Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis)
Abstract
:1. Introduction
2. Results
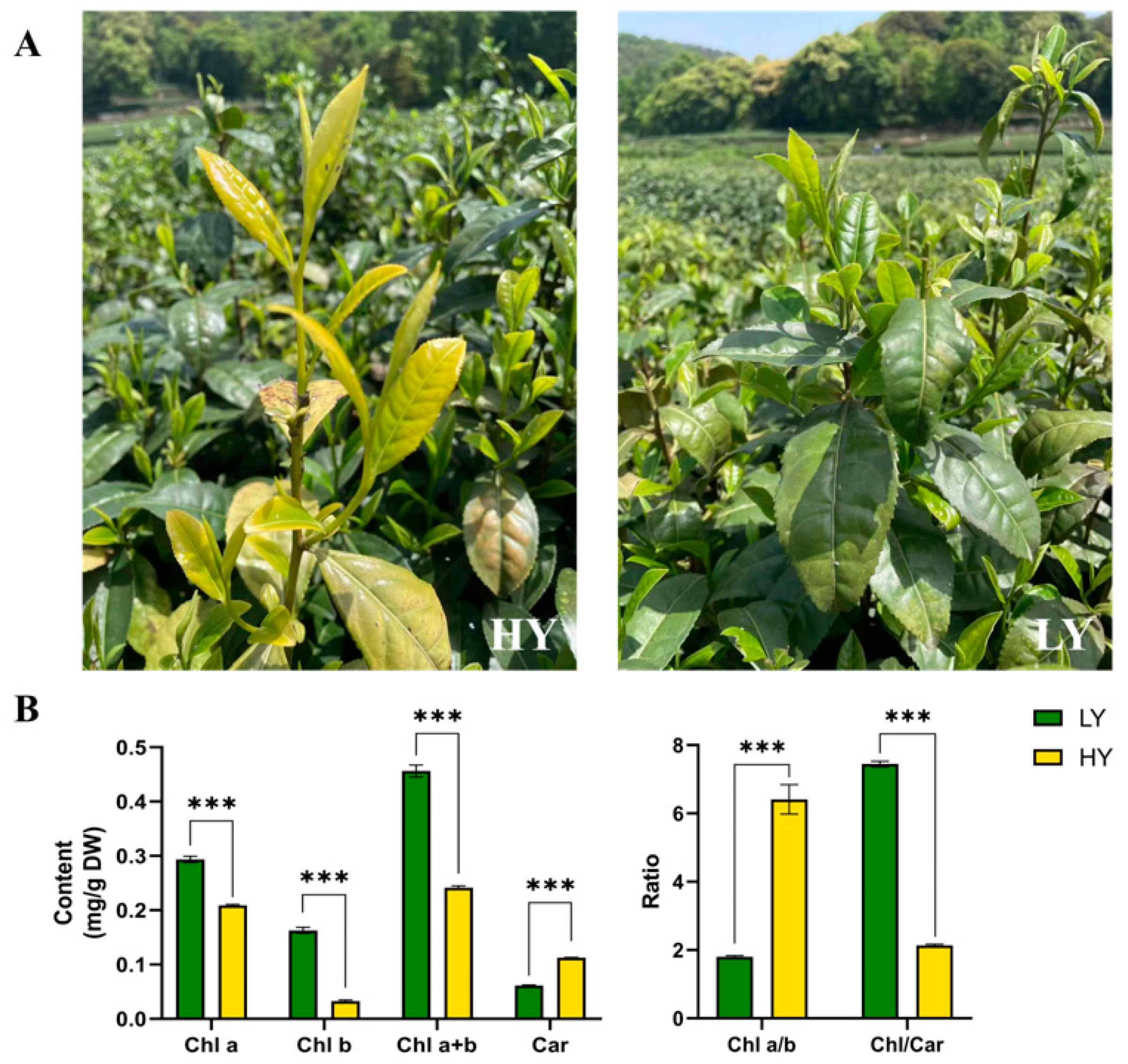
2.1. Morphological Features and Pigment Contents’ Comparison in HY and LY
2.2. Transcriptome Profiling of HY and LY
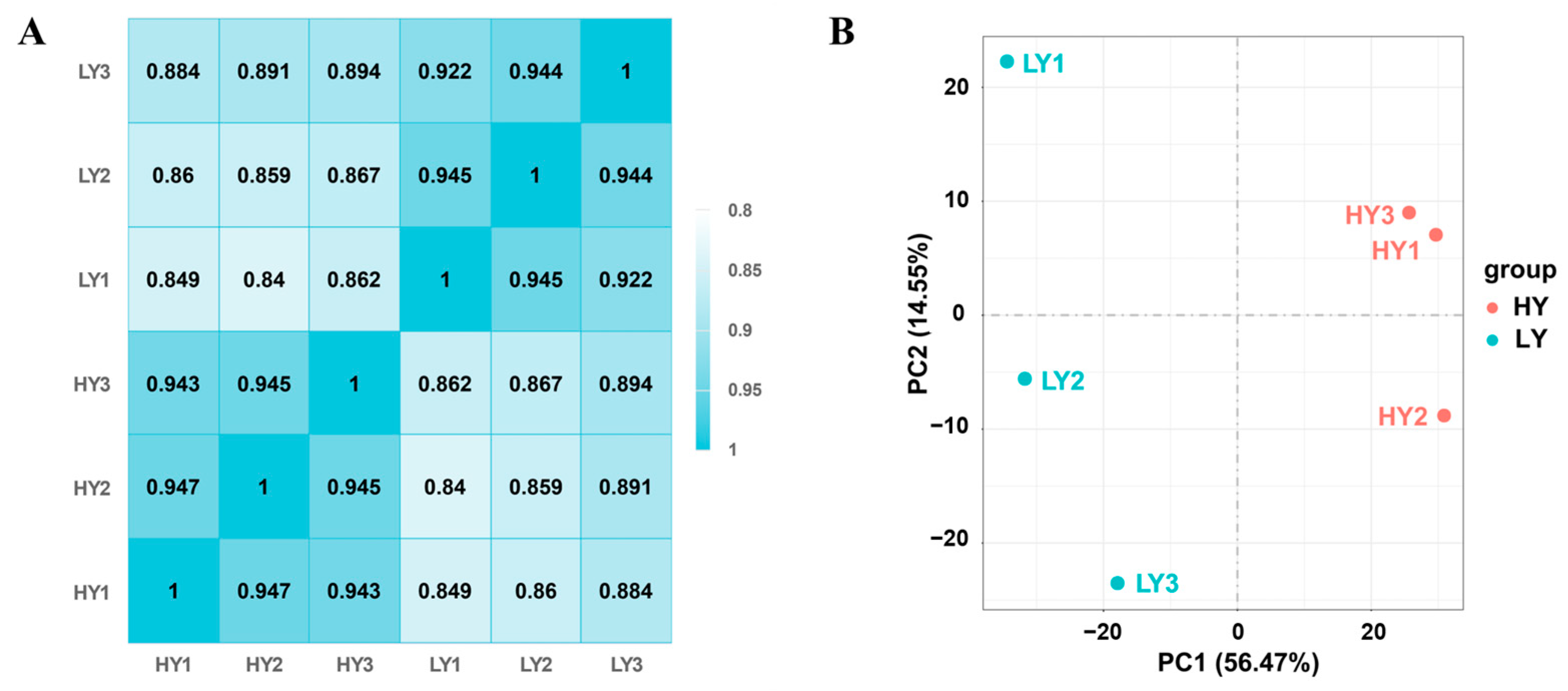
2.2.1. Evaluation of the Transcriptome Quality
2.2.2. DEGs Analysis of HY and LY
2.2.3. DEGs in Photosynthesis
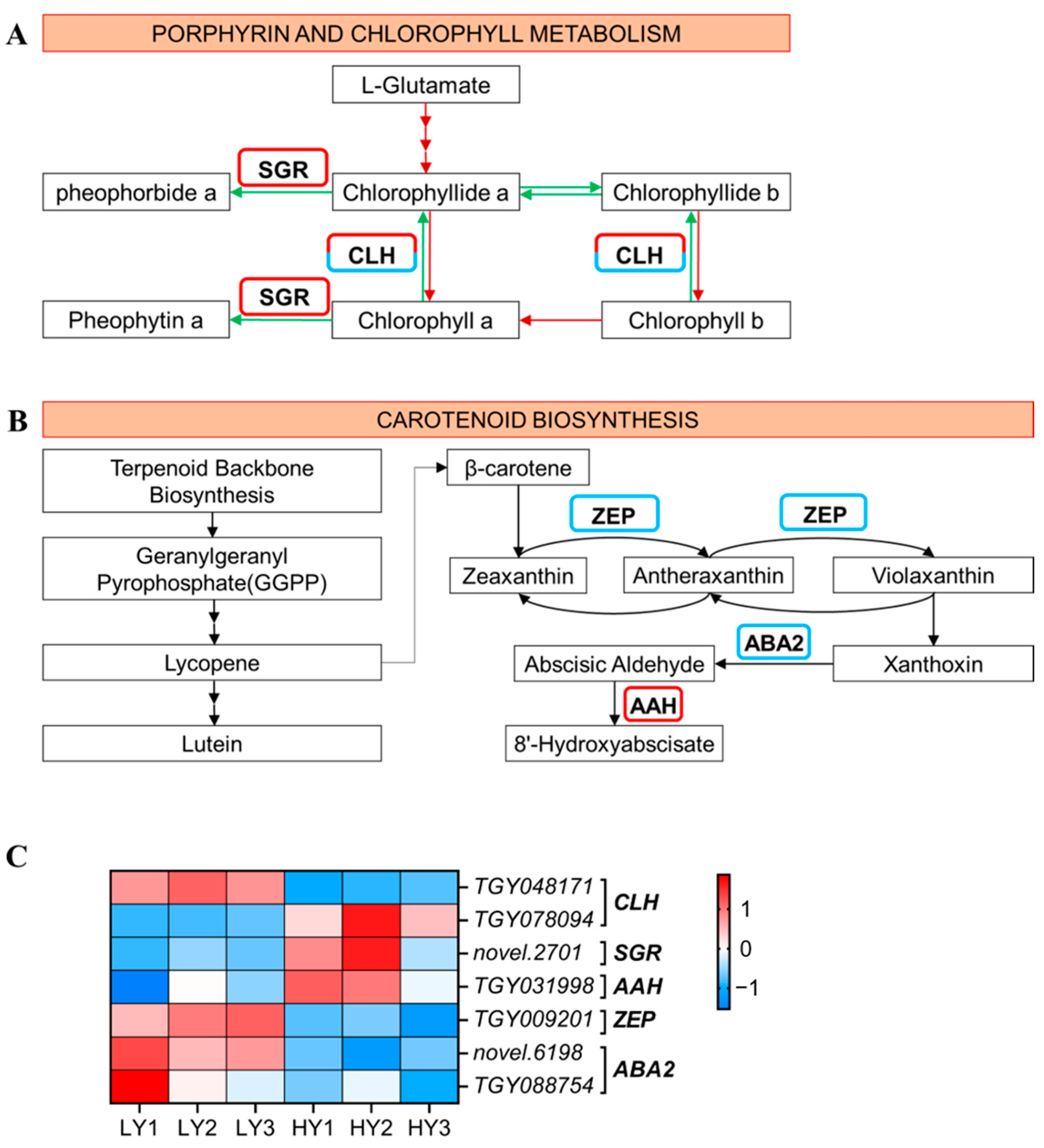
2.2.4. DEGs in Pigment Biosynthesis
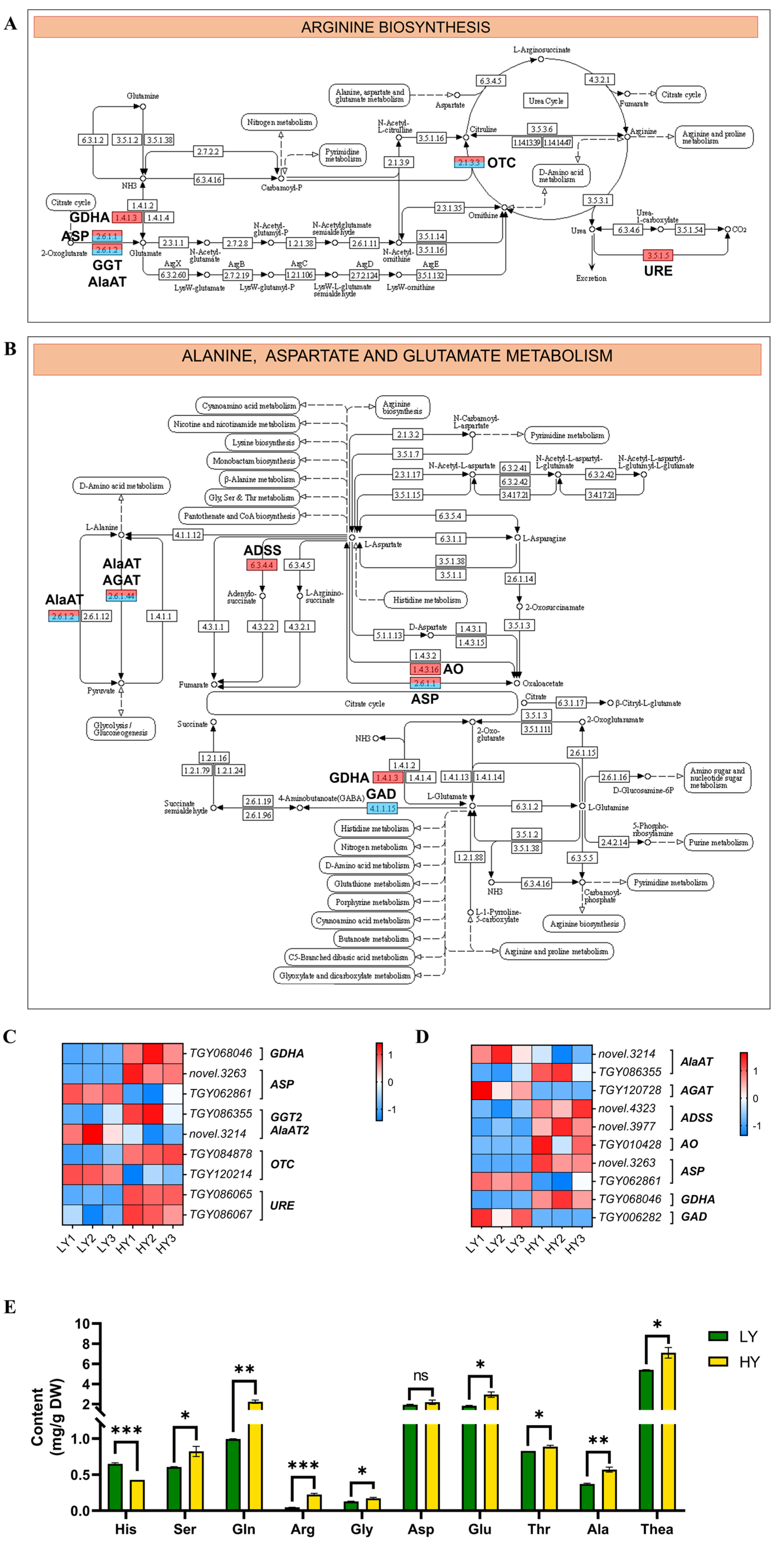
2.2.5. DEGs in Flavonoid and Amino Acid Biosynthesis
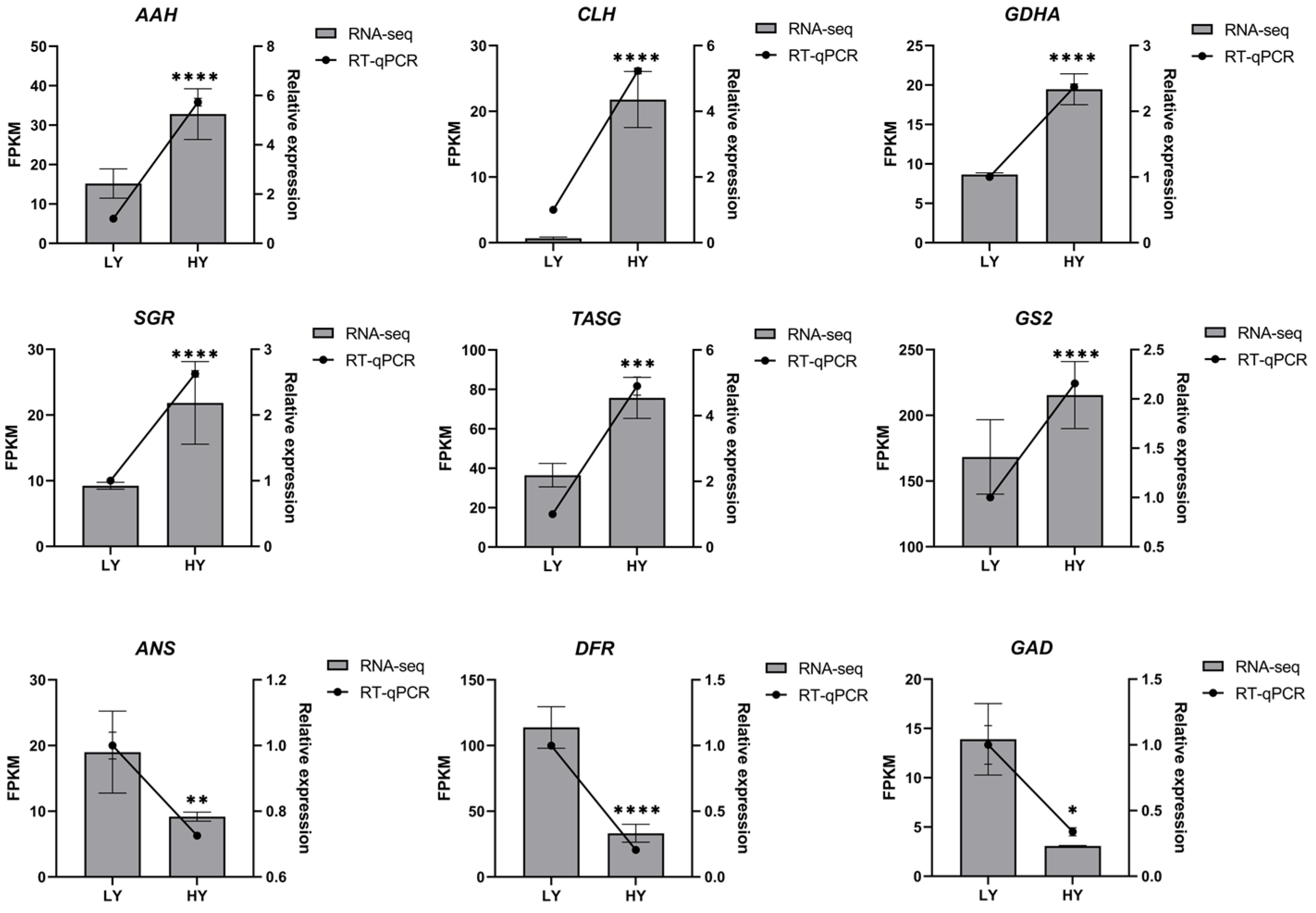
2.3. Quantitative Real-Time PCR (RT-qPCR) Analysis of Candidate Genes
3. Discussion
3.1. The Underlying Mechanism of the Chlorina Phenotype in HY
3.2. Differences in the Abundances of Quality-Related Biochemical Components between HY and LY
4. Materials and Methods
4.1. Plant Materials and Sample Preparation
4.2. RNA-seq
4.3. Enrichment Analysis of the DEGs
4.4. Quantitative Real-Time PCR (RT-qPCR) Analysis
4.5. Determination of Biochemical Compositions
4.5.1. Determination of Chlorophylls and Carotenoids
4.5.2. Determination of Free Amino Acids
4.5.3. Determination of Catechins
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mei, X.; Zhang, K.; Lin, Y.; Su, H.; Lin, C.; Chen, B.; Yang, H.; Zhang, L. Metabolic and Transcriptomic Profiling Reveals Etiolated Mechanism in Huangyu Tea (Camellia sinensis) Leaves. Int. J. Mol. Sci. 2022, 23, 15044. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Lin, C.; Wan, S.; Chen, B.; Wu, H.; Zhang, L. A Comparative Metabolomic Analysis Reveals Difference Manufacture Suitability in “Yinghong 9” and “Huangyu” Teas (Camellia sinensis). Front. Plant Sci. 2021, 12, 767724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, M.; Gao, X.; Zhou, F.; Shen, C.; Liu, Z. Multi-omics research in albino tea plants: Past, present, and future. Sci. Hortic. 2020, 261, 108943. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Chen, C.; Yue, C.; Hao, X.; Yang, Y.; Wang, X. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016, 130, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yue, C.; Cao, H.; Zhou, Y.; Zeng, J.; Yang, Y.; Wang, X. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Cao, J.; Li, J.; Zhou, B.; Tang, J.; Miao, A. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 2016, 6, 33369. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Chen, J.-D.; Wang, S.-L.; Chen, L.; Ma, C.-L.; Yao, M.-Z. Repressed Gene Expression of Photosynthetic Antenna Proteins Associated with Yellow Leaf Variation as Revealed by Bulked Segregant RNA-seq in Tea Plant Camellia sinensis. J. Agric. Food Chem. 2020, 68, 8068–8079. [Google Scholar] [CrossRef]
- Liu, Y.; Pang, D.; Jiang, H.; Chen, C.; Sun, Y.; Tian, Y.; Xu, Y.; Song, W.; Chen, L. Identifying key genes involved in yellow leaf variation in ‘Menghai Huangye’ based on biochemical and transcriptomic analysis. Funct. Integr. Genom. 2022, 22, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.Y.; Liang, Y.R.; Wang, H.; Wang, K.R.; Lu, J.L.; Zhang, G.H.; Lin, W.P.; Li, M.; Fang, Q.Y. A study on the chemical composition of albino tea cultivars. J. Hortic. Sci. Biotechnol. 2015, 81, 809–812. [Google Scholar] [CrossRef]
- Lu, M.; Han, J.; Zhu, B.; Jia, H.; Yang, T.; Wang, R.; Deng, W.-W.; Zhang, Z.-Z. Significantly increased amino acid accumulation in a novel albino branch of the tea plant (Camellia sinensis). Planta 2018, 249, 363–376. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-F.; Ma, J.-Q.; Huang, D.-J.; Ma, C.-L.; Jin, J.-Q.; Yao, M.-Z.; Chen, L. Comprehensive Dissection of Metabolic Changes in Albino and Green Tea Cultivars. J. Agric. Food Chem. 2018, 66, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gao, M.J.; Hou, R.Y.; Hu, X.Y.; Zhang, L.; Wan, X.C.; Wei, S. Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 2014, 155, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-X.; Yang, L.; Lei, Y.-S.; Ju, R.-N.; Miao, S.-G.; Jin, S.-H.; Coleman, H. Integrated transcriptome and amino acid profile analyses reveal novel insights into differential accumulation of theanine in green and yellow tea cultivars. Tree Physiol. 2022, 42, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Wang, J.Y.; Yao, M.Z.; Ma, C.L.; Chen, L. Quantitative trait loci mapping for free amino acid content using an albino population and SNP markers provides insight into the genetic improvement of tea plants. Hortic Res. 2022, 9, uhab029. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Qiao, S.; Li, M.; Cheng, H.; Ma, Q.; Wei, C.; Wan, X.; Zhang, Z. Strong biosynthesis and weak catabolism of theanine in new shoots contribute to the high theanine accumulation in Albino/etiolated tea plant (Camellia sinensis). Beverage Plant Res. 2023, 3, 23. [Google Scholar] [CrossRef]
- Dong, F.; Shi, Y.; Liu, M.; Fan, K.; Zhang, Q.; Ruan, J. iTRAQ-Based Quantitative Proteomics Analysis Reveals the Mechanism Underlying the Weakening of Carbon Metabolism in Chlorotic Tea Leaves. Int. J. Mol. Sci. 2018, 19, 3943. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Dong, X.; Yuan, X.; Hong, Y.; Zhang, L.; Zhang, X.; Chen, S. Identification and characterization of CsSRP43, a major gene controlling leaf yellowing in cucumber. Hortic. Res. 2022, 9, uhac212. [Google Scholar] [CrossRef]
- Smith, W.K.; Harrington, J.B. WHEAT ALBINOS. J. Hered. 1929, 20, 19–22. [Google Scholar] [CrossRef]
- MASCIA, P.N.; ROBERTSON, D.S. Genetic studies of the chlorophyll biosynthetic mutants of maize. J. Hered. 1980, 71, 19–24. [Google Scholar] [CrossRef]
- Wu, H.; Shi, N.; An, X.; Liu, C.; Fu, H.; Cao, L.; Feng, Y.; Sun, D.; Zhang, L. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling. Int. J. Mol. Sci. 2018, 19, 1594. [Google Scholar] [CrossRef]
- Zhao, M.-H.; Li, X.; Zhang, X.-X.; Zhang, H.; Zhao, X.-Y. Mutation Mechanism of Leaf Color in Plants: A Review. Forests 2020, 11, 851. [Google Scholar] [CrossRef]
- Matile, P.; Hörtensteiner, S.; Thomas, H. CHLOROPHYLL DEGRADATION. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 67–95. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Yang, F.; Shi, S.; Li, D.; Wang, Z.; Liu, H.; Huang, D.; Wang, C. Transcriptome Characterization of Cymbidium sinense ‘Dharma’ Using 454 Pyrosequencing and Its Application in the Identification of Genes Associated with Leaf Color Variation. PLoS ONE 2015, 10, e0128592. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, S.W.; Lim, G.H.; Lyu, J.I.; Choi, H.I.; Jo, Y.D.; Kang, S.Y.; Kang, B.C.; Kim, J.B. Transcriptome analysis to identify candidate genes associated with the yellow-leaf phenotype of a Cymbidium mutant generated by γ-irradiation. PLoS ONE 2020, 15, e0228078. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bai, P.P.; Gu, K.J.; Yang, S.Z.; Lin, H.Y.; Shi, C.G.; Zhao, Y.P. Dynamic transcriptome and network-based analysis of yellow leaf mutant Ginkgo biloba. BMC Plant Biol. 2022, 22, 465. [Google Scholar] [CrossRef]
- Nie, L.; Zheng, Y.; Zhang, L.; Wu, Y.; Zhu, S.; Hou, J.; Chen, G.; Tang, X.; Wang, C.; Yuan, L. Characterization and transcriptomic analysis of a novel yellow-green leaf wucai (Brassica campestris L.) germplasm. BMC Genom. 2021, 22, 258. [Google Scholar] [CrossRef]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s Green Cotyledon Gene, Encodes Magnesium-Dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K.; Shimoda, Y.; Tanaka, A.; Ito, H. Chlorophyll a is a favorable substrate for Chlamydomonas Mg-dechelatase encoded by STAY-GREEN. Plant Physiol. Biochem. 2016, 109, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Tang, Y.; Zhang, H.; Wu, P.; Chen, Y.; Li, M.; Wu, G.; Jiang, H. The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J. Plant Physiol. 2013, 170, 1367–1373. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Kim, D.; Kim, Y.S.; Hortensteiner, S.; Paek, N.C. Arabidopsis STAYGREEN-LIKE (SGRL) promotes abiotic stress-induced leaf yellowing during vegetative growth. FEBS Lett. 2014, 588, 3830–3837. [Google Scholar] [CrossRef]
- Liang, Y.; Ma, W.; Lu, J.; Wu, Y. Comparison of chemical compositions of Ilex latifolia Thumb and Camellia sinensis L. Food Chem. 2001, 75, 339–343. [Google Scholar] [CrossRef]
- Narukawa, M.; Kimata, H.; Noga, C.; Watanabe, T. Taste characterisation of green tea catechins. Int. J. Food Sci. Technol. 2010, 45, 1579–1585. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. The aroma, taste, color and bioactive constituents of tea. J. Med. Plants Res. 2011, 5, 2110–2124. [Google Scholar] [CrossRef]
- Alcázar, A.; Ballesteros, O.; Jurado, J.M.; Pablos, F.; Martín, M.J.; Vilches, J.L.; Navalón, A. Differentiation of Green, White, Black, Oolong, and Pu-erh Teas According to Their Free Amino Acids Content. J. Agric. Food Chem. 2007, 55, 5960–5965. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Duan, J.; Jiang, Y.; Shi, J.; Peng, L.; Xue, S.; Kakuda, Y. Production, Quality, and Biological Effects of Oolong Tea (Camellia sinensis). Food Rev. Int. 2011, 27, 1–15. [Google Scholar] [CrossRef]
- Wei, K.; Yu, S.; Quan, Q.; Aktar, S.; He, M.; Zhang, Y.; Fang, H.; Wu, R.; Ruan, L.; Mao, P.; et al. An integrative analysis of metabolomics, DNA methylation and RNA-Seq data reveals key genes involved in albino tea ‘Haishun 2’. Beverage Plant Res. 2022, 2, 2. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.-Y.; Zhou, J.; He, W.; Zeng, J.-M.; Jiang, Y.-W.; Cheng, H. Comparison of catechins and purine alkaloids in albino and normal green tea cultivars (Camellia sinensis L.) by HPLC. Food Chem. 2012, 130, 720–724. [Google Scholar] [CrossRef]
- Cheng, S.; Fu, X.; Liao, Y.; Xu, X.; Zeng, L.; Tang, J.; Li, J.; Lai, J.; Yang, Z. Differential accumulation of specialized metabolite L-theanine in green and albino-induced yellow tea (Camellia sinensis) leaves. Food Chem. 2019, 276, 93–100. [Google Scholar] [CrossRef]
- Pang, D.; Liu, Y.; Sun, Y.; Tian, Y.; Chen, L. Menghai Huangye, a novel albino tea germplasm with high theanine content and a high catechin index. Plant Sci. 2021, 311, 110997. [Google Scholar] [CrossRef]
- Satou, M.; Enoki, H.; Oikawa, A.; Ohta, D.; Saito, K.; Hachiya, T.; Sakakibara, H.; Kusano, M.; Fukushima, A.; Saito, K.; et al. Integrated analysis of transcriptome and metabolome of Arabidopsis albino or pale green mutants with disrupted nuclear-encoded chloroplast proteins. Plant Mol. Biol. 2014, 85, 411–428. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Shi, J.C.; Wang, D.J.; Liang, X.; Wei, F.; Gong, C.M.; Qiu, L.J.; Zhou, H.C.; Folta, K.M.; Wen, Y.Q.; et al. A point mutation in the gene encoding Mg-chelatase subunit I influences strawberry leaf color and metabolism. Plant Physiol. 2023, 192, 2737–2755. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, M.; Ruan, J. Integrated Transcriptome and Metabolic Analyses Reveals Novel Insights into Free Amino Acid Metabolism in Huangjinya Tea Cultivar. Front. Plant Sci. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, D.; Liu, M.; Ruan, J. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2018, 231, 272–281. [Google Scholar] [CrossRef]
- Wang, P.; Yu, J.; Jin, S.; Chen, S.; Yue, C.; Wang, W.; Gao, S.; Cao, H.; Zheng, Y.; Gu, M.; et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 2021, 8, 107. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. [34] Chlorophylls and Carotenoids: Pigments of Photosynthetic Biomembranes. Meth. Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- GB/T 8313-2018; Zhou, W. GB/T 8313-2018; Determination of Total Polyphenols and Catechins Content in Tea. Standards Press of China: Beijing, China, 2018.

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhang, C.; Ma, C.; Chen, L.; Yao, M. Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 15070. https://doi.org/10.3390/ijms242015070
Li Y, Zhang C, Ma C, Chen L, Yao M. Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis). International Journal of Molecular Sciences. 2023; 24(20):15070. https://doi.org/10.3390/ijms242015070
Chicago/Turabian StyleLi, Yuanyuan, Chenyu Zhang, Chunlei Ma, Liang Chen, and Mingzhe Yao. 2023. "Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis)" International Journal of Molecular Sciences 24, no. 20: 15070. https://doi.org/10.3390/ijms242015070
APA StyleLi, Y., Zhang, C., Ma, C., Chen, L., & Yao, M. (2023). Transcriptome and Biochemical Analyses of a Chlorophyll-Deficient Bud Mutant of Tea Plant (Camellia sinensis). International Journal of Molecular Sciences, 24(20), 15070. https://doi.org/10.3390/ijms242015070







