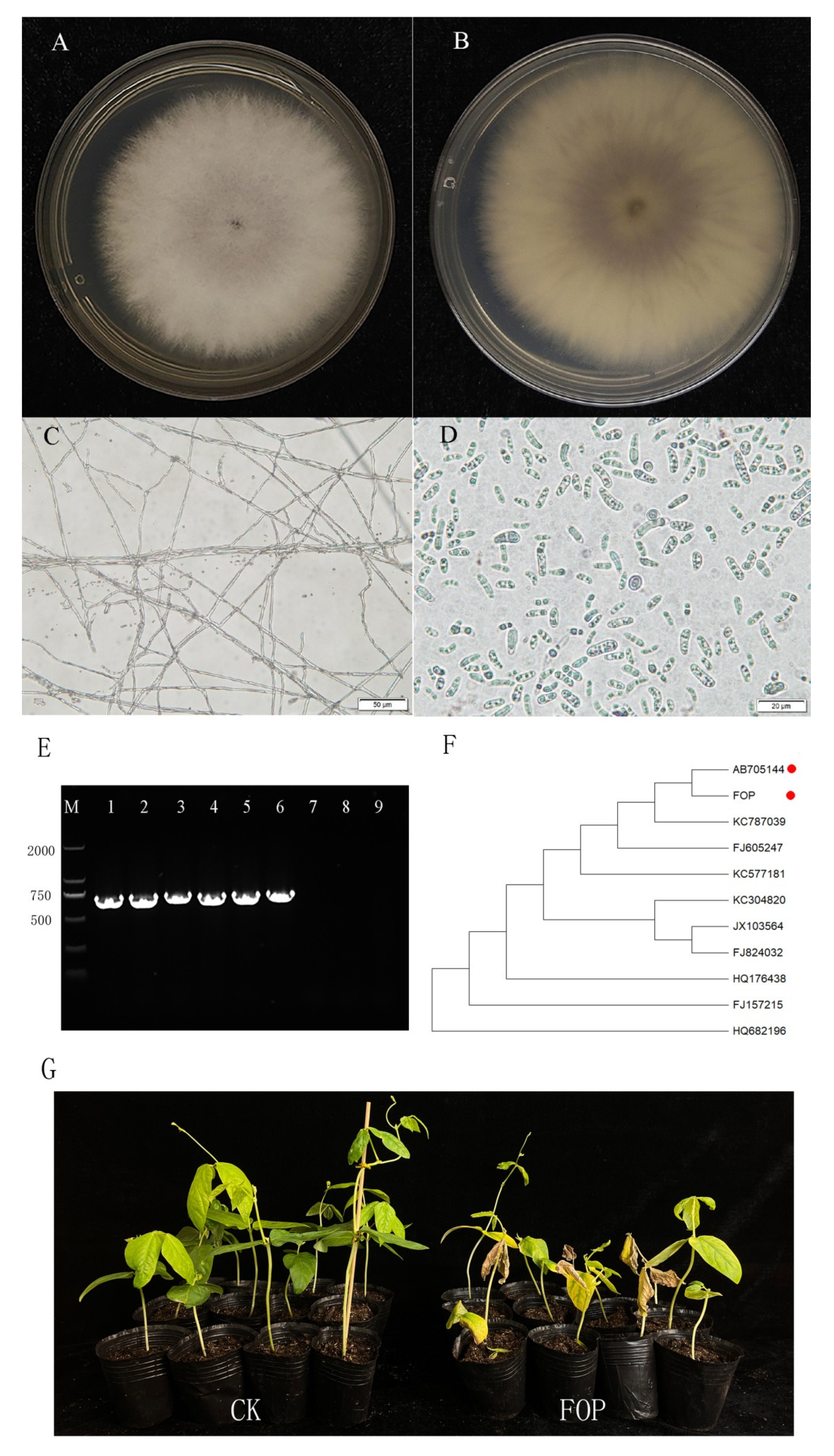
2.1. Morphological and Molecular Identification of the Strain Used in This Study
2.2. Phylogenetic Evolution Analysis of FOP
2.3. FOP Genome Assembly, Annotation and Comparative Genome Analysis
2.4. Comparative Analyses of Effectors among Five FO Genomes
3. Discussion
4. Materials and Methods
4.1. The Morphology and Pathogenicity Test for the FOP Strain
4.2. The Molecular Identification and Construction of Phylogenetic Trees for FOP
4.3. Genome Assembly and Annotation
4.4. Effector Annotation and Comparative Genome Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edel-Hermann, V.; Lecomte, C. Current Status of Fusarium oxysporum Formae Speciales and Races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, P.E. Life Cycle and Epidemiology of Fusarium oxysporum—ScienceDirect. In Fungal Wilt Diseases of Plants; Elsevier: Amsterdam, The Netherlands, 1981; pp. 51–80. [Google Scholar]
- Sasseron, G.R.; Benchimol-Reis, L.L.; Perseguini, J.M.K.C.; Paulino, J.F.C.; Bajay, M.M.; Carbonell, S.A.M.; Chiorato, A.F. Fusarium oxysporum f. sp. phaseoli genetic variability assessed by new developed microsatellites. Genet. Mol. Biol. 2020, 43, e20190267. [Google Scholar] [CrossRef] [PubMed]
- de Vega-Bartol, J.J.; Martín-Dominguez, R.; Ramos, B.; García-Sánchez, M.A.; Díaz-Mínguez, J.M. New virulence groups in Fusarium oxysporum f. sp. phaseoli: The expression of the gene coding for the transcription factor ftf1 correlates with virulence. Phytopathology 2011, 101, 470–479. [Google Scholar] [CrossRef] [Green Version]
- de Carvalho Paulino, J.F.; de Almeida, C.P.; Barbosa, C.C.F.; de Moraes, G.; Gonçalves, C.; Bueno, C.J.; Harakava, R.; Carbonell, S.A.M.; Chiorato, A.F.; Benchimol-Reis, L.L. Molecular and pathogenicity characterization of Fusarium oxysporum species complex associated with Fusarium wilt of common bean in Brazil. Trop. Plant Pathol. 2022, 47, 485–494. [Google Scholar] [CrossRef]
- Leitão, S.T.; Malosetti, M.; Song, Q.; van Eeuwijk, F.; Rubiales, D.; Vaz Patto, M.C. Natural Variation in Portuguese Common Bean Germplasm Reveals New Sources of Resistance Against Fusarium oxysporum f. sp. phaseoli and Resistance-Associated Candidate Genes. Phytopathology 2020, 110, 633–647. [Google Scholar] [CrossRef]
- Lonardi, S.; Muñoz-Amatriaín, M.; Liang, Q.; Shu, S.; Wanamaker, S.I.; Lo, S.; Tanskanen, J.; Schulman, A.H.; Zhu, T.; Luo, M.C.; et al. The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J. 2019, 98, 767–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, M.; Zeng, X.P.; Yan, W.R.; Ji, X.C.; Wang, H.F.; Zhao, Z.X. Pathogenic Identification of Cowpea Fusarium Wilt and Preliminary Research of Its Biological Characteristics in Hainan. Genom. Appl. Biol. 2015, 34, 345–349. [Google Scholar]
- Sun, J.Y.; Zhang, W.Z. Application status and breeding technology of new varieties of long bean in China. J. Chang. Jiang Veg. 2007, 10, 25–27. [Google Scholar]
- Reddy, M.V.; Sharma, S.B.; Nene, Y.L. Pigeonpea: Disease management. In The Pigeonpea; Nene, Y.L., Hall, S.D., Sheila, V.K., Eds.; CAB International: Wallingford, UK, 1990; pp. 303–347. [Google Scholar]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Corby Kistler, H.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar] [CrossRef]
- Hudson, O.; Waliullah, S.; Fulton, J.C.; Ji, P.; Dufault, N.S.; Keinath, A.; Ali, M.E. Marker Development for Differentiation of Fusarium Oxysporum f. sp. Niveum Race 3 from Races 1 and 2. Int. J. Mol. Sci. 2021, 22, 822. [Google Scholar] [CrossRef]
- Dobbs, J.T.; Kim, M.S.; Dudley, N.S.; Klopfenstein, N.B.; Yeh, A.; Hauff, R.D.; Jones, T.C.; Dumroese, R.K.; Cannon, P.G.; Stewart, J.E. Whole genome analysis of the koa wilt pathogen (Fusarium oxysporum f. sp. koae) and the development of molecular tools for early detection and monitoring. BMC Genom. 2020, 21, 764. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Song, S.Q.; Xu, J.; Zhou, J.; Zheng, S.X.; Xie, J.; Wang, X.; Peng, S.; Zhu, X.Q.; Song, R. First report of Fusarium oxysporum causing stem spots on Polygonatum odoratum in China. Plant Dis. 2022; online ahead of print. [Google Scholar]
- Khan, G.; Gleason, M.L.; Irshad, G.; Naz, F.; Mayfield, D.; Shokanova, A. First report of Fusarium sporotrichioides causing Fusarium fruit rot on peaches in Pakistan. Plant Dis. 2022; online ahead of print. [Google Scholar]
- da Silva Ribeiro, A.; Polonio, J.C.; Dos Santos Oliveira, J.A.; Ferreira, A.P.; Alves, L.H.; Mateus, N.J.; Mangolin, C.A.; de Azevedo, J.L.; Pamphile, J.A. Retrotransposons and multilocus sequence analysis reveals diversity and genetic variability in endophytic fungi-associated with Serjania laruotteana Cambess. Braz. J. Microbiol. 2021, 52, 2179–2192. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yu, H.; Ma, L.J. Accessory Chromosomes in Fusarium oxysporum. Phytopathology 2020, 110, 1488–1496. [Google Scholar] [CrossRef] [PubMed]
- Sabahi, F.; Banihashemi, Z.; de Sain, M.; Rep, M. Genome sequences of 38 Fusarium oxysporum strains. BMC Res. Notes 2022, 15, 229. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Sharma, M.; Thatcher, L.F.; Azam, S.; Hane, J.K.; Sperschneider, J.; Kidd, B.N.; Anderson, J.P.; Ghosh, R.; Garg, G.; et al. Comparative genomics and prediction of conditionally dispensable sequences in legume-infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genom. 2016, 17, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dam, P.; Rep, M. The Distribution of Miniature Impala Elements and SIX Genes in the Fusarium Genus is Suggestive of Horizontal Gene Transfer. J. Mol. Evol. 2017, 85, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Batson, A.M.; Fokkens, L.; Rep, M.; du Toit, L.J. Putative Effector Genes Distinguish Two Pathogenicity Groups of Fusarium oxysporum f. sp. spinaciae. Mol. Plant Microbe Interact. 2021, 34, 141–156. [Google Scholar] [CrossRef]
- Rep, M.; van der Does, H.C.; Meijer, M.; van Wijk, R.; Houterman, P.M.; Dekker, H.L.; de Koster, C.G.; Cornelissen, B.J. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef]
- Gawehns, F.; Houterman, P.M.; Ichou, F.A.; Michielse, C.B.; Hijdra, M.; Cornelissen, B.J.; Rep, M.; Takken, F.L. The Fusarium oxysporum effector Six6 contributes to virulence and suppresses I-2-mediated cell death. Mol. Plant Microbe Interact. 2014, 27, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Houterman, P.M.; Gawehns, F.; Cao, L.; Sillo, F.; Richter, H.; Clavijo-Ortiz, M.J.; Schmidt, S.M.; Boeren, S.; Vervoort, J.; et al. The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. New Phytol. 2015, 208, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Jangir, P.; Mehra, N.; Sharma, K.; Singh, N.; Rani, M.; Kapoor, R. Secreted in Xylem Genes: Drivers of Host Adaptation in Fusarium oxysporum. Front. Plant Sci. 2021, 12, 628611. [Google Scholar] [CrossRef] [PubMed]
- Ponukumati, S.V.; Elliott, M.L.; Des Jardin, E.A. Comparison of Secreted in Xylem (SIX) genes in two fusarium wilt pathogens of ornamental palms. Plant Pathol. 2019, 68, 1663–1681. [Google Scholar] [CrossRef]
- Schmidt, S.M.; Houterman, P.M.; Schreiver, I.; Ma, L.; Amyotte, S.; Chellappan, B.; Boeren, S.; Takken, F.L.; Rep, M. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genom. 2013, 14, 119. [Google Scholar] [CrossRef] [Green Version]
- van Dam, P.; Fokkens, L.; Schmidt, S.M.; Linmans, J.H.; Kistler, H.C.; Ma, L.J.; Rep, M. Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 2016, 18, 4087–4102. [Google Scholar] [CrossRef]
- Lv, H.H.; Fang, Z.Y.; Yang, L.M.; Xie, B.Y.; Liu, Y.M.; Zhuang, M.; Zhang, Y.Y.; Yang, Y. Research on screening of resistant resources to Fusarium wilt and inheritance of the resistant gene in cabbage. Acta Hortic. Sin. 2011, 38, 875–885. [Google Scholar]
- Zhang, Y.; Zheng, J.Q.; Wu, X.H.; Shi, Y.C.; Gu, P.Y.; Li, J.Q. Investigation of occurrences and damage of cabbage wilt in Yanqing country of Beijing. Chin. Agric. Sci. Bull. 2007, 23, 315–320. [Google Scholar]
- Magdama, F.; Monserrate-Maggi, L.; Serrano, L.; García Onofre, J.; Jiménez-Gasco, M.D.M. Genetic Diversity of Fusarium oxysporum f. sp. cubense, the Fusarium Wilt Pathogen of Banana, in Ecuador. Plants 2020, 9, 1133. [Google Scholar] [CrossRef]
- Wagner, T.A.; Gu, A.; Duke, S.E.; Bell, A.A.; Magill, C.; Liu, J. Genetic Diversity and Pathogenicity of Verticillium dahliae Isolates and Their Co-occurrence with Fusarium oxysporum f. sp. vasinfectum Causing Cotton Wilt in Xinjiang, China. Plant Dis. 2021, 105, 978–985. [Google Scholar] [CrossRef]
- Bell, A.A.; Kemerait, R.C.; Ortiz, C.S.; Prom, S.; Quintana, J.; Nichols, R.L.; Liu, J. Genetic Diversity, Virulence, and Meloidogyne incognita Interactions of Fusarium oxysporum Isolates Causing Cotton Wilt in Georgia. Plant Dis. 2017, 101, 948–956. [Google Scholar] [CrossRef] [Green Version]
- Halpern, H.C.; Qi, P.; Kemerait, R.C.; Brewer, M.T. Genetic Diversity and Population Structure of Races of Fusarium oxysporum Causing Cotton Wilt. G3 (Bethesda) 2020, 10, 3261–3269. [Google Scholar] [CrossRef] [PubMed]
- Alves-Santos, F.M.; Cordeiro-Rodrigues, L.; Sayagues, J.M.; Martin-Dominguez, R.; Garcia-Benavides, P.; Crespo, M.C.; D’ıaz-Minguez, J.M.; Eslava, A.P. Pathogenicity and race characterization of Fusarium oxysporum f.sp. phaseoli isolates from Spain and Greece. Plant Pathol. 2002, 51, 605–611. [Google Scholar] [CrossRef]
- Westphal, K.R.; Wollenberg, R.D.; Herbst, F.A.; Sørensen, J.L.; Sondergaard, T.E.; Wimmer, R. Enhancing the Production of the Fungal Pigment Aurofusarin in Fusarium graminearum. Toxins 2018, 10, 485. [Google Scholar] [CrossRef] [Green Version]
- Blin, K.; Wolf, T.; Chevrette, M.G.; Lu, X.; Schwalen, C.J.; Kautsar, S.A.; Suarez Duran, H.G.; de Los Santos, E.L.C.; Kim, H.U.; Nave, M.; et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017, 45, W36–W41. [Google Scholar] [CrossRef] [PubMed]
- Hoogendoorn, K.; Barra, L.; Waalwijk, C.; Dickschat, J.S.; van der Lee, T.A.J.; Medema, M.H. Evolution and Diversity of Biosynthetic Gene Clusters in Fusarium. Front. Microbiol. 2018, 9, 1158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Xing, M.; Kong, C.; Fang, Z.; Yang, L.; Zhang, Y.; Wang, Y.; Ling, J.; Yang, Y.; Lv, H. Genetic Diversity, Virulence, Race Profiling, and Comparative Genomic Analysis of the Fusarium oxysporum f. sp. conglutinans Strains Infecting Cabbages in China. Front. Microbiol. 2019, 10, 1373. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.; Hill, C.M.; Wu, S.; Ruan, J.; Ma, Z.S. DBG2OLC: Efficient Assembly of Large Genomes Using Long Erroneous Reads of the Third Generation Sequencing Technologies. Sci Rep. 2016, 6, 31900. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, M.; Baldwin-Brown, J.G.; Long, A.D.; Emerson, J.J. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016, 44, e147. [Google Scholar]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Ling, J.; Mao, Z.; Zhai, M.; Zeng, F.; Yang, Y.; Xie, B. Transcriptome profiling of Cucumis metuliferus infected by Meloidogyne incognita provides new insights into putative defense regulatory network in Cucurbitaceae. Sci Rep. 2017, 7, 3544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed] [Green Version]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Delcher, A.L.; Salzberg, S.L.; Phillippy, A.M. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinform. 2003, 10, 3.1–3.18. [Google Scholar] [CrossRef]



| Statistics of Genome Assembly | FOP |
|---|---|
| length of genome assembly (Mb) | 53.70 |
| number of contigs | 106 |
| N50 of contigs (Mb) | 4.32 |
| total length of retrotransposons (Mb) | 28.14 |
| number of annotated genes | 14,694 |
| average gen length (bp) | 1547 |
| average CDS length (bp) | 1203 |
| average protein length (bp) | 401 |
| genome completeness (BUSCO) | 94.31% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Y.; Li, Y.; Ping, X.; Yang, Q.; Mao, Z.; Zhao, J.; Lu, X.; Xie, B.; Yang, Y.; Ling, J. The Genome of Fusarium oxysporum f. sp. phaseoli Provides Insight into the Evolution of Genomes and Effectors of Fusarium oxysporum Species. Int. J. Mol. Sci. 2023, 24, 963. https://doi.org/10.3390/ijms24020963
Hao Y, Li Y, Ping X, Yang Q, Mao Z, Zhao J, Lu X, Xie B, Yang Y, Ling J. The Genome of Fusarium oxysporum f. sp. phaseoli Provides Insight into the Evolution of Genomes and Effectors of Fusarium oxysporum Species. International Journal of Molecular Sciences. 2023; 24(2):963. https://doi.org/10.3390/ijms24020963
Chicago/Turabian StyleHao, Yali, Yan Li, Xingxing Ping, Qihong Yang, Zhenchuan Mao, Jianlong Zhao, Xiaofei Lu, Bingyan Xie, Yuhong Yang, and Jian Ling. 2023. "The Genome of Fusarium oxysporum f. sp. phaseoli Provides Insight into the Evolution of Genomes and Effectors of Fusarium oxysporum Species" International Journal of Molecular Sciences 24, no. 2: 963. https://doi.org/10.3390/ijms24020963
APA StyleHao, Y., Li, Y., Ping, X., Yang, Q., Mao, Z., Zhao, J., Lu, X., Xie, B., Yang, Y., & Ling, J. (2023). The Genome of Fusarium oxysporum f. sp. phaseoli Provides Insight into the Evolution of Genomes and Effectors of Fusarium oxysporum Species. International Journal of Molecular Sciences, 24(2), 963. https://doi.org/10.3390/ijms24020963






