Pharmacological Activation of YAP/TAZ by Targeting LATS1/2 Enhances Periodontal Tissue Regeneration in a Murine Model
Abstract
1. Introduction
2. Results
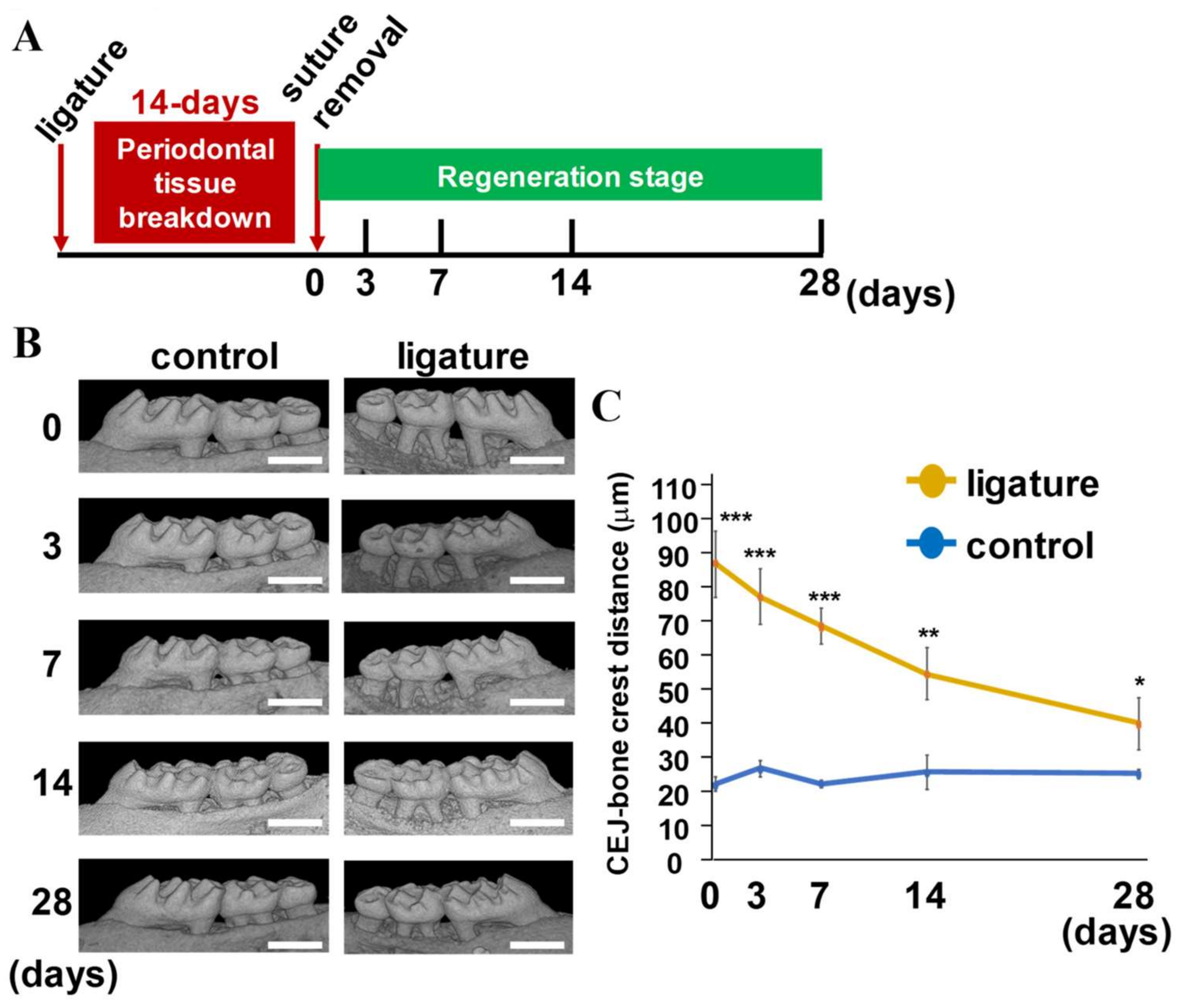
2.1. Evaluation of Sequential Alveolar Bone Recovery and Cementum Remodeling during Periodontal Tissue Regeneration
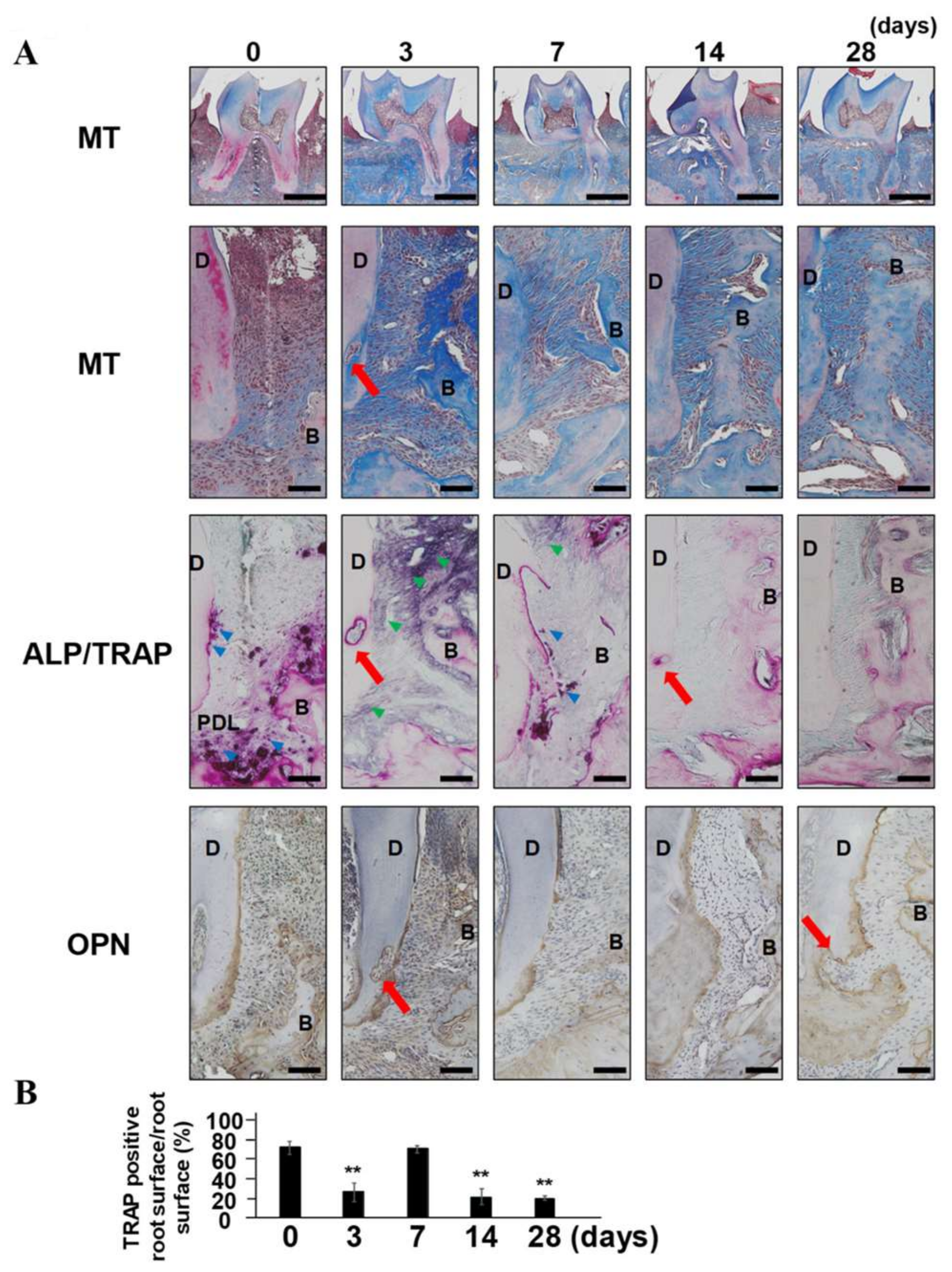
2.2. The Reappearance of TRAP-Positive Area along with Root Surface and External Root Absorption
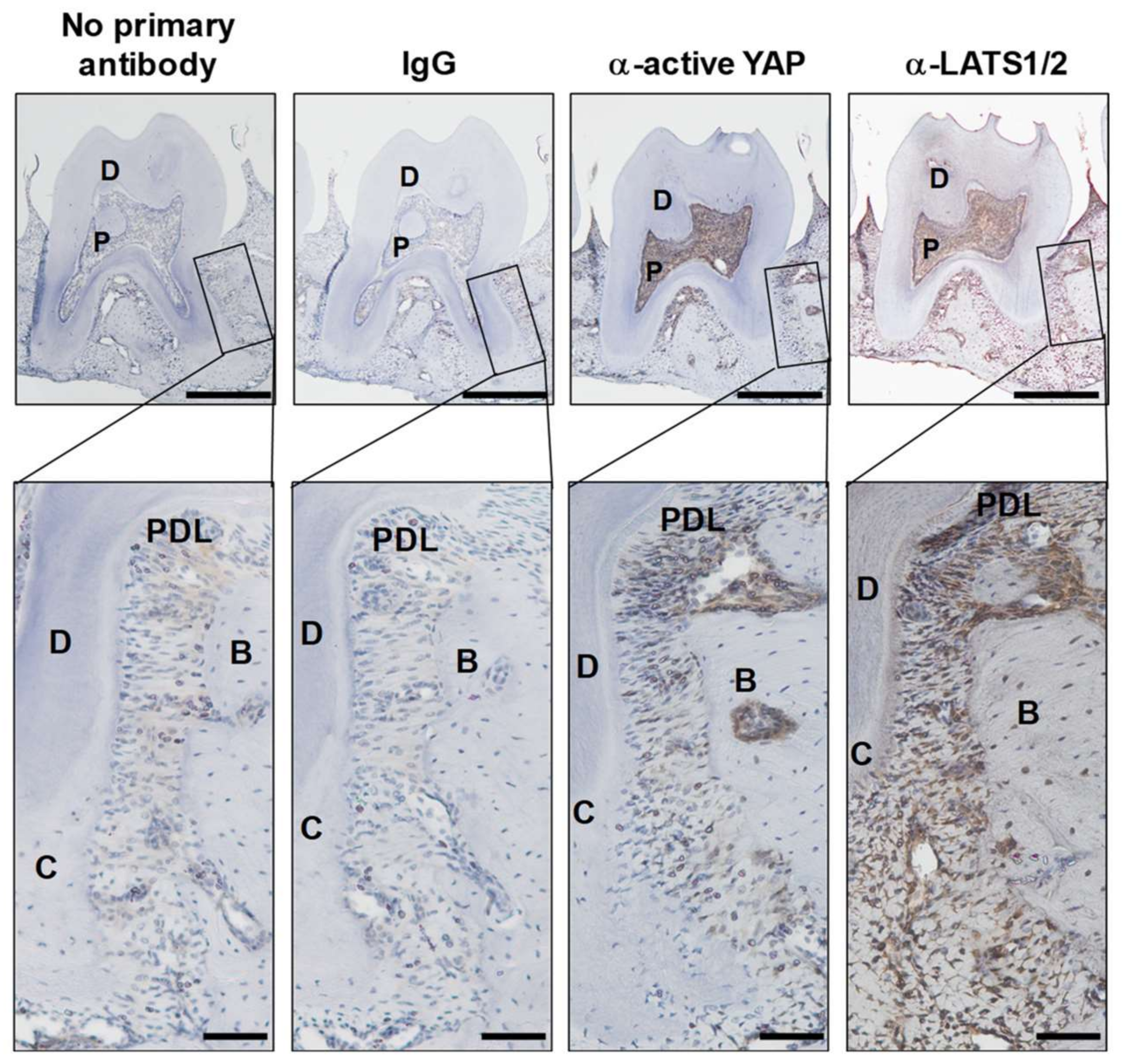
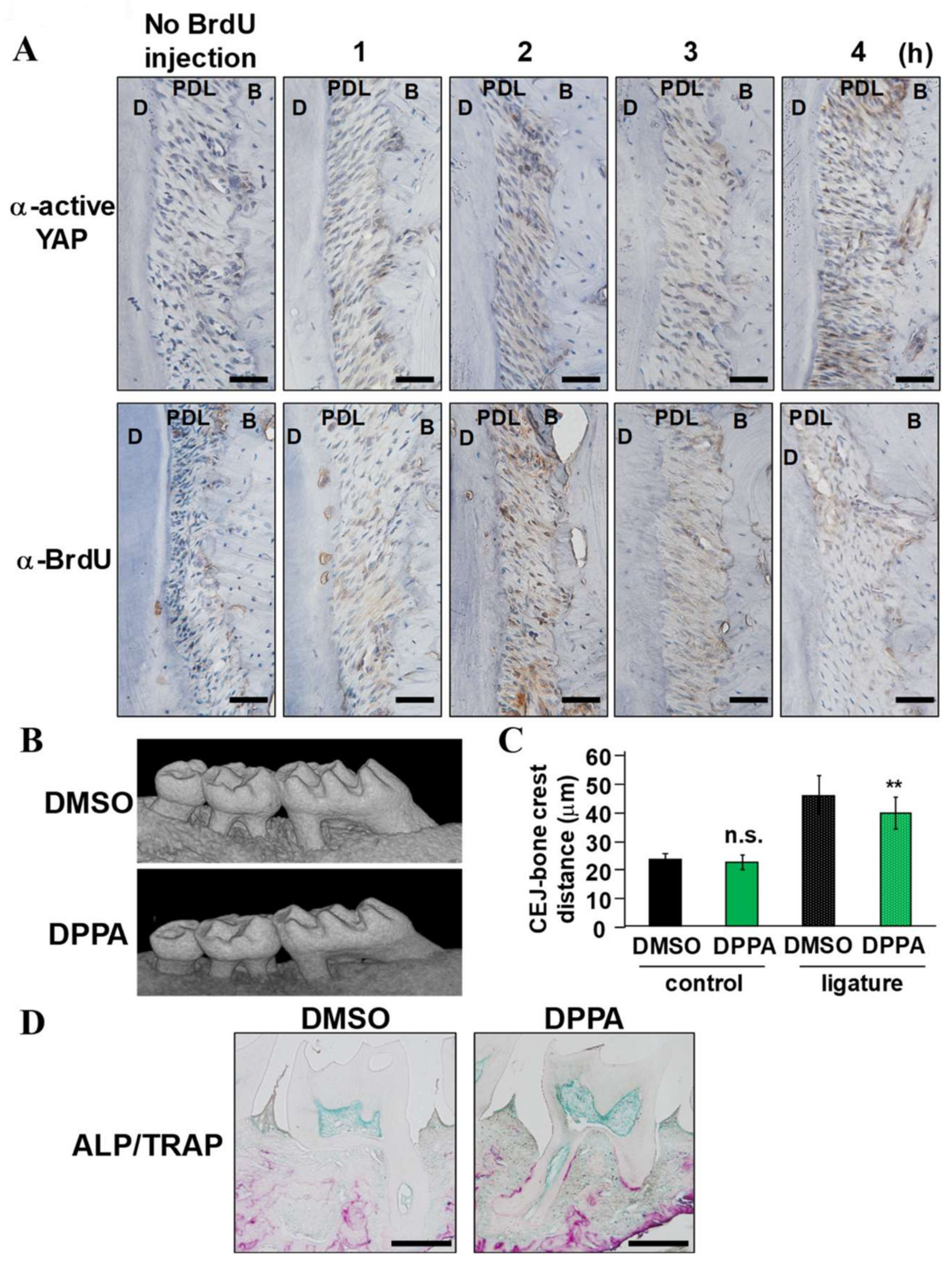
2.3. Expression of Active YAP and Its Regulator LATS1/2 in Periodontal Tissue
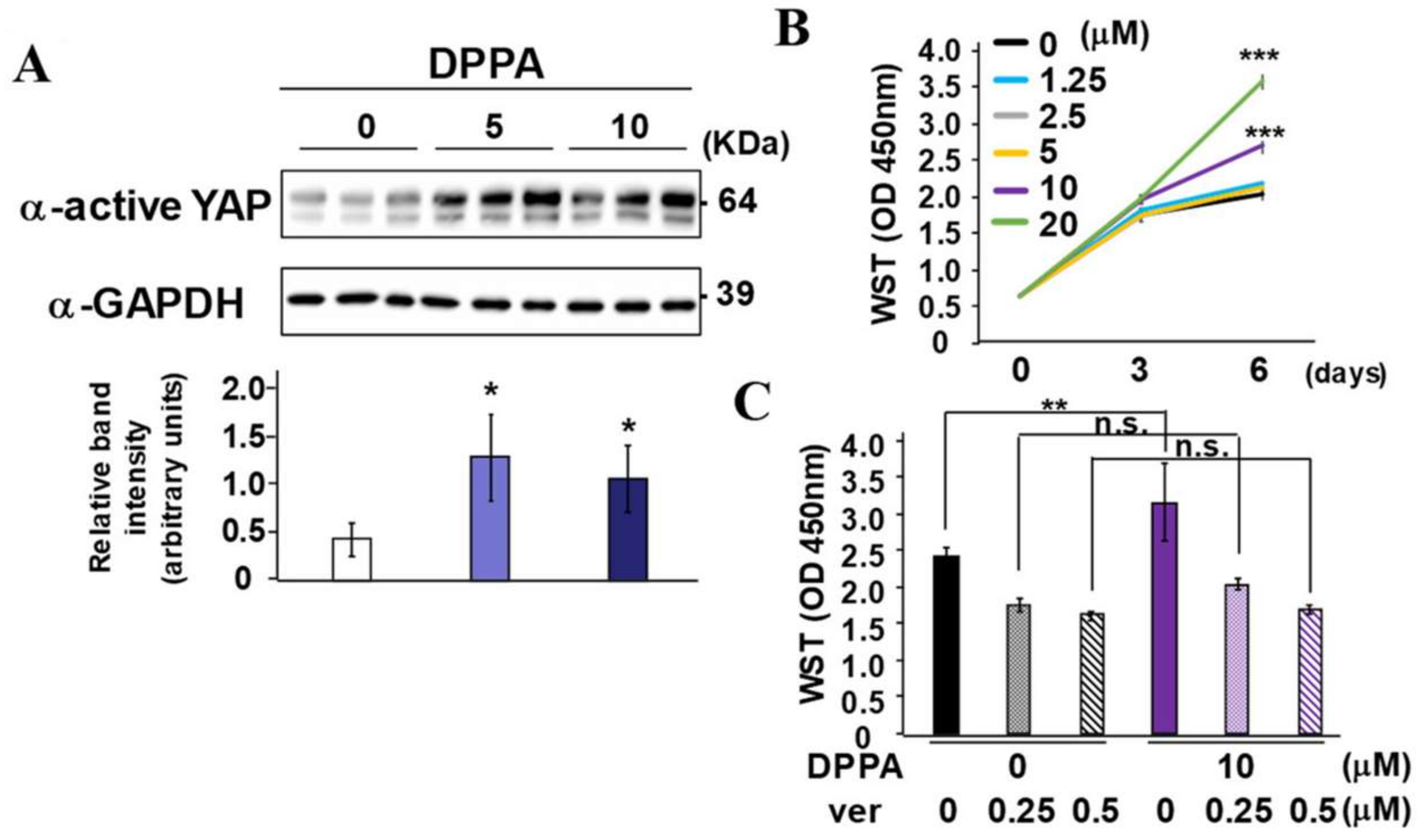
2.4. DPPA Acceleration of Periodontal Regeneration
2.5. DPPA Stimulation of PDLF Proliferation Dependence on YAP/TAZ Activity
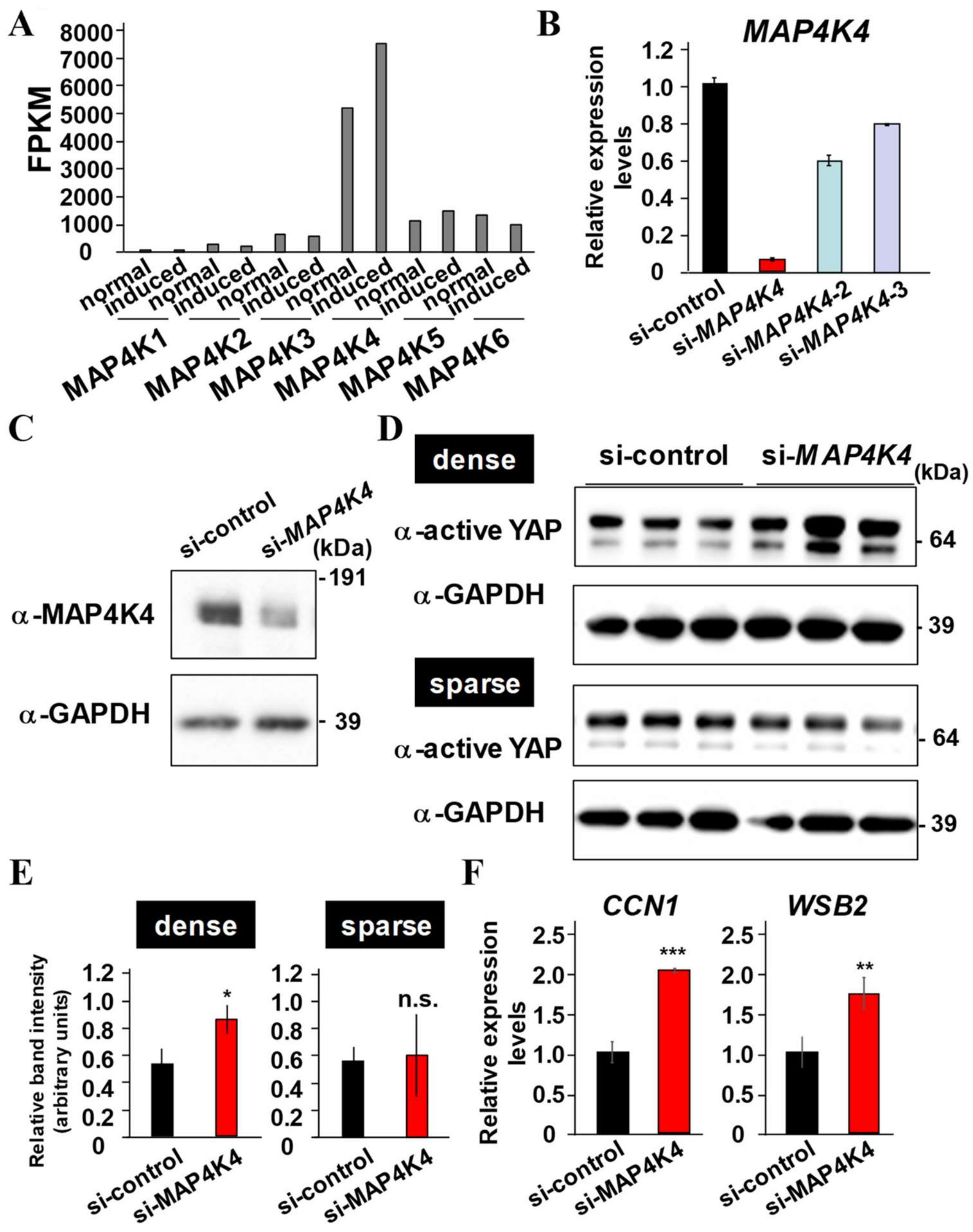
2.6. MAP4K4 Suppression Activates YAP
2.7. MAP4K4 Kinase Inhibition Enhances Proliferation and Osteo/Cementogenic Differentiation of PDLF by Dependently on YAP/TAZ
3. Discussion
4. Material and Methods
4.1. Reagents
4.2. Experimental Animals
4.3. Micro-Computed Tomography
4.4. Histology
4.5. Cell Culture and Stable Cell Generation
4.6. Quantitative PCR (qPCR) Analysis
4.7. Transient Transfection of siRNA
4.8. Immunoblotting
4.9. ALP Activity
4.10. Alizarin Red S Staining
4.11. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
Appendix A
| Primer Name | Direction | Sequence |
|---|---|---|
| MAP4K4 | forward | AATCGTCCGCCAGAGTACAG |
| reverse | AGTGGAGGAGGAATGGCTTT | |
| CCN1 | forward | ACTTCATGGTCCCAGTGCTC |
| reverse | TGCATTTCTTGCCCTTTTTC | |
| WSB2 | forward | CCAGAAGGCTTGTACCTTCG |
| reverse | TGACTCCACCATGTGGAAAA | |
| ALPL | forward | GACAAGAAGCCCTTCACTGC |
| reverse | AGACTGCGCCTGGTAGTTGT | |
| COL1A1 | forward | GTGCTAAAGGTGCCAATGGT |
| reverse | ACCAGGTTCACCGCTGTTAC | |
| RUNX2 | forward | CGCATTCCTCATCCCAGTAT |
| reverse | GCCTGGGGTCTGTAATATGA | |
| AM-tag | forward | CTTACGGGTGCCAAGATCC |
| reverse | CCAGAGGCCACTTGTGTAGC | |
| HPRT | forward | TGGCGTCGTGATTAGTGATG |
| reverse | CGAGCAAGACGTTCAGTCCT |
| Target Sequences for siRNA Duplex | |||
|---|---|---|---|
| Oligo Name | Sense Strand (5′→3′) | Antisense Strand (5′→3′) | Location |
| si-MAP4K4 | CACUAUUCUGAGGGAAAAUCU | AUUUUCCCUCAGAAUAGUGCA | CDS |
| si-MAP4K4-2 | UGUAUGAGUCAUACAAAUGUU | CAUUUGUAUGACUCAUACAUA | 3UTR |
| si-MAP4K4-3 | AAUUCAUGUCAUAGAUAACGA | GUUAUCUAUGACAUGAAUUCC | 3UTR |
| si-control | GUACCGCACGUCAUUCGUAUC | UACGAAUGACGUGCGGUACGU | - |
References
- Usui, M.; Onizuka, S.; Sato, T.; Kokabu, S.; Ariyoshi, W.; Nakashima, K. Mechanism of alveolar bone destruction in periodontitis—Periodontal bacteria and inflammation. Jpn. Dent. Sci. Rev. 2021, 57, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Saltiel, A.R.; Olefsky, J.M. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Investig. 2017, 127, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Korniluk, A.; Koper, O.; Kemona, H.; Piekarska, V.D. From inflammation to cancer. Ir. J. Med. Sci. 2017, 186, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, G.; Descarpentrie, J.; Evrard, S.; Khatib, A.M. Proprotein convertases: Key players in inflammation-related malignancies and metastasis. Cancer Lett. 2020, 473, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Jimi, E.; Takakura, N.; Hiura, F.; Nakamura, I.; Hirata-Tsuchiya, S. The Role of NF-κB in Physiological Bone Development and Inflammatory Bone Diseases: Is NF-κB Inhibition “Killing Two Birds with One Stone”? Cells 2019, 8, 1636. [Google Scholar] [CrossRef]
- Bassir, S.H.; Wisitrasameewong, W.; Raanan, J.; Ghaffarigarakani, S.; Chung, J.; Freire, M.; Andrada, L.C.; Intini, G. Potential for Stem Cell-Based Periodontal Therapy. J. Cell. Physiol. 2016, 231, 50–61. [Google Scholar] [CrossRef]
- Tsai, S.J.; Ding, Y.W.; Shih, M.C.; Tu, Y.K. Systematic review and sequential network meta-analysis on the efficacy of periodontal regenerative therapies. J. Clin. Periodontol. 2020, 47, 1108–1120. [Google Scholar] [CrossRef]
- Mikami, R.; Sudo, T.; Fukuba, S.; Takeda, K.; Matsuura, H.; Kariya, T.; Takeuchi, S.; Ochiai, A.; Kawamoto, S.; Toyoshima, K.; et al. Prognostic factors affecting periodontal regenerative therapy using recombinant human fibroblast growth factor-2: A 3-year cohort study. Regen. Ther. 2022, 21, 271–276. [Google Scholar] [CrossRef]
- Iwayama, T.; Sakashita, H.; Takedachi, M.; Murakami, S. Periodontal tissue stem cells and mesenchymal stem cells in the periodontal ligament. Jpn. Dent. Sci. Rev. 2022, 58, 172–178. [Google Scholar] [CrossRef]
- Dasgupta, I.; McCollum, D. Control of cellular responses to mechanical cues through YAP/TAZ regulation. J. Biol. Chem. 2019, 294, 17693–17706. [Google Scholar] [CrossRef]
- Lehoux, S.; Tedgui, A. Signal transduction of mechanical stresses in the vascular wall. Hypertension 1998, 32, 338–345. [Google Scholar] [CrossRef]
- Suki, B.; Sato, S.; Parameswaran, H.; Szabari, M.V.; Takahashi, A.; Suki, E.B. Emphysema and mechanical stress-induced lung remodeling. Physiology 2013, 28, 404–413. [Google Scholar] [CrossRef]
- Nitsan, i.; Drori, S.; Lewis, Y.E.; Cohen, S.; Tzlil, S. Mechanical communication in cardiac cell synchronized beating. Nat. Phys. 2016, 12, 472–477. [Google Scholar] [CrossRef]
- Wang, L.; You, X.; Zhang, L.; Zhang, C.; Zou, W. Mechanical regulation of bone remodeling. Bone Res. 2022, 10, 16. [Google Scholar] [CrossRef]
- Wang, Y.; Shi, J.; Tong, X. Cross-Talk between Mechanosensitive Ion Channels and Calcium Regulatory Proteins in Cardiovascular Health and Disease. Int. J. Mol. Sci. 2021, 22, 8782. [Google Scholar] [CrossRef]
- Coutiño, B.C.; Mayor, R. Mechanosensitive ion channels in cell migration. Cells Dev. 2021, 166, 203683. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Pavie, V.M.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.S.; Lu, W.; Lu, S.; et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6, 8357. [Google Scholar] [CrossRef]
- Chuang, H.; Wang, X.; Tan, T. MAP4K Family Kinases in Immunity and Inflammation. Adv. Immunol. 2016, 129, 277–314. [Google Scholar] [CrossRef]
- Pandya, M.; Gopinathan, G.; Tillberg, C.; Wang, J.; Luan, X.; Diekwisch, T. The Hippo Pathway Effectors YAP/TAZ Are Essential for Mineralized Tissue Homeostasis in the Alveolar Bone/Periodontal Complex. J. Dev. Biol. 2022, 10, 14. [Google Scholar] [CrossRef]
- Wu, H.; Taya, Y.; Kuraji, R.; Ito, H.; Soeno, Y.; Numabe, Y. Dynamic microstructural changes in alveolar bone in ligature-induced experimental periodontitis. Odontology 2020, 108, 339–349. [Google Scholar] [CrossRef]
- Lin, P.; Niimi, H.; Ohsugi, Y.; Tsuchiya, Y.; Shimohira, T.; Komatsu, K.; Liu, A.; Shiba, T.; Aoki, A.; Iwata, T.; et al. Application of Ligature-Induced Periodontitis in Mice to Explore the Molecular Mechanism of Periodontal Disease. Int. J. Mol. Sci. 2021, 18, 8900. [Google Scholar] [CrossRef]
- Lemaitre, M.; Monsarrat, P.; Blasco-Baque, V.; Loubières, P.; Burcelin, R.; Casteilla, L.; Planat-Bénard, V.; Kémoun, P. Periodontal Tissue Regeneration Using Syngeneic Adipose-Derived Stromal Cells in a Mouse Model. Stem Cells Transl. Med. 2017, 6, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Suzuki, S.; Hirata-Tsuchiya, S.; Sato, A.; Nemoto, E.; Saito, M.; Shiba, H.; Yamada, S. PPARγ-Induced Global H3K27 Acetylation Maintains Osteo/Cementogenic Abilities of Periodontal Ligament Fibroblasts. Int. J. Mol. Sci. 2021, 22, 8646. [Google Scholar] [CrossRef] [PubMed]
- Arzate, H.; David, M.Z.; Celis, G.M. Cementum proteins: Role in cementogenesis, biomineralization, periodontium formation and regeneration. Periodontol. 2000 2015, 67, 211–233. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Foster, B.L.; Bonewald, L.F. The Cementocyte-An Osteocyte Relative? J. Dent. Res. 2016, 95, 734–741. [Google Scholar] [CrossRef]
- Graziani, F.; Laurell, L.; Tonetti, M.; Gottlow, J.; Berglundh, T. Periodontal wound healing following GTR therapy of dehiscence-type defects in the monkey: Short-, medium- and long-term healing. J. Clin. Periodontol. 2005, 32, 905–914. [Google Scholar] [CrossRef]
- Anzai, J.; Nagayasu-Tanaka, T.; Terashima, A.; Asano, T.; Yamada, S.; Nozaki, T.; Kitamura, M.; Murakami, S. Long-term Observation of Regenerated Periodontium Induced by FGF-2 in the Beagle Dog 2-Wall Periodontal Defect Model. PLoS ONE 2016, 11, e0158485. [Google Scholar] [CrossRef]
- Boopathy, G.T.K.; Hong, W. Role of Hippo Pathway-YAP/TAZ Signaling in Angiogenesis. Front. Cell Dev. Biol. 2019, 7, 49. [Google Scholar] [CrossRef]
- Low, B.C.; Pan, C.Q.; Shivashankar, G.V.; Bershadsky, A.; Sudol, M.; Sheetz, M. YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett. 2014, 588, 2663–2670. [Google Scholar] [CrossRef]
- Kim, M.K.; Jang, J.W.; Bae, S.C. DNA binding partners of YAP/TAZ. BMB Rep. 2018, 51, 126–133. [Google Scholar] [CrossRef]
- Han, H.; Qi, R.; Zhou, J.J.; Ta, A.P.; Yang, B.; Nakaoka, H.J.; Seo, G.; Guan, K.L.; Luo, R.; Wang, W. Regulation of the Hippo Pathway by Phosphatidic Acid-Mediated Lipid-Protein Interaction. Mol. Cell. Biol. 2018, 72, 328–340. [Google Scholar] [CrossRef]
- Heng, B.C.; Zhang, X.; Aubel, D.; Bai, Y.; Li, X.; Wei, Y.; Fussenegger, M.; Deng, X. Role of YAP/TAZ in Cell Lineage Fate Determination and Related Signaling Pathways. Front. Cell Dev. Biol. 2020, 8, 735. [Google Scholar] [CrossRef]
- Yuan, H.; Suzuki, S.; Terui, H.; Hirata-Tsuchiya, S.; Nemoto, E.; Yamasaki, K.; Saito, M.; Shiba, H.; Aiba, S.; Yamada, S. Loss of IκBζ Drives Dentin Formation via Altered H3K4me3 Status. J. Dent. Res. 2022, 101, 951–961. [Google Scholar] [CrossRef]
- Cantley, M.D.; Zannettino, A.C.W.; Bartold, P.M.; Fairlie, D.P.; Haynes, D.R. Histone deacetylases (HDAC) in physiological and pathological bone remodeling. Bone 2017, 95, 162–174. [Google Scholar] [CrossRef]
- Suzuki, S.; Yamada, S. Epigenetics in susceptibility, progression, and diagnosis of periodontitis. Jpn. Dent. Sci. Rev. 2022, 58, 183–192. [Google Scholar] [CrossRef]
- Li, H.; Wu, B.K.; Kanchwala, M.; Cai, J.; Wang, L.; Xing, C.; Zheng, Y.; Pan, D. YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis. Nat. Cell Biol. 2022, 24, 373–383. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Meng, Z.; Qiu, Y.; Lin, K.C.; Kumar, A.; Placone, J.K.; Fang, C.; Wang, K.C.; Lu, S.; Pan, M.; Hong, A.W.; et al. RAP2 mediates mechanoresponses of the Hippo pathway. Nature 2018, 560, 655–660. [Google Scholar] [CrossRef]
- Machida, N.; Umikawa, M.; Takei, K.; Sakima, N.; Myagmar, B.E.; Taira, K.; Uezato, H.; Ogawa, Y.; Kariya, K. Mitogen-activated Protein Kinase Kinase Kinase Kinase 4 as a Putative Effector of Rap2 to Activate the c-Jun N-terminal Kinase*. J. Biol. Chem. 2004, 279, 15711–15714. [Google Scholar] [CrossRef]
- Carballo, E.R.; Gámez, B.; Ventura, F. p38 MAPK Signaling in Osteoblast Differentiation. Front. Cell Dev. Biol. 2016, 4, 40. [Google Scholar] [CrossRef]
- Suzuki, S.; Yuan, H.; Hirata-Tsuchiya, S.; Yoshida, K.; Sato, A.; Nemoto, E.; Shiba, H.; Yamada, S. DMP-1 promoter-associated antisense strand non-coding RNA, panRNA-DMP-1, physically associates with EGFR to repress EGF-induced squamous cell carcinoma migration. Mol. Cell. Biochem. 2021, 476, 1673–1690. [Google Scholar] [CrossRef] [PubMed]
- Hirata-Tsuchiya, H.; Suzuki, S.; Okamoto, K.; Saito, N.; Yuan, H.; Yamada, S.; Jimi, E.; Shiba, H.; Kitamura, C. A small nuclear acidic protein (MTI-II, Zn2+-binding protein, parathymosin) attenuates TNF-α inhibition of BMP-induced osteogenesis by enhancing accessibility of the Smad4-NF-κB p65 complex to Smad binding element. Mol. Cell. Biochem. 2020, 469, 133–142. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, A.; Suzuki, S.; Yuan, H.; Fahreza, R.R.; Wang, X.; Nemoto, E.; Saito, M.; Yamada, S. Pharmacological Activation of YAP/TAZ by Targeting LATS1/2 Enhances Periodontal Tissue Regeneration in a Murine Model. Int. J. Mol. Sci. 2023, 24, 970. https://doi.org/10.3390/ijms24020970
Sato A, Suzuki S, Yuan H, Fahreza RR, Wang X, Nemoto E, Saito M, Yamada S. Pharmacological Activation of YAP/TAZ by Targeting LATS1/2 Enhances Periodontal Tissue Regeneration in a Murine Model. International Journal of Molecular Sciences. 2023; 24(2):970. https://doi.org/10.3390/ijms24020970
Chicago/Turabian StyleSato, Akiko, Shigeki Suzuki, Hang Yuan, Rahmad Rifqi Fahreza, Xiuting Wang, Eiji Nemoto, Masahiro Saito, and Satoru Yamada. 2023. "Pharmacological Activation of YAP/TAZ by Targeting LATS1/2 Enhances Periodontal Tissue Regeneration in a Murine Model" International Journal of Molecular Sciences 24, no. 2: 970. https://doi.org/10.3390/ijms24020970
APA StyleSato, A., Suzuki, S., Yuan, H., Fahreza, R. R., Wang, X., Nemoto, E., Saito, M., & Yamada, S. (2023). Pharmacological Activation of YAP/TAZ by Targeting LATS1/2 Enhances Periodontal Tissue Regeneration in a Murine Model. International Journal of Molecular Sciences, 24(2), 970. https://doi.org/10.3390/ijms24020970






